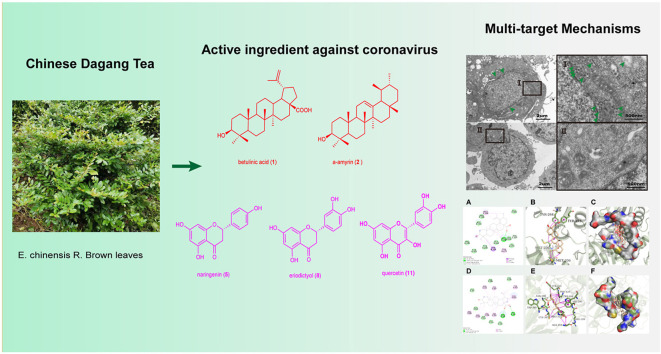
Abstract
Ethnopharmacological relevance
The leaves of Eurya chinensis(Chinese Dagang Tea)have been consumed as herbal tea for centuries in Guangdong, China, and have also been used to prevent influenza and treat colds and fevers in traditional Chinese medicine. However, there are no reports on the chemical profile and efficacy of its leaves for the treatment of fever and viral infections.
Materials and methods
The chemical constituents of Eurya chinensis leaves were isolated and identified by phytochemical study and spectroscopic data, E. chinensis extracts and compounds were evaluated for their antiviral activities by cytopathic effect (CPE) reduction and antibody-based EC50 assay. The antiviral effect of the main component was confirmed by immunofluorescence and transmission electron microscopy. Virtual screening and docking enzyme inhibition experiments were performed to analyze the anti-coronavirus mechanisms of the compounds from E. chinensis leaves.
Results
In this study, we found for the first time that E. chinensis leaf extract exhibited inhibitory effects against coronaviruses HCoV-OC43 in vitro. Among 23 monomer compounds isolated from E. chinensis leaf extract, the triterpenoids (betulinic acid, α-amyrin) and the flavonoids (naringenin, eriodictyol and quercetin) showed marked antiviral activity. Microscopic optical analyses further demonstrated that betulinic acid can remove virus particles from HCoV-OC43 infected cells. Virtual screening and docking analysis towards the coronavirus in vogue revealed that betulinic acid was able to bind well to PLpro and Nsp14N7-MTase, and that the flavonoids prefer to bind with PLpro, Nsp3MES, NspP14N7-MTase, Nsp16GTA, and Nsp16SAM. The enzyme inhibition experiments demonstrated that betulinic acid (1) exhibited significant inhibition of PLpro and N7-MTase activity of SARS-CoV-2.
Conclusion
This study proposes E. chinensis and its triterpenoids and flavonoids as promising potential treatments for coronaviruses.
Graphical abstract
1. Introduction
The ongoing outbreak of coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) (Zhu et al., 2020), constitutes an unprecedented threat to global public health (https://covid19.who.int/). The SARS-CoV-2 Omicron variant (B.1.1.529) identified by the World Health Organization (WHO) as the fifth “variant of concern” is characterized by a large number of mutations in the spike protein and extraordinary immune evasive ability (Cao et al., 2021; Planas et al., 2021). Increasing evidence shows the Omicron variant has strong immune escape potential that weakens the effectiveness of serums from vaccine recipients or convalescents as well as therapeutic monoclonal antibodies (Cele et al., 2021; C. Ma et al., 2022). Considering the intrinsically high possibility of mutations to beta coronavirus we must face, it is important to be proactive in developing broad-spectrum coronavirus vaccines and small molecule antiviral drugs to combat the Omicron variant and other potential new mutants of SARS-CoV-2, or even new coronaviruses in the future.
Notably, as members of the same beta coronavirus genus, human coronavirus OC43 (HCoV-OC43) and SARS-CoV-2 are closely related genetically and share several functional properties (Lu et al., 2020). HCoV-OC43 is considered to be the most prevalent subtype of HCoV (Zhang et al., 2015), causing 30% of respiratory infections in humans and leading to lifelong recurrent infections (Marra et al., 2003; Myint, 1995). Moreover, HCoV-OC43 is associated with fatal encephalitis (Kasereka and Hawkes, 2021; Morfopoulou et al., 2016). Due to its similarities to SARS-CoV-2, HCoV-OC43 has been used as a surrogate for these emerging viral strains to be studied without the requirement for biosafety level 3 (BSL-3) facilities(Kim et al., 2019; Ordonez et al., 2022; Park et al., 2021; Schirtzinger et al., 2022).
