Abstract
Recent improvements in molecular treatment and gene therapy led to discovering novel cancer remedies. Antisense LNA GapmeRs is a state-of-the-art molecular research field for diagnosing and treating various cancer types. Acute myeloid leukemia (AML) is a heterogeneous hematopoietic malignancy defined by the rapid accumulation and malignant proliferation of immature myeloid progenitors. SOX12 is a new potential target for acute myeloid leukemia. In this study, SOX12 was blocked by antisense LNA GapmeRs (ALG) in human AML cell lines (KG1 and M07e). Cells were transfected with Gapmer anti-SOX12 at 24, 48, and 72 h post-transfection. Transfection efficiency was assessed by a fluorescent microscope. Furthermore, evaluation of SOX12, TWIST1, CTNNB1, CASP3, and CASP9 expression was performed by quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR). Cell viability was determined by MTT assay. SOX12 expression was decreased remarkably in the ALG group. Moreover, SOX12 knockdown was associated with a decrease in TWIST1 and CTNNB1 expression. Besides, downregulation of SOX12 in both cell lines could induce apoptosis, probably through upregulation of CASP3 and CASP9. The findings reveal that SOX12 knockdown could be a new target for reducing AML cells proliferation through antisense therapy approach.
Key Words: Acute myeloblastic leukemia, antisense LNA GapmeRs, SOX12, TWIST1, CTNNB1, apoptosis
Leukemias are a group of progressive, malignant hematological diseases that consist of abnormally differentiated oligoclonal expansions and sometimes poorly differentiated hematopoietic cells that could invade the blood and other extramedullary tissues (1). The highest percentage of patients with acute myeloid leukemia (AML) belong to adults (2). Although induction chemotherapy is an effective treatment, relapse is one of the severe causes of treatment failure (3). Based on the French–American–British (FAB) classification, AML has nine subtypes (M0, M1, M2, M3, M4, M4E0, M5, M6, and M7) according to cytochemical staining, morphological charact-eristics of the cancerous cells (2). Mutations in different genes such as nucleophosmin 1 (NPM1), CCAAT enhancer binding protein alpha (CEBPA), runt-related transcription factor 1 (RUNX1), KIT receptor tyrosine kinase, and fms-like tyrosine kinase 3 (FLT3) are sometimes a cause of the incidence of AML (4).
Dysfunction of some specific transcription factors due to mutations is also an important cause of AML. New treatments for AML that target the appropriate transcription machinery are potentially effective (5, 6). Since the functions and activities of most transcription factors in AML remain elusive, the identification of these factors could give visions into the diagnosis and treatment of AML. The SRY-related high mobility group (HMG) box (SOX) gene family encodes a group of transcription factors having critical roles in the primary cellular processes during development (7). Structural abnormalities in SOX genes have been correlated with different types of cancer (8). SOX genes are divided into A-H sub-groups based on the HMG box. SOXC group consists of SOX4, SOX11, and SOX12. Among them, SOX4 has a pivotal role in regulating the normal function of hematopoietic stem cells, and its overexpression is a joint event in AML malignancy (9). SOX11 expression is illustrated in lymphoblastic lymphoma, some Burkitt lymphomas, and T-prolymphocytic leukemia (10). SOX12 (previously named SOX22) is expressed in several fetal and adult tissues and organs, and has an indispensable role in the differentiation and maintenance of different cell types. It is a direct target for forkhead box Q1 (FOXQ1) that promotes metastasis (11). The role of the canonical WNT/β-catenin signaling pathway is essential for regulating tissue development and maintaining homeostasis. As a result, the dysfunction of this pathway can cause different diseases, including cancer. β-catenin (CTNNB1) is a critical transcriptional co-activator in the canonical WNT signaling pathway and can firmly regulate cell-to-cell adhesion (12). The SOX genes' functions are mostly associated with the Wnt/β-catenin (WNT/CTNNB1) pathway by regulating β-catenin expression or its binding to WNT gene promoters as a target to control protein stability and nuclear translocation. Dysfunction in this pathway has a defined role in the AML incidence (13). Twist family bHLH transcription factor 1 (TWIST1) is a crucial factor that induces epithelial-mesenchymal transition (EMT). Several experiments showed that TWIST1 raises tumor metastasis and demonstrated poor prognosis in various human cancers (14). On the other hand, studies reported that the expression of several metastatic-related genes such as TWIST1 increased by up-regulation of SOX12 (15).
