Abstract
Strains of insect-pathogenic fungi with high virulence toward certain pest insects have great potential for commercial biological control applications. Identifying such strains has been a central theme in using fungi for biological control. This theme is supported by a persistent paradigm in insect pathology which suggests that the host insect is the predominant influence on the population genetics of insect-pathogenic fungi. In this study, a population genetics analysis of the insect-pathogenic fungus Metarhizium anisopliae from forested and agricultural habitats in Ontario, Canada, showed a nonrandom association of alleles between two distinct, reproductively isolated groups (index of multilocus association = 1.2). Analyses of the mitochondrial DNA showed no differences between the groups. The two groups were associated with different habitat types, and associations with insect hosts were not found. The group from forested areas showed an ability for cold-active growth (i.e., 8°C), while the group from the agricultural area showed an ability for growth at high temperatures (i.e., 37°C) and resilience to UV exposure. These results represent a significant paradigm shift; habitat selection, not host insect selection, drives the population structure of these insect-pathogenic deuteromycetous fungi. With each group we observed recombining population structures as well as clonally reproducing lineages. We discuss whether these groups may represent cryptic species. Worldwide, M. anisopliae may be an assembly of cryptic species, each adapted to certain environmental conditions. The association of fungal genotypes with habitat but not with host insects has implications on the criteria for utility of this, and perhaps other, fungal biocontrol agents.
Insect-pathogenic fungi have genetic features related to insect infection (17), and the population genetics of these fungi are also assumed to be influenced primarily by host insect taxa (5, 6, 14, 19, 20, 24, 28, 33, 35). Metarhizium anisopliae is an insect-pathogenic, haploid, deuteromycetous fungus that is assumed to reproduce clonally, and it is also assumed that certain genotypes are related to an insect host (5, 6, 14, 19, 20, 24, 28, 33, 35). It also has the potential for parasexual reproduction (36), although an analysis of clonality versus recombination has not been undertaken. One of the distinctive features of a clonal population is the widespread occurrence of identical genotypes (23). Here we have undertaken to determine the population structure of M. anisopliae and to test the paradigm that certain genotypes are related to insect hosts.
M. anisopliae is a recognized pathogen of more than 200 insect species, including several major pests (29). It is a recurrent paradigm in the literature that the insect host drives the population structure, i.e., that there are fungal isolates or genotypes adapted for pathogenesis toward certain species or taxa of insects (5, 6, 14, 19, 20, 24, 28, 32, 35). Because this fungus offers an environmentally safe alternative to chemical pesticides, it is of growing interest in the control of agriculturally important pests (29) and more virulent strains are being sought for commercial biological-control applications. In the context of isolating more virulent strains and tracking a strain released into the environment, an analysis of the population structure is critical. If the fungus is strictly clonal, isolation, application, and tracking of the strain would be relatively straightforward. But if recombination occurs, the effectiveness of a particular strain may be questionable and tracking becomes more complicated.
Here we applied tools of population genetics to analyze the genetic structure of M. anisopliae isolates from Ontario, Canada. Statistical analysis was used to establish the extent of linkage disequilibrium amongst loci and therein provide some indication of the population structure of this entomopathogenic fungus and potential association of fungi with an insect host or habitat.
MATERIALS AND METHODS
Fungal isolates.
M. anisopliae was equally sampled from agricultural and forested soils across Ontario, Canada (3). The isolates were grown on potato dextrose agar slants at 25°C. Conidiospores from these isolates were inoculated into 150 ml of 0.1% (wt/vol) yeast extract–2% peptone–1% dextrose in flasks and shaken at 200 rpm at 25°C for 3 to 4 days. The mycelia were then removed by vacuum filtration onto filter paper and stored at −80°C. The samples were then crushed in liquid nitrogen by using a mortar and pestle, and DNA was extracted from them.
Analysis of polymorphisms.
We assessed three measures of genetic variability: allozymes, random amplified polymorphic DNAs (RAPDs), and restriction fragment length polymorphisms (RFLP) of a subtilisin-like protease-encoding gene, pr1 (19) in 83 isolates of M. anisopliae.
Horizontal starch gel electrophoresis techniques were used to score allozyme variation (22). This technique has previously been applied to M. anisopliae (33), so only polymorphic loci were assayed. All isolates were examined for the enzymes glutamate dehydrogenase (GDH), glucosephosphate isomerase, glutathione reductase (GR), malate dehydrogenase (MDH), superoxide dismutase (SOD), and phosphogluconate dehydrogenase (PGDH).
Primers and sequences for RAPDs were OPE-8, 5′-AACGGCGACA; OPE-13, 5′-AGGACTGCCA; and OPE-16, 5′-GGTGAACGCT. These primers were chosen, since they showed polymorphic banding patterns for the isolates used. Amplifications were run in a thermal cycler (mini-cycler; MJ Research), under previously described conditions (4). Each different RAPD banding pattern was treated as a single allelic variant.
Primers, amplification conditions, and restriction enzyme digests (RsaI and DdeI) for pr1 were similar to those previously used (19).
Portions of the mitochondrial DNA (mtDNA), including the large ribosomal DNA (rDNA), were amplified with previously outlined primers and protocols (37).
Data analysis.
Using NTSYS (30), a similarity matrix was calculated for each pairwise comparison of fungal isolates, based on the simple-matching coefficient method (31). The matrix was analyzed for hierarchical clustering by the method unweighted pair group method with arithmetic averages (31). Allele mismatch distributions for each pairwise comparison were calculated (STATISTICA for Windows, StatSoft, Inc., Tulsa, Okla.).
The observed population was compared to 10,000 computer-generated theoretical populations, each with an n of 83. The haplotype of each theoretical fungal isolate was generated by random sampling of alleles at each locus using allele frequencies found in the observed population. These theoretical populations approximate random association of alleles found in a sexually reproducing population. Allele mismatch distributions were also determined for each pairwise comparison (3,403 pairwise comparisons [n = 83]) in each of the 10,000 computer-generated populations.