Despite their diversity of species, coronaviruses share critical nonstructural proteins (Nsps) necessary for viral replication and transcription, suggesting the possibility of developing broad-spectrum therapeutic agents to deal with the current epidemic and manage possible future outbreaks(Davies et al., 2020; Milani et al., 2021). Main protease (Mpro), also named as chymotrypsin-like protease (3CLpro), cleaves most of the sites in the polyproteins into Nsps. Papain-like protease (PLpro) works with Mpro to cleave the Nsp1/2, Nsp2/3 and Nsp3/4 polyproteins into Nsps. RNA-dependent RNA polymerase (RdRp, Nsp12/7/8) is important in the replication and transcription of the viral genome. Nsp14 in coronaviruses is important for viral replication and transcription, and the C-terminal domain functions as a guanine-N7 methyltransferase (N7-MTase) for mRNA capping. Nsp16 as S-adenosylmethionine (SAM) dependent nucleoside-2′-O methyltransferase is a conserved structure and essential for viral RNA stability and expression for coronaviruses.
Natural products are considered to be remarkable sources for developing anti-coronavirus drugs (Mani et al., 2020). Various compounds and preparations from traditional herbal medicines have yielded promising results in treating COVID-19 either alone or in combination with conventional medicines (Chen et al., 2021; Runfeng et al., 2020). Eurya chinensis R.Br. is a plant of the genus Eurya (family Pentaphylacaceae), mainly found in the valley and hilly areas of southern China. E. chinensis has been used in traditional Chinese medicine to treat influenza, fever, jaundice and traumatic injuries for more than six hundred years (J. Song et al., 2016). The flowers of E. chinensis bloom in winter and are a source of nectar for that season. In Jiangxi Province, its stems have been used traditionally to make rice cakes as an “ashwagandha” (immunity enhancer). In Guangdong Province, E. chinensis leaves have been used for hundreds of years as herbal “Dagang Tea” in order to cool down and prevent heat stroke.
Our group previously found that the main chemical constituents of E. chinensis stems include mainly terpenoids, coumarins and flavonoids (J. Song et al., 2018a, 2018b) possessing anti-inflammatory and antibacterial activity (J. L. Song et al., 2017). However, there are no reports on the chemical profile and efficacy of its leaves, which were widely used as “Dagang Tea” for clinical care and the treatment of fever and viral infections. Here, we showed that E. chinensis leaf extract exhibited anti-coronavirus activity. We further revealed the chemical composition of E. chinensis leaves responsible for the anti-coronavirus activity. Our study demonstrated that the compounds from E. chinensis leaves hold great promise for the treatment of coronaviruses.
2. Materials and methods
2.1. Plant material
The green leaves of E. chinensis R. Br. were harvested from Nankang District in Gangzhou City of Jiangxi Province of China in 2016. The voucher specimen (No.SCBG-NPL-201617) has been deposited in the Laboratory of Natural Product Chemistry Biology, South China Botanical Garden (SCBG).
2.2. Extraction, separation, and structure analysis
The air-dried leaves of E. chinensis (23.00 kg) were crushed and extracted with 95% EtOH for 48 h (30 L × 3) at room temperature. All the extracts were combined and evaporated under reduced pressure to obtain brown ethanolic crude extract (4.03 kg), which was then suspended in water (3 L) and then extracted sequentially with n-hexane (3 L × 3) and ethyl acetate (3 L × 3) in order to obtain the n-hexane-soluble (0.06 kg), EtOAc-soluble (1.11 kg) and water-soluble (0.53 kg) fractions. The EtOAc-soluble fraction was subjected to silica gel column chromatography with a gradient of n-hexane/ethyl acetate (from 20: 1 to 0:1) to yield the subfractions (Er1–Er4). The subfractions were subjected to silica gel (200–300 mesh, Qingdao Marine Chemical Inc., Qingdao, China), Sephadex LH-20 gel (CHP20P, 75–150 μm, Mitsubishi Chemical Industries Ltd, Tokyo, Japan), C18 reversed-phase silica gel (150–200 mesh, Merck, Darmstadt, Germany) column chromatography to obtain the pure compounds by following similar procedures as we described previously (J. Song et al., 2018a, 2018b) with minor modification. 1D and 2D NMR spectra were recorded on a Bruker Advance-500 spectrometer with TMS as internal standard (Bruker BioSpin AG, Fallanden, Switzerland). Spectroscopic data were presented in Supplementary materials.