Antisense LNA GapmeRs are a new generation of DNA antisense oligonucleotides that contain locked nucleic acid (LNA) and induce the degradation of target sequence via RNase H-dependent mechanism (16). To date, not many studies have been reported on the potential function of SOX12 in AML. In this study, we investigated the potential role of SOX12 in AML by knocking down its expression in two AML cell lines by using antisense LNA GapmeR, and by assessing the expression of related genes, TWIST1 and CTNNB1, in the cell populations. Also, expressions of CASP3 and CASP9 as apoptotic factors were investigated.
Materials and methods
Cell culture
KG1 cell line (human acute myeloid leukemia, AML M6) and M07e cell line (human megakaryocytic cell line) were purchased from the National Cell Bank of Iran (Pasteur Institute, Tehran, Iran). Complete medium including RPMI-1640 medium (Gibco, Paisley, UK) supplemented with 10% v/v fetal bovine serum (FBS; Gibco, UK) and 1% penicillin/streptomycin (Sigma-Aldrich, Saint Louis, MO, USA), were used to culture KG1cells. Additionally, for M07e cells, this media was complemented with five ng/ml granulocyte macrophage-colony stimulating factor (GM-CSF; R&D Systems, Minneapolis, MN, USA). Cells were cultured in a 25-cm2 cell culture flask (Nunc, Roskilde, Denmark) and incubated at 37 °C with a 5% CO2 atmosphere. The cells were passaged once a week to maintain the exponential phase.
The local ethics committee approved this study of Isfahan University of Medical Sciences (IRAN), and the study has been approved by the appropriate institutional and national research ethics committee. It has been performed by the ethical standards as laid down in the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Cell transfection
The LNA targeting sequence of the SOX12 gene was acquired from a reputable site: http://www.Incrnadb.org as taCTTGTAGTCCGG GTAAtc. Antisense LNA GapmeRs (ALG) and antisense LNA GapmeR negative control oligonucleotides (ALGNC) for SOX12 were purchased from Eurofins. ALG and ALGNC were labeled at their 5'end with 6-FAM (6-carboxyfluorescein), which is a fluorescent dye. KG1 and M07e cells were transfected by using the PolyFect™ transfection reagent kit (Qiagen, Germany) pursuant to the manufacturer's guidance. In summary, a total number of about 5 × 105 cells of each line, in the exponential phase, were seeded into six-well culture plates (Nunc, Denmark) containing 1.8 mL RPMI 1640 per well without antibiotics and FBS. Two different microtubes were labeled as G (GapmeR) and T (Transfect). Each cell well contained 100 µL RPMI. 6 µL LNA GapmeRper from G microtube and 3 µL of transfection reagent from T microtube were added to each cell well. After pipetting well, the transfection reagent was added into antisense LNA GapmeR and incubated at room temperature in the dark for 15 min, then 200 µL of this reagent was added to each cell well while the plates were rotated to ensure that all the reagent and cell culture media were mixed thoroughly. We seeded 3 wells for each cell line labeled as GapmeR, Negative, and control (without LNA GapmeR and ALGNC, containing only the cells). After 6 h incubation, 100 µL FBS and 10 µL antibiotics were added to each well. Finally, the cells were incubated for 24, 48, and 72 h. Transfected cells were measured by fluorescence microscopy and flow cytometry due to the fluorescent dye (6-FAM) (17).
Real-time PCR
Real-time PCR was used to assess the efficiency of the SOX12 inhibition by ALG. Briefly, total RNA of both KG1 and M07e cells were extracted with RNA extraction kit (Biofact, Korea) at 24, 48, and 72 h after transfection. RNA concentration and purity were determined on a spectrophotometer by calculating the optical density ratio (OD) at 260 and 280 nm wavelengths. Complementary DNA (cDNA) was then synthe-sized using the universal cDNA Synthesis Kit (Biofact, Korea). The SYBR green master mix kit was used for real-time PCR analysis (Biofact, Korea). The reference gene for normalization of the expression level was GAPDH, and all assays were carried out in triplicate. The forward and reverse primers for the selected genes were listed in Table 1. The annealing temperature was 60 oC for all studied genes and 40 amplification cycles were performed for each real-time PCR experiment in the ABI Step One Plus (ABI, USA) instrument, and the relative expression levels were analyzed by the ΔΔCt method.