A frequency distribution of the number of allelic differences between isolates in the observed population and in theoretical populations was produced, and the variance was calculated (STATISTICA for Windows). Using these variances, an index of multilocus association (IA), which indicates the extent of linkage disequilibrium amongst loci in the population, was calculated (7). The observed variance is divided by the expected variance, and 1 is then subtracted from this value [(Vobs/Vexp) − 1]. An IA value near 0 indicates random association of alleles.
Fungal bioassays.
Four insect species, tobacco hornworm larvae (Manduca sexta), waxworm larvae (Galleria mellonella), mealworm beetle larvae (Tenebrio molitor), and cricket (Gryllus pennsylvanicus), were bioassayed against all fungal isolates. Ten microliters of a conidial suspension (107 conidia/ml) was applied to the insect surface, and the insects were housed separately in plastic vials and incubated at 25°C. Insects were fed and checked daily for mortality. Twenty insects were bioassayed for each of the 83 fungal isolates.
Assessment of cold activity, thermal activity, and UV resilience.
Physiological tests were performed after defining the population genetic structure, and these data were not used in determining genetic groups. Fungal growth under various conditions was assessed spectrophotometrically in a 96-well, flat-bottomed cell culture plate filled with 100 μl of potato dextrose agar. Each well was inoculated with 3 μl of a conidial suspension (approximately 105 conidia) in 0.01% Triton X-100. Cold-active growth was assessed at 8°C, and an A630 of >0.25 after 14 days was chosen as indicative of cold-active growth. Thermal-active growth was assessed at 37°C, and an A630 of >0.25 after 5 days was chosen as indicative of thermal-active growth. UV resilience was assessed after exposing the fungal conidia to the equivalent of UV light at 2,200 mW m−2 for 1 h. The cell culture plates were then incubated at 25°C, and an A630 of >0.25 after 6 days was chosen as indicative of UV resilience. All assays were done in triplicate.
RESULTS AND DISCUSSION
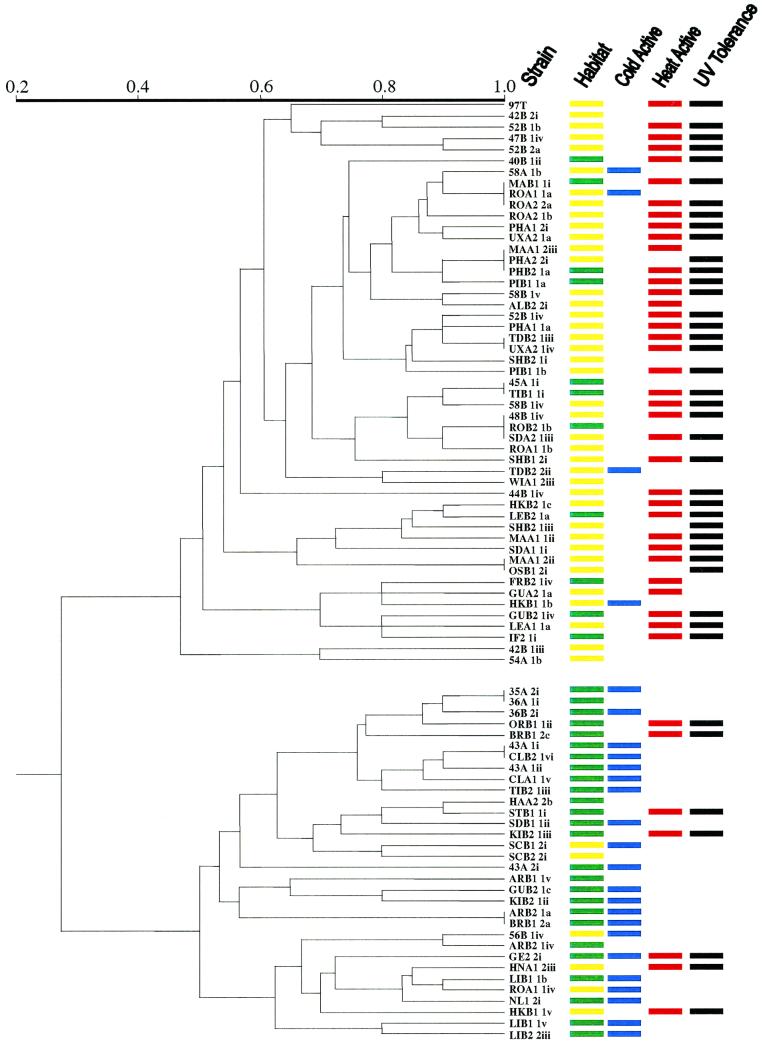
Table 1 shows the phenotypes for each of the 10 markers assayed for each of the isolates. Based on the phenotype of each of the measures of variability (allozymes, RAPDs, and RFLP of pr1) in 83 isolates of M. anisopliae and using a simple matching-coefficient method, a similarity matrix was calculated for each pairwise comparison of the fungal isolates. Using all informative characters, a cluster analysis revealed two distinctive, deeply rooted groups (Fig. 1). Using only allozyme data, 14 genotypic classes were observed and again two distinct groups were observed. Cluster analysis of the RAPD data concurred, and again the results revealed the same deeply rooted groups but showed a higher degree of variability between isolates. The use of RAPDs alone as markers in defining fungal population structures may be problematic, since they are dominant markers and not all bands may segregate in crosses. However, in this particular study, the use of RAPDs was supported by the allozyme and RFLP data as diagnostic markers for delineating the two groups.
TABLE 1.