2.3. Chemical fingerprint of E. chinensis leaf extract
Chemical fingerprinting of E. chinensis leaf extract was performed on a Thermo UltiMate 3000 instrument, coupled with a Thermo Orbitrap Elite spectrometer, with an ACQUITY UPLC HSS T3 column (Waters, 100 × 2.1 mm, 1.8 μm. EtOH crude extracts and EtOAc fractions were analyzed by LC-MS with a gradient program of MeCN–H2O (0.1% formic acid) (0–25 min, 5–95%, 25–30 min 95%, 30–35 min 5%; 0.2 mL/min; MS scan 100–1200 Da) with an automated full dependent MS/MS scan. The analysis of betulinic acid was performed on the same LC-MS spectrometer and UPLC column using the following linear gradient of MeCN–H2O (0.1% formic acid) (0–5min, 80–100%, 5–8 min 100%, 8.1–12.1 min 80%; 0.2 mL/min).
2.4. Preparation of cells and viruses
HCoV-OC43 and Human ileocecal adenocarcinoma (HRT-18) (ATCC CCL-244) were kindly donated by Guangzhou Medical University. HRT-18 cells were cultured in RPMI 1640 (containing L-glutamine) medium supplemented with 10% fetal bovine serum (FBS), 1% Penicillin/Streptomycin (P/S), 1% sodium pyruvate, 1% non-essential amino acids (NEAA), 0.025M HEPES at 37 °C incubator with 5% CO2.
HCoV-OC43 was expanded in HRT-18 cells in RPMI 1640 (with L-glutamine) medium supplemented with 2% FBS, 1% P/S, 1% sodium pyruvate, 1% NEAA, 0.025M HEPES at 37 °C incubator with 5% CO2 for 4–6 days until 50% of cells developed hornet's nest-like lesions. After one freeze/thaw cycle the virus was collected and filtered with a 0.22-μm Stericup® Filter Unit (Millipore, USA), half of the tissue culture infective dose (TCID50) of the virus was determined according to the Reed-Muench method.
2.5. Cytotoxicity assay
Cell Counting Kit-8 (CCK-8, Beyotime Biotechnology) was applied for cell viability detection according to the manufacturer's protocol.
2.6. Cytopathic effect (CPE) inhibition assay
Antiviral activity was evaluated by CPE inhibition assay. After the cells had grown into a monolayer in the 96-well plate, the cells were infected with 100 TCID50 of HCoV-OC43 and treated with two-fold serial dilutions of the compounds. At 72 h post-infection (hpi), CPE (cell lysis, cell rounding and vacuolization) were observed and visualized with the phase contrast inverted microscope (BDS400, Optec, China). No CPE confirmed the presence of antiviral activity and remdesivir as a positive control.
2.7. Antibody-based EC50 assay
Antibody-based EC50 assay was performed as described previously (Schirtzinger et al., 2022). HCoV-OC43 Nucleocapsid were detected with HCoV-OC43 Nucleocapsid antibody (Sino Biological, China).
2.8. Immunofluorescence (IF) analysis
Cells were incubated in monolayers on 24 well plates lined with climbing sheets, inoculated with 100 TCID50 of HCoV-OC43 at 37 °C for 2 h. The virus inoculum was then removed and replaced with RPMI1640 medium supplemented with 2% FBS containing desired sample concentration. At 48 hpi, the cells were fixed with 4% PFA/PBS, HCoV-OC43 nucleocapsid and the nucleus were detected with HCoV-OC43 nucleocapsid antibody (Sino Biological) and DAPI (Sigma) respectively. Fluorescence detection was performed using a laser scanning confocal fluorescence microscope (TCS SP8 STED 3X, Leica).