Table 1.
Sequences of primer pairs used for real-time RT-PCR method
| Primer name | Sequences (5′–3′) |
|---|---|
|
SOX12
-F
SOX12 -R TWIST1 -F TWIST1 -R CTNNB1 -F CTNNB1 -R CASP3 -F CASP3 -R CASP9 -F CASP9 -R GAPDH -F GAPDH -R |
CTGGAGTGGTGGGATTGGTC GGGTGTCAGAGGGACAAAGG TCTCGGTCTGGAGGATGGAG AATGACATCTAGGTCTCCGGC CAACCAAGAAAGCAAGCTCATC CAGATAGCACCTTCAGCACTC TCCACAGCACCTGGTTATTATTC ACTCAAATTCTGTTGCCACCTTTC ATTTGGTGATGTCGGTGCTC TCACGGCAGAAGTTCACATTG TGCACCACCAACTGCTTAGC GGCATGGACTGTGGTCATGAG |
Cell viability assay
Cell viability of KG1 and M07e cells was assessed using the tetrazolium (MTT) colorimetric assay. According to MTT reduction with the production of water-insoluble purple formazan by mitochondrial dehydrogenase in the active cells, this method is straightly correlated with the number of living cells. The MTT assay was
carried out at three-time points after the start of the transfection. First, 200 µl MTT (Sigma-Aldrich, USA) with 50 mg/ml concentration was added to 5 ×105 KG1 and M07e cells suspension in 2 ml RPMI 1640 medium and was then incubated 4 h at 37 oC under 5% CO2 in the darkness. Subsequently, 200 µl dimethyl sulfoxide (DMSO) (Sigma-Aldrich, USA) was added to each well followed by shaking for dissolving formazan salt. The OD was measured at 570 nm by using a spectrophotometer (PG Instruments T80, PG Instrument, England). Cell viability was determined as absorption of the aliquot of transfected cells normalized to the absorption of control cells maintained in complete medium.
Statistical analysis
All tests were performed three times, and the data were analyzed using SPSS version 25 software. Also, GraphPad Prism version 8.3.0.538 was used to draw graphs. To evaluate differences between groups, data were analyzed by a two-way ANOVA test. The data are represented as mean ± standard deviation (mean ± SD). For all statistical analyzes, P<0.001 was considered statistically significant.
Results
Antisense LNA GapmeRs decreased SOX12 expression
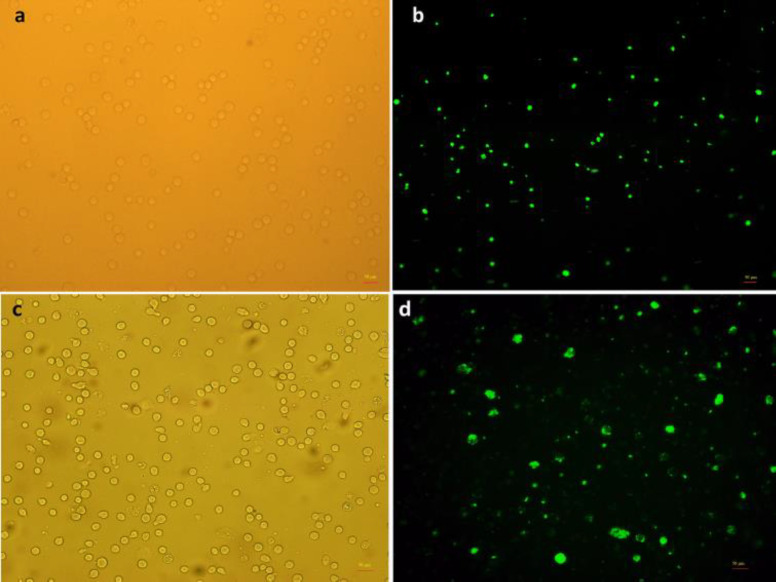
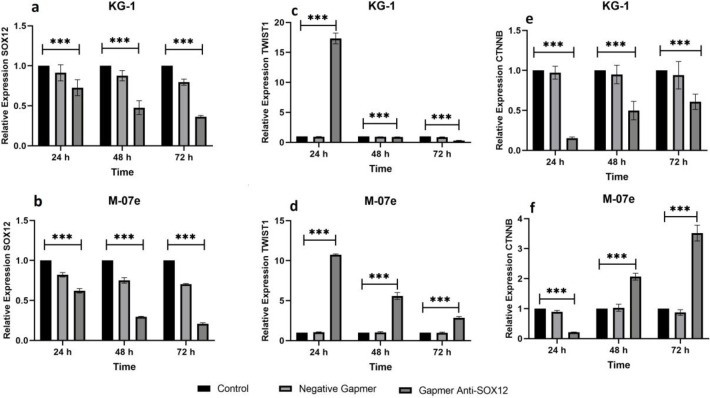
The transfection efficiency of antisense LNA GapmeRs transfected cells was determined by fluorescence microscopy due to the oligonucleotide strands' FAM group (Figure 1). Additionally, flow cytometry confirmed the transfection level by more than 90% (data not shown). QRT-PCR was performed in two human acute myeloid cell lines, including KG1 and M07e, to measure the expression of SOX12 in all groups (control, ALGNC, antisense LNA GapmeRs) at three-time points after transfection. SOX12 expression was decreased dramatically in the antisense LNA GapmeRs group at all time points in comparison with other groups in both cell lines (P <0.001) (Figure 2a, 2b).