Presumed phenotypes using RAPDs, RFLP analysis of pr1 with RsaI and DdeI, and five allozyme loci for 83 isolates of M. anisopliae collected in Ontario, Canadaab
| Isolate | RAPD phenotype for:
|
pr1 phenotype for:
|
Allozyme phenotype for:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OPE-8 | OPE-13 | OPE-16 | RsaI | DdeI | GDH | GR | MDH | PGDH | SOD | |
| 97T | 1 | 6 | 1 | 3 | 2 | 2 | 2 | 1 | 2 | 2 |
| 35A2i | 2 | 4 | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 |
| 36A1i | 2 | 4 | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 |
| 36B2i | 2 | 4 | 1 | 2 | 1 | 1 | 2 | 1 | 1 | 1 |
| 40B1ii | 4 | 8 | 1 | 1 | 2 | 2 | 1 | 1 | 2 | 2 |
| 42B2i | 4 | 2 | 3 | 3 | 2 | 2 | 2 | 1 | 2 | 2 |
| 42B1iii | 5 | 8 | 2 | 2 | 2 | 1 | 2 | 1 | 2 | 2 |
| 43A1I | 2 | 4 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 1 |
| 43A1ii | 2 | 4 | 2 | 2 | 1 | 1 | 2 | 2 | 1 | 1 |
| 43A2i | 1 | 4 | 3 | 1 | 1 | 1 | 2 | 2 | 2 | 1 |
| 44B1iv | 2 | 3 | 4 | 1 | 2 | 2 | 2 | 1 | 1 | 2 |
| 45A1i | 3 | 2 | 3 | 1 | 2 | 2 | 1 | 1 | 2 | 2 |
| 47B1iv | 5 | 2 | 4 | 1 | 2 | 2 | 2 | 1 | 2 | 2 |
| 48B1iv | 3 | 2 | 4 | 1 | 2 | 2 | 1 | 1 | 2 | 2 |
| 52B1b | 4 | 3 | 3 | 1 | 2 | 2 | 2 | 1 | 2 | 2 |
| 52B2a | 5 | 2 | 1 | 1 | 2 | 2 | 2 | 1 | 2 | 2 |
| 52B1iv | 4 | 2 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 2 |
| 54A1b | 0 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 |
| 56B1iv | 2 | 4 | 1 | 1 | 1 | 1 | 2 | 1 | 2 | 2 |
| 58A1b | 5 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 2 | 2 |
| 58B1v | 1 | 1 | 3 | 1 | 2 | 2 | 1 | 1 | 2 | 2 |
| ALB22i | 4 | 1 | 3 | 1 | 2 | 2 | 1 | 1 | 2 | 2 |
| ARB11v | 1 | 2 | 1 | 1 | 2 | 1 | 2 | 2 | 1 | 1 |
| ARB21a | 4 | 1 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 1 |
| ARB21iv | 2 | 5 | 1 | 1 | 1 | 1 | 2 | 1 | 2 | 2 |
| BRB12a | 4 | 1 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 1 |
| BRB12c | 2 | 4 | 3 | 2 | 1 | 1 | 2 | 1 | 2 | 1 |
| CLA11v | 2 | 6 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 1 |
| CLB21vi | 2 | 4 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 1 |
| FRB21iv | 4 | 5 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 2 |
| GE22i | 2 | 1 | 1 | 2 | 2 | 1 | 2 | 1 | 1 | 1 |
| GUB21iv | 2 | 6 | 2 | 2 | 1 | 2 | 1 | 1 | 2 | 2 |
| GUA21a | 3 | 5 | 2 | 1 | 1 | 2 | 1 | 1 | 2 | 2 |
| GUB21c | 2 | 2 | 4 | 2 | 2 | 1 | 2 | 2 | 2 | 1 |
| HAA22b | 2 | 5 | 3 | 2 | 1 | 1 | 2 | 2 | 1 | 1 |
| HKB11b | 4 | 5 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 2 |
| HKB11v | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 2 | 1 |
| HKB21c | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 2 |
| HNA12iii | 2 | 7 | 1 | 1 | 2 | 1 | 2 | 1 | 2 | 1 |
| IF21i | 1 | 6 | 2 | 1 | 1 | 2 | 1 | 1 | 2 | 2 |
| SHB21iii | 3 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 2 |
| KIB21ii | 2 | 2 | 4 | 1 | 2 | 1 | 2 | 2 | 3 | 1 |
| KIB21iii | 2 | 5 | 2 | 1 | 1 | 1 | 2 | 2 | 2 | 1 |
| LEA11a | 1 | 5 | 2 | 2 | 1 | 2 | 1 | 1 | 2 | 2 |
| LEB21a | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 2 |
| LIB11b | 2 | 1 | 1 | 1 | 2 | 1 | 2 | 1 | 2 | 1 |
| LIB11v | 2 | 6 | 2 | 1 | 3 | 1 | 2 | 1 | 2 | 1 |
| LIB22iii | 2 | 6 | 1 | 2 | 3 | 1 | 2 | 1 | 2 | 1 |
| MAA12ii | 1 | 6 | 2 | 2 | 3 | 2 | 1 | 1 | 1 | 2 |
| MAA11ii | 1 | 1 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 2 |
| MAA12iii | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 2 |
| MAB11i | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 2 | 2 |
| NL12i | 2 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 2 | 1 |
| ORB11ii | 2 | 6 | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 |
| OSB12i | 1 | 6 | 2 | 2 | 3 | 2 | 1 | 1 | 1 | 2 |
| PIB11a | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 2 |
| PIB11b | 1 | 2 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 2 |
| PHA11a | 3 | 2 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 2 |
| PHA12i | 1 | 3 | 1 | 1 | 2 | 2 | 1 | 1 | 2 | 2 |
| PHA22i | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 2 |
| PHB21a | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 2 |
| ROA11a | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 2 | 2 |
| ROA11b | 3 | 2 | 4 | 1 | 2 | 2 | 1 | 1 | 1 | 2 |