2.9. Transmission electron microscopy (TEM)
Monolayers of HRT-18 cells were incubated with 100 TCID50 HCOV-OC43 for 2 h at 37 °C. The virus inoculum was then removed and replaced with RPMI1640 medium supplemented with 2% FBS containing desired sample concentration. At 48 h pi, virus particles were visualized by negative staining as described previously and then observed under transmission electron microscopy (Tecnai G2 Spirit Bio TWIN, FEI).
2.10. Preparation of ligands
3D SDF files corresponding to 23 individual compounds were downloaded from PubChem and ZINC databases. Compounds for which no 3D SDF files could be searched in any database were manually drawn and converted to 3D SDF format using Chemoffice software and optimized using the default force field. Energy minimization and optimization of all 3D SDF files were performed during the virtual screening process (H. Li, Leung, Wong and Ballester, 2016).
2.11. Molecular docking and visualization
3D SDF files corresponding to 23 individual compounds were uploaded to COVID-19 Docking Server (https://ncov.schanglab.org.cn/index.php) for small molecule docking. The predicted binding energy of Top 1 ranked binding mode for each compounds were shown in Table 1 . Papain-like protease (PLpro) (PDB ID 6WUU) with the peptide inhibitor VIR250, Nonstructural protein 14 C-terminal guanine-N7 methyltransferase (Nsp14N7-Mtase) (PDB ID 5C8S) with the inhibitor SAH were obtained from the protein database (PDB) (Kong et al., 2020; Y. Ma et al., 2015; Rut et al., 2020)
Table 1.
Docking results for 23 small molecule compounds against the targets of SARS-CoV-2 (Mpro, PLpro, Nsp3AMP, Nsp3MES, RdRp RTP, Nsp14N7-MTase, Nsp16GTA, Nsp16MGP, Nsp16SAM).
| Compou.No. | M pro | PL pro | Nsp3 AMP | Nsp3 MES | RDRP RTP | Nsp14N7 -MTase |
Nsp16 GTA | Nsp16 MGP | Nsp16 SAM |
|---|---|---|---|---|---|---|---|---|---|
| 1 | −7.1 | −9.6 | −7.8 | −7.3 | −6.9 | −11.2 | −7.7 | −7.7 | −7.1 |
| 2 | −7.3 | −8.8 | −7.5 | −8.1 | −7.5 | −8 | −8.5 | −7.6 | −7.8 |
| 3 | −7.2 | −9.3 | −7.8 | −8 | −7.5 | −9.1 | −8.5 | −8.3 | −8.5 |
| 4 | −7.9 | −7.9 | −7.2 | −8.5 | −7 | −8.4 | −8.2 | −7.1 | −8.2 |
| 5 | −7.1 | −7.5 | −7.6 | −6.9 | −6.6 | −7.8 | −7.8 | −6.59 | −7.2 |
| 6 | −7.4 | −8 | −7.3 | −8.5 | −7.2 | −8.3 | −8.4 | −6.9 | −8.5 |
| 7 | −7.3 | −8.1 | −7.3 | −9.1 | −7.3 | −8.4 | −8.8 | −7.2 | −8.7 |
| 8 | −7.4 | −8.1 | −7.1 | −9.1 | −7.1 | −8.7 | −8.7 | −7 | −8.7 |
| 9 | −7.4 | −8 | −7.1 | −8.5 | −7.1 | −8.5 | −8.4 | −6.9 | −8.6 |
| 10 | −7.4 | −8 | −6.8 | −8.3 | −7 | −8.6 | −8.1 | −7 | −8.1 |
| 11 | −7.8 | −8.5 | −7.7 | −8.7 | −7.7 | −8.8 | −8.2 | −6.9 | −8.3 |
| 12 | −7.6 | −8.3 | −7.2 | −8.9 | −7.4 | −8.7 | −8.5 | −6.9 | −8.4 |