Fig. 1.
Transfection of AML cell lines with antisense LNA GapmeRs. KG1 and M07e cells have been transfected with 6-FAM labeled antisense LNA GapmeRs. To evaluate the transfection efficiency of both cell lines, which have been assessed by a fluorescent microscope. a, b: phase contrast (a) and fluorescent (b) images of the same field of KG1 cells; c, d: phase contrast (c) and fluorescent (d) images of the same field of M07e cells. The majority of cells in both cell lines were transfected. Scale bars: 50 µm
Fig. 2.
Evaluation of the SOX12, TWIST 1, and CTNNB1expression in AML cell lines. a, b: expression of SOX12 in KG1 (a) and M07e (b) cells; c, d: expression of TWIST1 in KG1 (c) and M07e (d) cells ; e, f: expression of CTNNB1 in KG1 (e) and M07e (f) cells. All genes expression assessment was performed by real-time PCR at 24, 48, and 72 h after GapmeR anti-SOX12 transfection. Data analysis was performed by ∆∆Ct method, and the untreated group (control) was reflected as a reference for comparison with other groups. The data are presented as mean ± SD of triple independent tests (***P < 0.001)
Knockdown of SOX12 decreased the expression of TWIST1
The qRT-PCR assay was used to evaluate the effect of downregulation of SOX12 on TWIST1 expression in AML cell lines at 24, 48, and 72 h post-transfection. In all groups, gene expression of TWIST1 was decreased considerably after transfection. The expression of TWIST1 was gradually decreasing over time as its expression was at the lowest level 72 h post-transfection in KG1 (Figure 2c) and M07e (Figure 2d) (P<0.001). However, the downregulation of TWIST1 in the KG1 cell line was more remarkable than its expression in the M07e cell line.
Assessment of CTNNB1 expression in KG1 and M07e cell lines
The qRT-PCR assay was performed to determine the effect of downregulation of SOX12 on CTNNB1 expression in AML cell lines at 24, 48, and 72 h post-transfection. The results in KG1 cells demonstrated that CTNNB1 expression was decreased in GapmeR anti-SOX12 in comparison with the two other groups at each time, and its expression was at the lowest level 24 h after transfection (P<0.001) (Figure 2e). In M07e cells, although the most significant reduction was observed 24 h after transfection, CTTNB1 expression increased gradually over time (P<0.001) (Figure 2f).