| ROA11iv | 2 | 1 | 1 | 1 | 2 | 1 | 2 | 1 | 2 | 2 |
| ROA21b | 1 | 1 | 4 | 1 | 2 | 2 | 1 | 1 | 2 | 2 |
| ROA22a | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 2 | 2 |
| ROB21b | 3 | 2 | 4 | 1 | 2 | 2 | 1 | 1 | 2 | 2 |
| SCB12i | 2 | 5 | 5 | 1 | 2 | 1 | 2 | 2 | 3 | 1 |
| SCB22i | 2 | 5 | 5 | 2 | 2 | 1 | 2 | 2 | 1 | 1 |
| SDA11i | 1 | 1 | 1 | 2 | 1 | 2 | 1 | 1 | 1 | 1 |
| SDB11ii | 2 | 5 | 6 | 1 | 1 | 1 | 2 | 2 | 1 | 1 |
| SHB12i | 3 | 2 | 4 | 2 | 2 | 2 | 1 | 1 | 1 | 2 |
| SHB21i | 1 | 2 | 3 | 1 | 2 | 2 | 1 | 1 | 1 | 2 |
| SDA21iii | 3 | 2 | 4 | 1 | 2 | 2 | 1 | 1 | 2 | 2 |
| STB11i | 2 | 5 | 1 | 2 | 1 | 1 | 2 | 2 | 1 | 1 |
| TIB11i | 3 | 2 | 3 | 1 | 2 | 2 | 1 | 1 | 2 | 2 |
| TIB21iii | 2 | 4 | 4 | 2 | 1 | 1 | 2 | 2 | 1 | 1 |
| TDB22ii | 1 | 2 | 2 | 1 | 2 | 1 | 1 | 1 | 2 | 2 |
| TDB21iii | 1 | 2 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 2 |
| UXA21a | 1 | 2 | 1 | 1 | 2 | 2 | 1 | 1 | 2 | 2 |
| UXA21iv | 1 | 2 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 2 |
| WIA12iii | 1 | 2 | 1 | 1 | 2 | 1 | 2 | 1 | 2 | 2 |
| 58B1iv | 1 | 2 | 3 | 1 | 2 | 2 | 1 | 1 | 2 | 2 |
For RAPDs and RFLP, each number refers to a different major banding pattern. For allozymes, each number refers to a single band with a different electrophoretic mobility.
The first two letters or first two numbers of isolate refer to location: 97, Peterborough; 35 and 36, Long Lake; 40, Kirby; 42, Elizabethville; 43, Bewdley; 44 and 45, Parkhill Rd.; 47, Frazerville; 48, South Monaghan; 52, Mathers Corners; 54, East of Assumption; 56, Norwood; 58, Dummer Lake; AL, Alliston; AR, Arthur; BR, Bracebridge; CL, Cloyne; FR, Frankford; GE, Geraldton; GU, Guelph; HA, Haldimand; HK, Havelock; HN, Haliburton; IF, Iroquois Falls; KI, Kinmount; LE, Leamington; LI, Lindsay; MA, Madoc; NL, New Liskeard; OR, Orillia; OS, Owen Sound; PI, Picton; PH, Parkhill; RO, Rodney; SC, St. Catharines; SD, Stratford; SH, Sharbot Lake; ST, St. Thomas; TI, Tillsonburg; TD, Toledo; UX, Uxbridge; WI, Wingham; WO, Woodstock.
FIG. 1.
Dendrogram illustrating the genetic relatedness amongst Ontario isolates of M. anisopliae. Two distinct, deeply rooted groups are observed. Bars to the right of the dendrogram represent the habitat location (yellow indicates agricultural habitat and green represents forested habitat), cold activity (blue represents ability to grow at 8°C), thermal activity (red represents ability to grow at 37°C), and UV tolerance (black represents ability to grow after UV exposure) of each of the isolates.
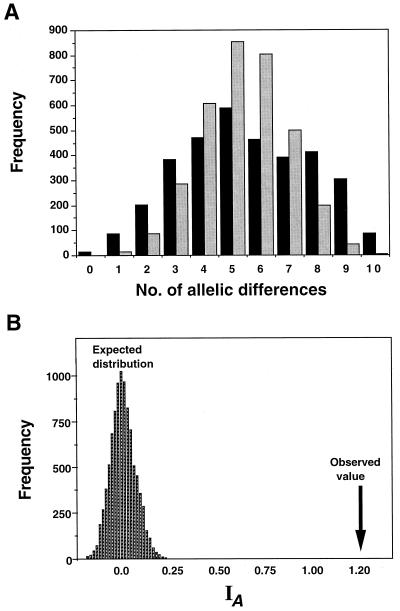
To test for gametic disequilibrium in the entire sample of isolates, we used IA (7). The IA values for the 10,000 theoretical populations clustered around a mean IA of 0 (Fig. 2). The IA of 1.2 in the observed population has a P value of <0.00001 of being within the distribution of the values calculated for random association of alleles (Fig. 2). We applied the same analysis, using only the allozyme data because of the potential problems of nonsegregating RAPD bands, and again the results showed an IA of 1.4 (P < 0.00001). Therefore, the two groups represent two distinctive, nonrecombining genetic lineages, Ontario group 1 (OG1) and Ontario group 2 (OG2).
FIG. 2.
(A) The allele mismatch distribution in Ontario isolates of M. anisopliae (gray bars) and the mean value of 10,000 computer-generated replicates by random sampling of alleles (black bars). The haplotype of each theoretical fungal isolate was generated by random sampling of alleles at each locus using allele frequencies found in the observed population. Allele mismatch distributions for each pairwise comparison were calculated, and the value for the observed population was compared to the mean value for the computer-generated theoretical populations (i.e., 3,403 pairwise comparisons [n = 83] in each of the 10,000 computer-generated populations). The computer-generated distribution is unimodal, while the observed population shows a bimodal distribution pattern and an inflated variance owing to linkage disequilibrium (B). The IA values were calculated for the 10,000 computer-generated populations. The arrow shows the calculated IA of 1.2 in the observed population.