| 13 | −7.1 | −7.6 | −7.8 | −7.3 | −7 | −7.9 | −7.9 | −6.8 | −7.3 |
| 14 | −5.7 | −6.6 | −5.8 | −7 | −6 | −7.4 | −6.3 | −7 | −6.2 |
| 15 | −5.2 | −5.8 | −5.6 | −6.7 | −5.7 | −6.7 | −5.9 | −5.3 | −6 |
| 16 | −5.2 | −5.5 | −5.4 | −6.1 | −6.6 | −6.2 | −5.6 | −4.9 | −5.6 |
| 17 | −5.5 | −5.9 | −5.4 | −6.4 | −5.7 | −6.6 | −5.9 | −5.1 | −5.9 |
| 18 | −4.4 | −4.9 | −4.7 | −5.2 | −5 | −5.1 | −5 | −4.4 | −4.9 |
| 19 | −4.7 | −5.5 | −5.1 | −5.7 | −5.1 | −5.2 | −6 | −4.8 | −6 |
| 20 | −4.9 | −5.3 | −5 | −6 | −5.2 | −5.4 | −5.6 | −4.8 | −5.4 |
| 21 | −4.8 | −5.4 | −6 | −5 | −5.2 | −5.5 | −5.9 | −5.1 | −5.9 |
| 22 | −6.1 | −6.6 | −6.9 | −7 | −5.2 | −6.3 | −6 | −5.3 | −6.1 |
| 23 | −5.2 | −6 | −5.7 | −6.2 | −5.2 | −5.7 | −5.5 | −5.7 | −5.4 |
| Liganda | −8.4 | −7.6 | −7.4 | −5.5 | −8.3 | −8.2 | −10.3 | −7.5 | −8.4 |
Origin Ligands of Mpro, PLpro, Nsp3AMP, Nsp3MES, RdRp, Nsp14N7-MTase, Nsp16GTA, Nsp16MGP, Nsp16SAM: N3, VIR250, Adenosine Monophosphate (AMP), 2-(N-morpholino)-ethanesulfonic acid (MES), Remdesivir(RTP), Uridine-5′-Monophosphate, S-adenosyl-L-homocysteine (SAH), 7-methyl-GpppA (GTA), 7-methyl-guanosine- 5′-triphosphate (MGP), S-Adenosylmethionine (SAM), respectively.
Using AutoDock Tools 1.5.6 software to remove water molecules, isolated receptors, and primitive ligands from the crystal structure, then add non-polar hydrogen to the target protein, calculate the Gasteiger charge, atom type “AD4 type ", etc. The same operations are performed on the molecule and saved in PDBQT format. Run AutoDock 4.2.6 software for docking and visualizing the results using PyMOL (Oleg & J., 2009).
2.12. Lipinski and ADMET analysis
The pharmacokinetics, drug similarity, and medicinal chemistry of the compounds in E.chinensis leaves were further examined using the SwissADME server (Daina et al., 2017). The ligand molecules were entered via SMILES format.
2.13. PLpro inhibition assay
An enzyme inhibition assay based on continuous fluorimetric measurements was established according to previous protocol with minor modifications(L. Li et al., 2022). Briefly, 50 μL of serially diluted compounds in 50 mM HEPES assay buffer with100 μM Z-RLRGG-AMC (Glpbio) was transferred to Corning 96-well black polystyrene plates. The reaction was initiated by the addition of 50 μL of recombinant SARS-CoV-2 PLpro (HY–P70122, MCE) at a final concentration of 0.10 μg/mL. The fluorescence intensity at ex/em 380/460 nm was recorded continuously by a microplate reader. The increase in fluorescence intensity during the initial linear phase of the reaction (first 5 min) was analyzed. The half-maximal Inhibitory concentration (IC50) was calculated through log (inhibitor) vs. response -- Variable slope (four parameters) equation in Prism V8 (GraphPad).