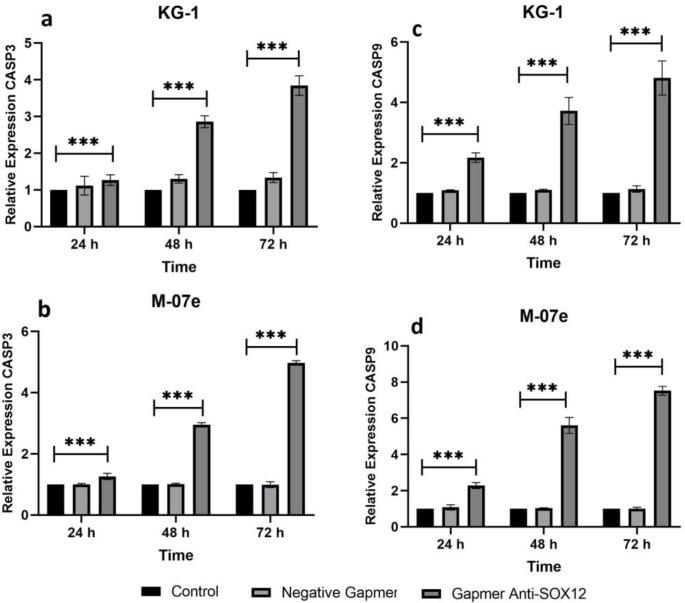
Expression level of CASP3 and CASP9 increa-sed after SOX12 blockage
The qRT-PCR assay was designed to detect the expression level of CASP3 and CASP9 in KG1 and M07e in all groups at three-time points (24, 48, and 72 h). The CASP3 and CASP9 expression were upper in the antisense LNA GapmeR group in comparison with the control groups at all three time points in the KG1 cell line (P < 0.001) (Figure 3a, 3c) and in M07e cell line (P < 0.001) (Figure 3b, 3d). The highest level of CASP3 was observed 72 h after transfection in the M07e cell line (Figure 3b). Also, CASP9 was overexpressed 72 h after transfection in the M07e cell line (Figure 3d). Together, these results demonstrated that knockdown of SOX12 significantly induced gene expression of apoptotic factors in AML cell lines.
Fig. 3.
Evaluation of CASP3 and CASP9 expression in AML cell lines. a, b) CASP3 expression level in KG1 (a) and M07e (b) cell; c, d: CASP9 expression level in KG1 (c) and M07e (d) cells. All genes expression assessment was performed by real-time PCR at 24, 48, and 72 h after GapmeR anti-SOX12 transfection. Data analysis was performed by ∆∆Ct method, and the untreated group (control) was considered as a reference for comparison with other groups. The data are displayed as mean ± SD of three independent experiments (***P < 0.001)
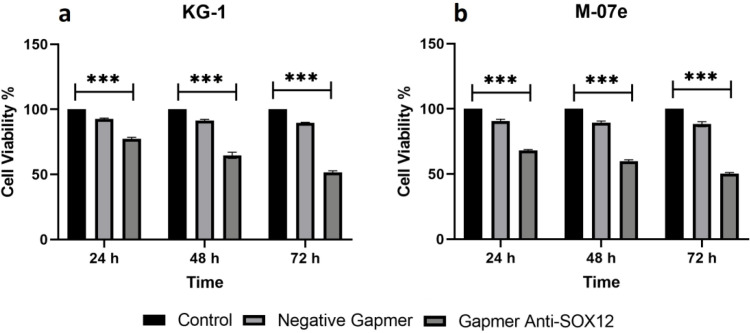
Cell viability reduction in KG1 and M07e cells by inhibition of SOX12
The MTT test at 24, 48 and, 72 h after transfection was performed to investigate the effects of SOX12 blockage on cell viability in KG1 and M07e cell lines. The cell viability in both cell lines was considerably decreased and was about 50% in 72 h after Gapmer anti-SOX12 transfection. Compared to the control group, the viability in ALG and ALGNC groups was reduced, but the reduction in the ALG group was notably significant (Figure 4a, 4b). The ALGNC group's reduction may have been due to the toxicity of transfection reagents or the cells' senescence (P <0.001).
Fig. 4.
AML cell lines viability after GapmeR anti- SOX12 transfection. a) KG1 cells; b) M07e cells. Cells viability was measured by MTT assay at 24, 48, and 72 h after GapmeR anti-SOX12 transfection. The viability of untreated cells (control group) at all three-time points has been considered 100%, and the viability of the other groups is represented as a percentage of untreated cells at the same time points. Data are mean ± SD of three independent experiments (***P <0.001).