Cluster analysis can create the appearance of groups even where none exist. To ensure that these were true groups, Fisher's exact test was applied to each variable. Seven of the 10 variables (SOD, GDH, GR, MDH, RFLP of pr1 using RsaI or DdeI, and RAPD patterns produced by OPE-8) were diagnostic of a group. Allozyme data indicated that of the two SOD alleles, the fast-migrating allele (SOD-f) was associated with OG1 and the slow migrating allele (SOD-s) was associated with OG2 (Fisher's exact test, P < 0.00001). Similarly, GDH-s was associated with OG1 and GDH-f with OG2 (Fisher's exact test, P < 0.00001); GR-s was associated with OG1 and GR-f with OG2 (Fisher's exact test, P < 0.00001); MDH-f was associated with OG1 and MDH-s with OG2 (Fisher's exact test, P < 0.00001). Two endonucleases which showed multiple polymorphisms with pr1, RsaI and DdeI, also showed RFLP patterns diagnostic of a group. Restriction patterns generated by the endonucleases produced six banding types; each enzyme produced three profile types. One RsaI banding pattern was associated with OG1, while two banding patterns were associated with OG2 (Fisher's exact test, P < 0.00001). Two DdeI banding patterns were characteristic of OG1, while one banding pattern was characteristic of OG2 (Fisher's exact test, P < 0.00001). Of the five distinctive banding patterns produced by RAPD primer OPE-8, two banding patterns were characteristic of OG2, while three were characteristic of OG1 (Fisher's exact test, P < 0.00001).
We found evidence of clonal lineages within each group, since isolates from disparate locations shared identical genotypes. For example, isolates MAB1 1i (from Madoc, Ont., Canada) and ROA1 1a (from Rodney, Ont., Canada) in OG1 shared identical genotypes and were collected ca. 400 km from each other. Isolates with identical genotypes in OG2, ARB21a (from Arthur, Ont., Canada) and BRB12a (from Bracebridge, Ont., Canada), were collected ca. 200 km from each other.
We also found evidence of recombination within each group. We grouped characters (using only the isozyme data since using RAPD and RFLP data can be problematic [34]) into monophyletic biallelic families for each of the five allozyme loci and compared these groupings for the 10 pairwise combinations of loci. The analysis demonstrated that all four combinations of characters were found for 3 of the 10 pairwise combinations of loci in OG1 (between GDH and GR, GDH and PGDH, and GR and PGDH). Three of four combinations of characters were found for three other pairwise combinations. In OG2, all four combinations of characters were found in one of the pairwise combinations of loci (MDH and PGDH), and three combinations of characters were found in two other pairwise combinations of loci.
The analysis of clonality or recombination within each group using the IA was ambiguous because the isolates were sampled from two reproductively isolated groups. Many of the alleles were nearly fixed for each group, and this could cause association of alleles even if recombination were the norm (34). The most common allozyme allele frequencies in OG1 were GDH-s (0.97), GR-s (1.00), MDH-f (0.59), PGDH-f (0.59), and SOD-f (1.00). In OG2 the most common allozyme allele frequencies were GDH-f (0.85), GR-f (0.74), MDH-s (1.00), PGDH-f (0.61), and SOD-s (0.98).
Evaluation of mtDNA by (i) RFLP analysis of total mtDNA and (ii) sequence analysis of the mitochondrial large rDNA from 15 isolates randomly chosen from each group also showed no nucleotide variation (data not shown). A qualitative summary of molecular methods for fungal systematics (8) showed that little variation should be observed at the population level for mitochondrial rDNA genes. Since the groups had no apparent differences in the mtDNA, we are certain that they are closely related.
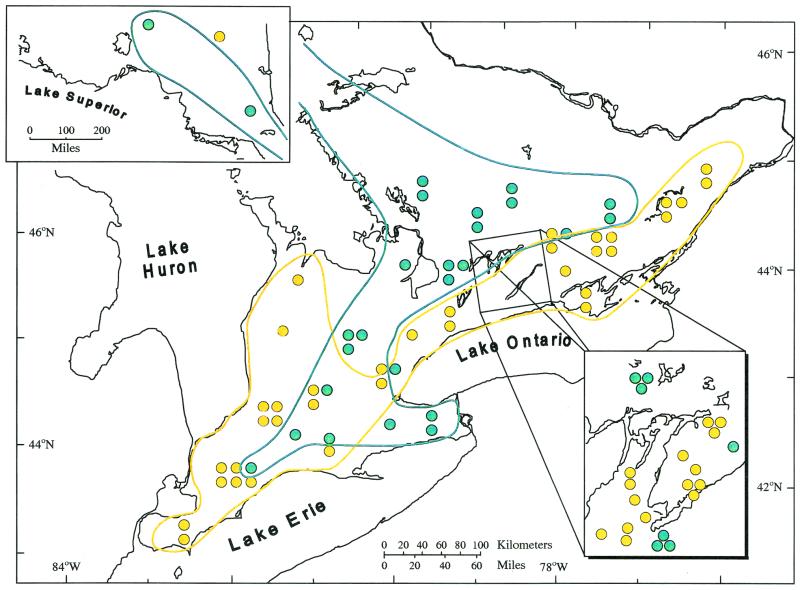
Once we had determined that we had defined two separate genetic groups, we analyzed the association of the genetic groups with their geographic origins. Figure 3 shows the association of the two M. anisopliae genetic groups and their geographic origins. We also assessed the habitat type from which the fungi were isolated within each geographic locale, and the genetic groups were strongly associated with habitat type. The isolates from agricultural soils, regardless of crop type (hay, alfalfa, corn, wheat, barley, beans, peaches, and grapes), belonged predominantly to OG1, while isolates in OG2 were from forested-soil samples (coniferous, deciduous, and mixed; χ2 = 28.35 degrees of freedom [df] = 1, P < 0.00001). Using an insect pest database, we listed potential insect hosts from each habitat and could not find any insect species that occurred in all of the crop types of the agricultural habitat or all of the biomes found in the forested habitat. Furthermore, several insect species were bioassayed for each of the M. anisopliae isolates and included a coleopteran (T. molitor), two lepidopterans (M. sexta and G. mellonella), and an orthopteran (G. pennsylvanicus). Bioassay results were variable, but no consistent patterns of virulence for each clonal group, assessed by time (in days) for 50% mortality (LT50 values), were noted.