2.14. Nsp14N7-MTase inhibition assay
The enzyme inhibition experiments were adapted from previously described methods with minor modifications(Aouadi et al., 2017). The enzymatic reaction was performed in 15 μl. 4.6 μl of mix (50 mM HEPES assay buffer and SARS-CoV-2 Nsp14N7 methyltransferase (HY–P70132, MCE) at 0.12 μg/mL final concentration) were added in the assay wells, containing previously dispensed inhibitors (0.4 μL), Reaction was started with 10 μl of a mix containing both GpppA (S1406S, NEB) and SAM at 8 μM and 2 μM final concentrations respectively and was incubated 20 min at 30 °C. For the detection of released SAH, the Methyltransferase Fluorometric Assay Kit (700150, cayman) was used according to the supplier protocols. Initiate the reactions by quickly adding 100 μl of Master Mixture and read the plate in a plate reader at 27 °C every minute for 30 min at ex/em 540/590 nm. The half-maximal Inhibitory concentration (IC50) was calculated through log (inhibitor) vs. response -- Variable slope (four parameters) equation in Prism V8 (GraphPad).
2.15. Statistical method
Statistical analyses were performed using Prism V8 (GraphPad). The experiments of TCID50, CC50, EC50, IC50 were carried out in triplicate and results were calculated as mean values through log (inhibitor) vs. response -- Variable slope (four parameters) equation in Prism V8. Selective index (SI) is defined as the ratio of CC50/EC50. A statistical analysis of immunofluorescence was conducted on at least two independent experiments, each with 10 images per condition.
3. Results
3.1. Anti-HCoV-OC43 and cytotoxicity of E. chinensis extracts
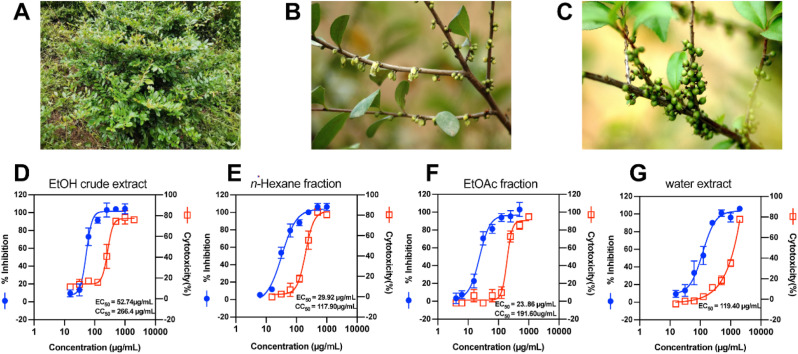
The cytotoxicity of E. chinensis extracts were evaluated on HRT-18 cell using CCK-8 assay. The results showed that the CC50 values of EtOH crude extract, n-hexane-soluble fraction, EtOAc-soluble fraction and water extract were 266.40 μg/mL, 117.90 μg/mL, 191.60 μg/mL, >1000 μg/mL respectively. CPE inhibition assay demonstrated that EtOH crude extract, n-hexane-soluble fraction, EtOAc-soluble fraction and water extract significantly inhibited CPE caused by HCoV-OC43 infection at a certain concentration range, respectively. Antibody-based EC50 assay demonstrated that EtOAc-soluble fraction showed the strongest antiviral activity against HCoV-OC43 (EC50 = 23.86 μg/mL, CC50 = 191.86 μg/mL, SI = 8.03) and water extract also showed antiviral activity against HCoV-OC43 (EC50 = 119.40 μg/mL) without significant cytotoxicity (Fig. 1 D–G)
Fig. 1.
(A) E. chinensis bushes; (B) E.chinensis flowers; (C) E. chinensis fruits; (D–G) EC50 for HCoV-OC43 replication and and CC50 for cell viability in HRT-18 cells of EtOH crude extract and fractions, water extract of E. chinensis.