Discussion
This is the first study to specifically explore the SOX12 inhibition with antisense LNA GapmeRs in AML. The results demonstrate that SOX12 expression was decreased remarkably in the ALG group. Moreover, SOX12 knockdown was associated with a decrease in TWIST1 and CTNNB1 expression. However, as mentioned in the results, maximum reduction for these two genes was observed at different time. SOX12 knockdown could affect apoptotic factors such as CASP3 and CASP9. Using the MTT assay, it was demonstrated that cell viability reduction was associated with the inhibition of SOX12. SOX12 is one of the SOXC family members. It is suggested that SOXC genes have a vital role in redundancy to control cell differentiation and expansion (18). Several SOXC family members, such as SOX4, SOX11, and SOX12, have been overexpressed in human cancer tissues (15, 19, 20). SOX12 has recently come into view as a novel oncogene. It is overexpressed in hepatocellular carcinoma with regional lymph nodes and remote metastases (21). Furthermore, SOX12 is a potential cancer-stem-cell-like marker for hepatocellular carcinoma that increases chemoresistance (22). Different studies indicated that the downregulation of SOX12 induces an antitumor effect in breast, lung, pancreatic, and colorectal cancer cells (19, 20, 23, 24). On the other hand, several studies were perfoemed on the association of SOX genes and AML (25, 26). In particular, a study illustrated that SOX12 was highly expressed in AML in comparison with normal hematopoietic stem and progenitor cells, and its knockdown in THP1 cells or primary AML cells with a lentivirus containing shRNAs was shown to inhibit cell proliferation and reduce the clonogenicity and leukemia propagation (11). Interestingly, in a recent study, SOX12 could have been blocked by antisense LNA GapmeR; thus, its expression decreased significantly.
To further confirm the transactivating of SOX12, we investigated TWIST1 expression as well as SOX12 expression as a direct transcriptional target of SOX12. It has been reported that the silencing of SOX12 remarkably decreased the mRNA and protein expression of TWIST1 in breast cancer cells. Moreover, the change in TWIST's mRNA levels indicates that SOX12 may bind to this factor's promoter to control its transcription (20). Another study has been shown that SOX12 promotes migration and invasion of hepatocellular cacinoma through upregulating TWIST1 and FGFBP1 (15). Moreover, knockdown of SOX12 in lung cancer cell lines, such as SPC-A-1 and A549 cells, led to a significant decrease in the mRNA and protein level of TWIST1. This study also demonstrated that SOX12 directly binds to the promoters of cyclin E and TWIST1 in A549 cells (19). Together with these studies, our data indicated that knockdown of SOX12 in both KG1 and M07e cells led to a significant decrease in the mRNA expression of TWIST1. Several studies demonstrated that the functions of the SOX genes and WNT/CTNNB1 pathway correlate with the regulation of β-catenin expression (13). In this study, we investigated the association between knocking down SOX12 and CTTNB1 expression, especially in AML diseases. As a result, in both cell lines, CTTNB1 expression decreased after 24 h, and gradually until 72 h, its expression increased, which can be due to the effect of other signaling pathways.
Apoptosis or programmed cell death is the primary mechanism for eliminating damaged, unwanted cells that cannot be restored (27). Two predominant signaling cascades have been delineated to initiate the apoptotic program in cells, the intrinsic (mitochondrial) and extrinsic (death ligand) pathways (28, 29). Both cascades lead to the activation of the executive caspase 3, 6, and 9, which are essential mediators of apoptosis (30). So, caspase plays a pivotal role in the executive phase and, its relevance in many cancer cells and cancer cell lines has been demonstrated (31-33). To investigate apoptotic factors' expression after the knockdown of SOX12, we determined the expression of CASP3 and CASP9. As expected, these factors increased in both KG1 and M07e cell lines.
In conclusion, we revealed that SOX12 expression and related genes, TWIST1 and CTNNB1, were downregulated in AML cell lines. Moreover, we showed that SOX12 plays a vital role in the programmed cell death of AML cell lines. Our results can be useful in translational medicine for subsequent research in AML treatment and generate cutting-edge drugs based on antisense therapy. This study suggested that SOX12 may be a novel biomarker for AML, and can be translated into novel therapeutic strategies for AML.