FIG. 3.
Geographic distribution of group 1 (yellow) and group 2 (green) in Ontario. Groups overlap in southwestern Ontario, which corresponds with a region with a high diversity of forested and agricultural habitats. Inset at top left shows northern Ontario, while inset at lower right shows the Kawartha Lakes region.
If M. anisopliae is an insect pathogen, why is habitat influencing its population genetics? In the absence of an insect host, M. anisopliae has the ability to survive in the soil, and viability is influenced by factors such as pesticides (27), organic content of the soil (13), desiccation (11), UV, and temperature (38). It is here, in the physical environment, that we speculate that selective factors mediate population genetics. The principal difference between soils of the forested and agricultural habitats was that the soils of the forested habitats were canopied, while the agricultural soils were continuously or intermittently exposed.
Although there are presumably many factors within these general habitats that could influence the population structure of M. anisopliae, at least three candidate factors (cold activity, ability to grow at high temperatures, and UV resilience) could act as selective forces in determining population distribution patterns (Fig. 1). These factors were not used to assess the population structure of these fungi but were included to show the potential association of physiological parameters with population genetics. We observed a strong association of M. anisopliae OG2 with the ability to grow at 8°C (χ2 = 31.19, df = 1, P < 0.00001). Recently, we have identified transcripts related to cold-active growth from several cold-active strains of M. anisopliae (J. N. A. De Croos and M. J. Bidochka, unpublished data). We also observed a strong association of M. anisopliae OG1 with the ability to grow at 37°C (χ2 = 18.69, df = 1, P < 0.00001) and with resilience to UV exposure (χ2 = 17.19, df = 1, P < 0.00001). We propose that it is the saprophytic phase, not the pathogenic phase, of this facultative pathogen's life cycle that is most important in shaping its population genetics. In a review article, Carruthers and Soper (10) insightfully stated that “…the pathogen may be limited in the field by environmental conditions and/or spatial and temporal interactions with its host rather than … the lack of pathogenicity.” The search for highly virulent isolates of this fungus directed at certain insect pests may be inherently flawed, since virulent isolates have comparable facility to infect susceptible insects but have large discrepancies in their abilities to tolerate certain environmental conditions.
The two groups were geographically isolated from one another with the exception of a small area in south-central Ontario (Fig. 3) possessing a relatively good mix of forested and agricultural habitats. The fungal isolates in this area contained all possible alleles, so that even when these fungi were in relative proximity to one another, there was no evidence of recombination between the groups. A plausible interpretation for the present geographic distribution of the two groups could be one of migration by individuals of one group into an unaltered habitat and then migration by another group into a habitat altered by agricultural practices in Ontario. The migration of separate groups into the same geographic area has been documented for certain clonal plant species (15). If interpreted in this way, it is possible that OG2 migrated into Ontario shortly after the time of the glacial retreat (ca. 8,000 years ago). The apparent success of OG2 in this forested habitat could correlate with its ability to grow at low temperatures. OG1, on the other hand, is probably a more southern group existing in the United States. There is evidence that genotypes similar to OG1 exist in North Carolina (i.e., strain 2575). It is possible that as land became cultivated in Ontario (in the early 1900s), conidia of OG1 were dispersed north into southern Ontario, where they were able to survive the open habitat in which relatively higher temperatures and UV exposure occur. Farther east in Ontario, OG2 does occur near OG1, though usually in more northern locations. Prevailing wind patterns and continuous forest could explain their presence in this region.
Conclusions.
M. anisopliae is a haploid, mitosporic, insect-infecting fungus and has no known teleomorphic stages. However, our analysis showed a recombining population structure in addition to clonality. M. anisopliae falls within the one-fifth of described fungi thought to be asexual but recently shown to have a recombinational component to their population structure (34). Many fungi do not have true sexual stages (i.e., many deuteromycetes) but are still capable of genetic recombination through parasexuality. Therefore, they may have a recombinational component to their population structure and are not considered to be totally clonal, as is Coccidioides immitis (9).
Modes of genetic recombination have been identified in M. anisopliae, but it can only anastomose with closely related, parasexually compatible isolates (2, 33). We suggest that individuals within a group have the capability to recombine. Several species of insect-pathogenic fungi grow as protoplasts in the hemolymph of an infected insect (25). Here there is the potential for protoplast fusion and genetic exchange. Recently, genetic recombination has been shown by isolates of M. anisopliae coinfecting an insect (21). With respect to the potential for sexual outcrossing, a teleomorph for M. anisopliae has not been described, but Cordyceps taii was shown to be the teleomorph to Metarhizium taii (39). However, neither parasexuality, sexual reproduction, nor perithecia have been demonstrated in nature for M. anisopliae.