3.2. Structure identification of the chemical constituents of E. chinensis leaf
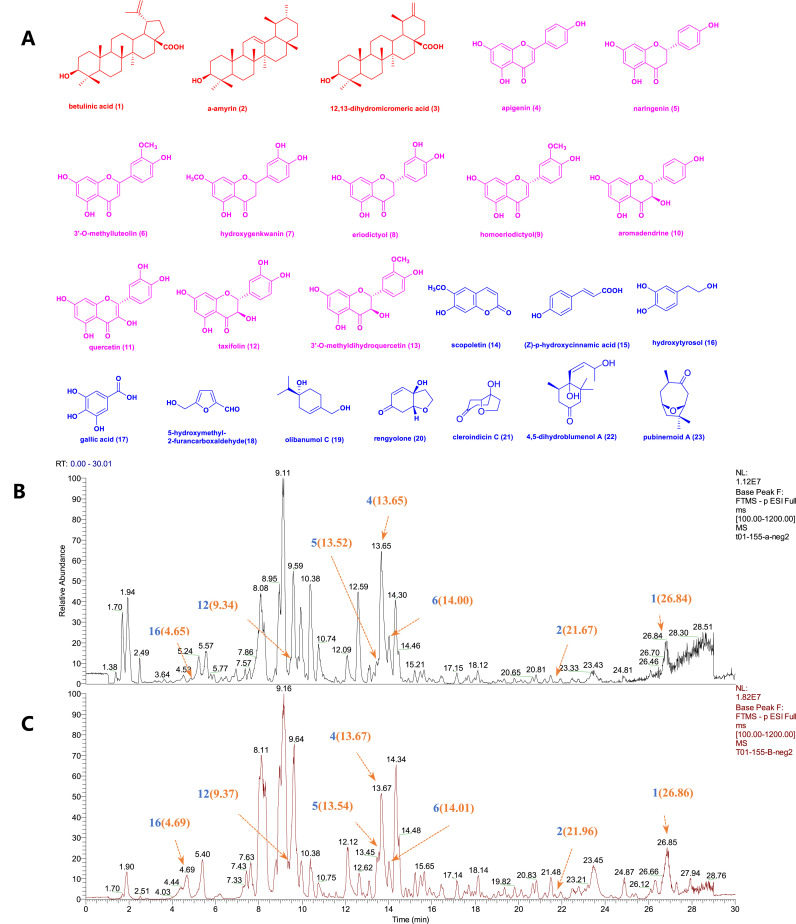
To further search for antiviral components, EtOAc fraction of E. chinensis leaves was isolated and purified to yield 23 compounds (Fig. 2 ) including three terpenoids: betulinic acid (1), α-amyrin (2), 12,13-dihydromicromeric acid (3), ten flavonoids: apigenin (4), naringenin (5), 3′-O-methylluteolin (6), hydroxygenkwanin (7), eriodictyol (8), homoeriodictyol (9), aromadendrine (10), quercetin (11), taxifolin (12), and 4′-O-methyltaxifolin (13), ten other types of compounds: scopoletin (14), (E)-p-hydroxycinnamic acid (15), hydroxytyrosol (16), gallic acid (17), 5-hydroxymethyl-2-furancarboxaldehyde (18), olibanumol C (19), rengyolone (20), cleroindicin C (21), 4, 5-dihydroblumenol A (22), and pubinernoid A (23). The structural characterization of these compounds was accomplished by comparing their respective NMR data with the corresponding values reported in literature (Supplementary materials).
Fig. 2.
(A). Structures of the compounds isolated from E. chinensis. Red represents triterpenes (1–3), pink represents flavonoids(4–13), and blue represents other types of compounds(14–23).(B–C). Chemical fingerprint of EtOH crude extracts (B) and EtOAc fractions (C). Blue numbers represent identified compounds. The position of peaks corresponding to compounds were indicated by orange arrows and number (retention time). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The chemical fingerprints of EtOH crude extract (Fig. 2B) and EtOAc fraction (Fig. 2C) were provided to understand the chemical composition and content by using liquid chromatography mass spectrometry (LC-MS). In the present study, two terpenoids [betulinic acid (1) and α-amyrin (2)], four flavonoids [apigenin (4), naringenin (5), 3′-O-methylluteolin (6), taxifolin (12)], and an additional hydroxytyrosol (16) were identified by comparison with their reference standards' retention time and MS2 data. The content of betulinic acid in EtOAc fraction was found to be 0.64 (%w/w) by LC-MS spectrometer and UPLC column with betulinic acid standard sample as control, which demonstrated that E. chinensis leaves contain a high content of betulinic acid.