Acknowledgments
This study was conducted with the financial support of Isfahan University of Medical Sciences (Iran); Grant number 198042.
Conflict of Interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Cruz-Miranda GM, Hidalgo-Miranda A, Barcenas-Lopez DA, et al. Long Non-Coding RNA and Acute Leukemia. Int J Mol Sci. 2019:20. doi: 10.3390/ijms20030735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Kouchkovsky I, Abdul-Hay M. 'Acute myeloid leukemia: a comprehensive review and 2016 update'. Blood Cancer J. 2016;6:e441. doi: 10.1038/bcj.2016.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374:2209–21. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dohner H, Weisdorf DJ, Bloomfield CD. Acute Myeloid Leukemia. N Engl J Med. 2015;373:1136–52. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- 5.Oellerich T, Mohr S, Corso J, et al. FLT3-ITD and TLR9 use Bruton tyrosine kinase to activate distinct transcriptional programs mediating AML cell survival and proliferation. Blood. 2015;125:1936–47. doi: 10.1182/blood-2014-06-585216. [DOI] [PubMed] [Google Scholar]
- 6.Fuchs O. Transcription factor NF-kappaB inhibitors as single therapeutic agents or in combination with classical chemotherapeutic agents for the treatment of hematologic malignancies. Curr Mol Pharmacol. 2010;3:98–122. doi: 10.2174/1874467211003030098. [DOI] [PubMed] [Google Scholar]
- 7.Tosic N, Petrovic I, Grujicic NK, et al. Prognostic significance of SOX2, SOX3, SOX11, SOX14 and SOX18 gene expression in adult de novo acute myeloid leukemia. Leuk Res. 2018;67:32–8. doi: 10.1016/j.leukres.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Dong C, Wilhelm D, Koopman P. Sox genes and cancer. Cytogenet Genome Res. 2004;105:442–7. doi: 10.1159/000078217. [DOI] [PubMed] [Google Scholar]
- 9.Zhang H, Alberich-Jorda M, Amabile G, et al. Sox4 is a key oncogenic target in C/EBPalpha mutant acute myeloid leukemia. Cancer Cell. 2013;24:575–88. doi: 10.1016/j.ccr.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meggendorfer M, Kern W, Haferlach C, et al. SOX11 overexpression is a specific marker for mantle cell lymphoma and correlates with t(11;14) translocation, CCND1 expression and an adverse prognosis. Leukemia. 2013;27:2388–91. doi: 10.1038/leu.2013.141. [DOI] [PubMed] [Google Scholar]
- 11.Wan H, Cai J, Chen F, et al. SOX12: a novel potential target for acute myeloid leukaemia. Br J Haematol. 2017;176:421–30. doi: 10.1111/bjh.14425. [DOI] [PubMed] [Google Scholar]
- 12.He S, Tang S. WNT/beta-catenin signaling in the development of liver cancers. Biomed Pharmacother. 2020;132:110851. doi: 10.1016/j.biopha.2020.110851. [DOI] [PubMed] [Google Scholar]
- 13.Man CH, Fung TK, Wan H, et al. Suppression of SOX7 by DNA methylation and its tumor suppressor function in acute myeloid leukemia. Blood. 2015;125:3928–36. doi: 10.1182/blood-2014-06-580993. [DOI] [PubMed] [Google Scholar]
- 14.Yang MH, Chen CL, Chau GY, et al. Comprehensive analysis of the independent effect of twist and snail in promoting metastasis of hepatocellular carcinoma. Hepatology. 2009;50:1464–74. doi: 10.1002/hep.23221. [DOI] [PubMed] [Google Scholar]
- 15.Huang W, Chen Z, Shang X, et al. Sox12, a direct target of FoxQ1, promotes hepatocellular carcinoma metastasis through up-regulating Twist1 and FGFBP1. Hepatology. 2015;61:1920–33. doi: 10.1002/hep.27756. [DOI] [PubMed] [Google Scholar]