It is clear from the level of nearly fixed alleles in the two groups that they have a long history of reproductive isolation and may be considered different biological species (1). Our interpretation is very similar to that found for cryptic species of Aspergillus flavus (16), and it would be interesting to determine whether different environmental tolerances are associated with the groups in this fungus. Sympatric occurrence between the M. anisopliae groups could reflect secondary contact due to the effects of agriculture. Again, a similar interpretation was forwarded for the distribution of A. flavus genotypes (16). On a worldwide level, St. Leger et al. (33) assessed genetic variation in 120 isolates of M. anisopliae and grouped them into 48 genotypic classes. By an earlier interpretation of their genetic similarity dendrogram (33), these genotypic classes broadly represent nine groups worldwide. A phylogenetic analysis of the genus Metarhizium based on rDNA sequence analysis revealed 10 distinct clades worldwide (12). These groups or clades of M. anisopliae may represent an aggregate of cryptic species. A more comprehensive population genetics investigation of a worldwide sample of isolates and how they are adapted to different environmental conditions may reveal patterns of allopatric speciation and secondary contact to which our data have only alluded. We do not believe that secondary contact can lead to genetic rehomogenation, since evidence suggests that individuals in different groups are vegetatively incompatible (2).
There is some circumstantial evidence and interpretation that M. anisopliae is genotypically differentiated according to insect host specificity (5, 6, 14, 19, 20, 24, 28, 33, 35). Habitat influences the distribution and abundance of many species of insects (26), and our data show that it also influences M. anisopliae distribution and abundance. Any evidence of insect host-related population structure within this fungus should be viewed primarily as a result of the habitat in which the insect and fungus cooccur. However, we do not dismiss that there may be other genotypes with evolutionary histories of insect host specificity, but this has not been entirely elucidated.
The interpretation of our data represents a paradigm shift in the perceived nature of the predominant factors influencing population genetics in M. anisopliae. There is no doubt that M. anisopliae has certain features that are adapted for insect pathogenesis (32), and up until now, this fact has been viewed as the predominant factor driving population structure. However, this is a facultative insect pathogen that has some ability to survive in the soil when not infecting a host insect. Habitat preferences should be considered as a feature for selecting fungal strains to be used in insect biocontrol efforts, particularly if fungal cycling and survival are key considerations. With respect to genetic engineering of biocontrol fungi, one can potentially elucidate most of the factors involved in virulence, theoretically engineer the most virulent fungus, but place the engineered genes into a fungal strain that survives poorly in a certain habitat. Pathogen infectivity and virulence on the one hand and survival outside the hosts and capacity to disperse on the other are crucial in biocontrol efforts.
ACKNOWLEDGMENT
This research was supported by an operating grant from the National Sciences and Engineering Research Council of Canada (NSERC) to M.J.B.
REFERENCES
- 1.Avise J C. Molecular markers, natural history and evolution. New York, N.Y: Chapman & Hall; 1994. [Google Scholar]
- 2.Bidochka M J, Melzer M J, Lavender T M, Kamp A M. Genetically related isolates for the entomopathogenic fungus Metarhizium anisopliae harbour homologous dsRNA viruses. Mycol Res. 2000;104:1094–1097. [Google Scholar]
- 3.Bidochka M J, Kasperski J E, Wild G A M. Occurrence of the entomopathogenic fungi Metarhizium anisopliae and Beauveria bassiana in soils from temperate and near-northern habitats. Can J Bot. 1998;76:1198–1204. [Google Scholar]
- 4.Bidochka M J, McDonald M A, St. Leger R J, Roberts D W. Differentiation of species and strains of entomopathogenic fungi by random amplification of polymorphic DNA (RAPD) Curr Genet. 1994;25:107–113. doi: 10.1007/BF00309534. [DOI] [PubMed] [Google Scholar]
- 5.Bridge P D, Prior C, Sagbohan J, Lomer C J, Carey M, Buddie A. Molecular characterization of isolates of Metarhizium from locusts and grasshoppers. Biodivers Conserv. 1997;6:177–189. [Google Scholar]
- 6.Bridge P D, Williams M A J, Prior C, Paterson R R M. Morphological, biochemical and molecular characteristics of Metarhizium anisopliae and M. flavoviride. J Gen Microbiol. 1993;139:1163–1169. [Google Scholar]
- 7.Brown A H D, Feldman M W, Nevo E. Multilocus structure of natural populations of Hordeum spontaneum. Genetics. 1980;96:523–536. doi: 10.1093/genetics/96.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruns T D, White T J, Taylor J W. Fungal molecular systematics. Annu Rev Ecol Syst. 1991;22:525–564. [Google Scholar]
- 9.Burt A, Carter D A, Koenig G L, White T J, Taylor J W. Molecular markers reveal cryptic sex in the human pathogen Coccidioides immitis. Proc Natl Acad Sci USA. 1996;93:770–773. doi: 10.1073/pnas.93.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carruthers R I, Soper R S. Fungal diseases. In: Fuxa J R, Tanada Y, editors. Epizootiology of insect diseases. New York, N.Y: John Wiley & Sons, Inc.; 1987. pp. 357–415. [Google Scholar]
- 11.Clerk G C, Madelin M F. The longevity of three insect-parasitizing Hyphomycetes. Trans Br Mycol Soc. 1965;48:193–209. [Google Scholar]