- 16.Parasramka MA, Maji S, Matsuda A, et al. Long non-coding RNAs as novel targets for therapy in hepatocellular carcinoma. Pharmacol Ther. 2016;161:67–78. doi: 10.1016/j.pharmthera.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghadiri A, Sharifi M, Mehrzad V, et al. Reduce proliferation of human bone marrow cells from acute myeloblastic leukemia with minimally differentiation by blocking lncRNA PVT1. Clin Transl Oncol. 2020;22:2103–10. doi: 10.1007/s12094-020-02360-4. [DOI] [PubMed] [Google Scholar]
- 18.Penzo-Mendez AI. Critical roles for SoxC transcription factors in development and cancer. Int J Biochem Cell Biol. 2010;42:425–8. doi: 10.1016/j.biocel.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Hu F, Shen S, et al. Knockdown of SOX12 expression inhibits the proliferation and metastasis of lung cancer cells. Am J Transl Res. 2017;9:4003–14. [PMC free article] [PubMed] [Google Scholar]
- 20.Ding H, Quan H, Yan W, et al. Silencing of SOX12 by shRNA suppresses migration, invasion and proliferation of breast cancer cells. Biosci Rep. 2016:36. doi: 10.1042/BSR20160053. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Yuan P, Meng L, Wang N. SOX12 upregulation is associated with metastasis of hepatocellular carcinoma and increases CDK4 and IGF2BP1 expression. Eur Rev Med Pharmacol Sci. 2017;21:3821–6. [PubMed] [Google Scholar]
- 22.Zou S, Wang C, Liu J, et al. Sox12 Is a Cancer Stem-Like Cell Marker in Hepatocellular Carcinoma. Mol Cells. 2017;40:847–54. doi: 10.14348/molcells.2017.0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du F, Chen J, Liu H, et al. SOX12 promotes colorectal cancer cell proliferation and metastasis by regulating asparagine synthesis. Cell Death Dis. 2019;10:239. doi: 10.1038/s41419-019-1481-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Wang Z, Huang L, et al. MiR-29b suppresses proliferation and mobility by targeting SOX12 and DNMT3b in pancreatic cancer. Anticancer Drugs. 2019;30:281–8. doi: 10.1097/CAD.0000000000000719. [DOI] [PubMed] [Google Scholar]
- 25.Leung RKC, Leung HC, Leung AYH. Diverse pathogenetic roles of SOX genes in acute myeloid leukaemia and their therapeutic implications. Semin Cancer Biol. 2020;67:24–9. doi: 10.1016/j.semcancer.2019.11.001. [DOI] [PubMed] [Google Scholar]
- 26.Pan C, Liang L, Wang Z, et al. Expression and significance of SOX B1 genes in glioblastoma multiforme patients. J Cell Mol Med. 2022;26:789–99. doi: 10.1111/jcmm.17120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ola MS, Nawaz M, Ahsan H. Role of Bcl-2 family proteins and caspases in the regulation of apoptosis. Mol Cell Biochem. 2011;351:41–58. doi: 10.1007/s11010-010-0709-x. [DOI] [PubMed] [Google Scholar]
- 28.Fesik SW. Promoting apoptosis as a strategy for cancer drug discovery. Nat Rev Cancer. 2005;5:876–85. doi: 10.1038/nrc1736. [DOI] [PubMed] [Google Scholar]
- 29.Andersen MH, Becker JC, Straten P. Regulators of apoptosis: suitable targets for immune therapy of cancer. Nat Rev Drug Discov. 2005;4:399–409. doi: 10.1038/nrd1717. [DOI] [PubMed] [Google Scholar]
- 30.Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770–6. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- 31.Kania J, Konturek SJ, Marlicz K, et al. Expression of survivin and caspase-3 in gastric cancer. Dig Dis Sci. 2003;48:266–71. doi: 10.1023/a:1021915124064. [DOI] [PubMed] [Google Scholar]
- 32.Soung YH, Lee JW, Kim SY, et al. Somatic mutations of CASP3 gene in human cancers. Hum Genet. 2004;115:112–5. doi: 10.1007/s00439-004-1129-3. [DOI] [PubMed] [Google Scholar]
- 33.Sharifi M, Moridnia A. Apoptosis-inducing and antiproliferative effect by inhibition of miR-182-5p through the regulation of CASP9 expression in human breast cancer. Cancer Gene Ther. 2017;24:75–82. doi: 10.1038/cgt.2016.79. [DOI] [PubMed] [Google Scholar]