- 12.Driver F, Milner R J, Trueman J W H. A taxonomic revision of Metarhizium based on a phylogenetic analysis of rDNA sequence data. Mycol Res. 2000;104:134–150. [Google Scholar]
- 13.Dutky S R. Insect microbiology. Adv Appl Microbiol. 1959;1:175–200. doi: 10.1016/s0065-2164(08)70479-9. [DOI] [PubMed] [Google Scholar]
- 14.Fegan M, Manners J M, Maclean D J, Irwin J A G, Samuels K D Z, Holdom D G, Li D P. Random amplified polymorphic DNA markers reveal a high degree of genetic diversity in the entomopathogenic fungus Metarhizium anisopliae var. anisopliae. J Gen Microbiol. 1993;139:2075–2081. doi: 10.1099/00221287-139-9-2075. [DOI] [PubMed] [Google Scholar]
- 15.Gabrielsen T M, Brochman C. Sex after all: high levels of diversity detected in the arctic clonal plant Saxifraga cernua using RAPD markers. Mol Ecol. 1998;7:1701–1708. [Google Scholar]
- 16.Geiser D M, Pitt J I, Taylor J W. Cryptic speciation and recombination in the aflotoxin-producing fungus Aspergillus flavus. Proc Natl Acad Sci USA. 1998;95:388–393. doi: 10.1073/pnas.95.1.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hajek A E, St. Leger R J. Interactions between fungal pathogens and insect hosts. Annu Rev Entomol. 1994;39:293–322. [Google Scholar]
- 18.Koufopanou V, Burt A, Taylor J W. Concordance of gene genealogies reveals reproductive isolation in pathogenic fungus Coccidioides immitis. Proc Natl Acad Sci USA. 1997;94:5478–5482. doi: 10.1073/pnas.94.10.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leal S C M, Bertioli D J, Butt T M, Carder J H, Burrows P R, Peberdy J F. Amplification and restriction endonuclease digestion of the Pr1 gene for the detection and characterization of Metarhizium strains. Mycol Res. 1997;101:257–265. [Google Scholar]
- 20.Leal S C M, Bertioli D J, Butt T M, Peberdy J F. Characterization of isolates of the entomopathogenic fungus Metarhizium anisopliae by RAPD-PCR. Mycol Res. 1994;98:1077–1081. [Google Scholar]
- 21.Leal-Bertioli S C M, Butt T M, Peberdy J F, Bertioli D J. Genetic exchange in Metarhizium anisopliae strains co-infecting Phaedon cochleariae is revealed by molecular markers. Mycol Res. 2000;104:409–414. [Google Scholar]
- 22.May B. Starch gel electrophoresis of allozymes. In: Hoelzel A R, editor. Molecular genetic analysis of populations: a practical approach. Oxford, United Kingdom: IRL Press; 1994. pp. 1–27. [Google Scholar]
- 23.Milgroom M G. Recombination and the multilocus structure of fungal populations. Annu Rev Phytopathol. 1996;34:457–477. doi: 10.1146/annurev.phyto.34.1.457. [DOI] [PubMed] [Google Scholar]
- 24.Neuveglise C, Brygoo Y, Riba G. 28S rDNA group-I introns: a powerful tool for identifying strains of Beauveria brongniartii. Mol Ecol. 1997;6:373–381. doi: 10.1046/j.1365-294x.1997.00202.x. [DOI] [PubMed] [Google Scholar]
- 25.Pendland J C, Hung S-Y, Boucias D G. Evasion of host defense by in vivo-produced protoplast-like cells of the insect mycopathogen Beauveria bassiana. J Bacteriol. 1993;175:5962–5969. doi: 10.1128/jb.175.18.5962-5969.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price P W. Insect ecology. New York, N.Y: John Wiley & Sons, Inc.; 1997. [Google Scholar]
- 27.Ramarajah Urs N V, Govindu H C, Shivashankara K S. The effect of certain insecticides on the entomogenous fungi Beauveria bassiana and Metarhizium anisopliae. J Invertebr Pathol. 1967;9:398–403. doi: 10.1016/0022-2011(67)90077-8. [DOI] [PubMed] [Google Scholar]
- 28.Riba G, Bouvier-Fourcade I, Caudal A. Isoenzymes polymorphism in Metarhizium anisopliae (Deuteromycotina: Hyphomycetes) entomogenous fungi. Mycopathology. 1986;96:161–169. [Google Scholar]
- 29.Roberts D W, Hajek A E. Entomopathogenic fungi as bioinsecticides. In: Leatham G F, editor. Frontiers of industrial mycology. New York, N.Y: Chapman & Hall; 1992. pp. 144–159. [Google Scholar]
- 30.Rohlf F J. NTSYS-PC. Numerical taxonomy and multivariate analysis system, version 1.80. Setauket, N.Y: Exeter Software; 1994. [Google Scholar]
- 31.Sneath P, Sokal R. Numerical taxonomy. W. H. San Francisco, Calif: Freeman and Company; 1973. [Google Scholar]
- 32.St. Leger R J. Biology and mechanisms of insect-cuticle invasion by deuteromycetous fungal pathogens. In: Beckage N C, Thompson S N, Federici B A, editors. Parasites and pathogens of insects. Vol. 2. New York, N.Y: Academic Press; 1993. pp. 211–229. [Google Scholar]
- 33.St. Leger R J, May B, Allee L L, Frank D C, Staples R C, Roberts D W. Genetic differences in allozymes and in formation of infection structures among isolates of the entomopathogenic fungus Metarhizium anisopliae. J Invertebr Pathol. 1992;60:89–101. [Google Scholar]
- 34.Taylor J W, Jacobson D J, Fisher M C. The evolution of asexual fungi: reproduction, speciation and classification. Annu Rev Phytopathol. 1999;37:197–246. doi: 10.1146/annurev.phyto.37.1.197. [DOI] [PubMed] [Google Scholar]
- 35.Tigano-Milani M S, Gomes A C M M, Sobral B W S. Genetic variability among Brazilian isolates of the entomopathogenic fungus Metarhizium anisopliae. J Invertebr Pathol. 1995;65:206–210. [Google Scholar]
- 36.Tinline R D, Noviello C. Heterokaryosis in the entomogenous fungus Metarrhizium anisopliae. Mycology. 1971;63:701–712. [PubMed] [Google Scholar]
- 37.White T J, Bruns T D, Lee S B, Taylor J W. Analysis of phylogenetic relationships by amplification and direct sequencing of ribosomal DNA genes. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. New York, N.Y: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 38.Zimmerman G. Effect of high temperatures and artificial sunlight on the viability of conidia of Metarhizium anisopliae. J Invertebr Pathol. 1982;40:36–40. [Google Scholar]
- 39.Zong-Qui L, Ai-Ying L, Jie-Ling L. A new species of the genus Cordyceps and its Metarhizium anamorph. Acta Mycol Sin. 1991;10:257–262. [Google Scholar]