3.3. Physicochemical, pharmacokinetic and pharmacological similarity properties of small molecule chemical constituents from E. chinensis leaves
Since the Lipinski's rule of five is a useful guide for predicting the potentiality of oral exposure for optimized chemical compounds, we examined physicochemical, pharmacokinetic and pharmacological similarity properties of small molecule chemical constituents in E. chinensis leaves by applying Lipinski's rule of five. Besides, compounds with 7 or fewer rotatable bonds reflect good prospects for oral bioavailability, and a lower total polar surface area (TPSA) value indicates good permeability or bioavailability of compounds(Hay et al., 2014).
STable1 collects the molecular properties and bioactivity scores of 23 small molecule chemical constituents in E. chinensis leaves. The molecular weights of the 23 small molecule chemical components ranged from 126 to 456 kDa. The flavonoids had sensational water solubility and gastrointestinal adsorption properties, which were found to satisfy the Lipinski's rules and lead-like criteria. The triterpenoids had poor water solubility and violated one of Lipinski's rules, but they have achieved a bioavailability of 0.85. The other type compounds satisfy the Lipinski's rules. Overall, the results demonstrated that 23 small molecule chemical constituents in E. chinensis leaves have good molecular and chemical properties and bioactivity scores, suggesting that these constituents can possibly be made orally bioavailable in humans.
3.4. Anti-HCoV-OC43 activity and cytotoxicity of compounds
Sixteen compounds (1, 2, 5, 8, 11–13, 14–19, 21–23) were evaluated for their antiviral activity against HCoV-OC43 via CPE. The results demonstrated that triterpenoids 1, 2 and flavonoids 5, 8, 11 showed antiviral activity at a range of concentrations by preventing the formation of CPE (hornet's nest lesions), but other types of compounds could not prevent CPE at all concentrations tested. The cytopathicity induced by HCoV-OC43 infection did not always work out (Schirtzinger et al., 2022). We eventually discovered that HCoV-OC43 shows a CPE dependent on the state of cell growth and accounted for it, but some compounds (3, 4, 6, 7, 9, 10, 20) were consumed in the assay prior to yielding meaningful data.
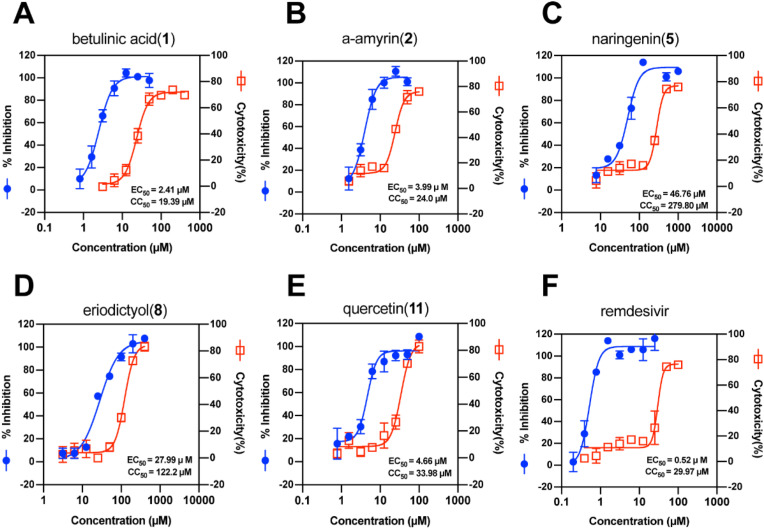
Triterpenoids 1, 2 and flavonoids 5, 8, 11 were subsequently subjected to antibody-based EC50 assays and CCK-8 assays (Fig. 3 ). Among tested five compounds, betulinic acid (1) had the strongest antiviral activity (EC50 = 2.41 μM, CC50 = 19.39 μM, SI = 8.04), followed by α-amyrin (2) (EC50 = 3.99 μM, CC50 = 24.0 μM, SI = 6.03). Among tested flavonoids, quercetin (11) also showed strong antiviral activity (EC50 = 4.66 μM, CC50 = 33.98 μM, SI = 7.29), followed by eriodictyol (8).
Fig. 3.
Compounds (A, betulinic acid; B, α-amyrin; C, naringenin; D, eriodictyol; E, quercetin; F,remdesivir))dose-response curves for HCoV-OC43 replication and for cell viability in HRT-18 cells.
3.5. IF and TEM analysis
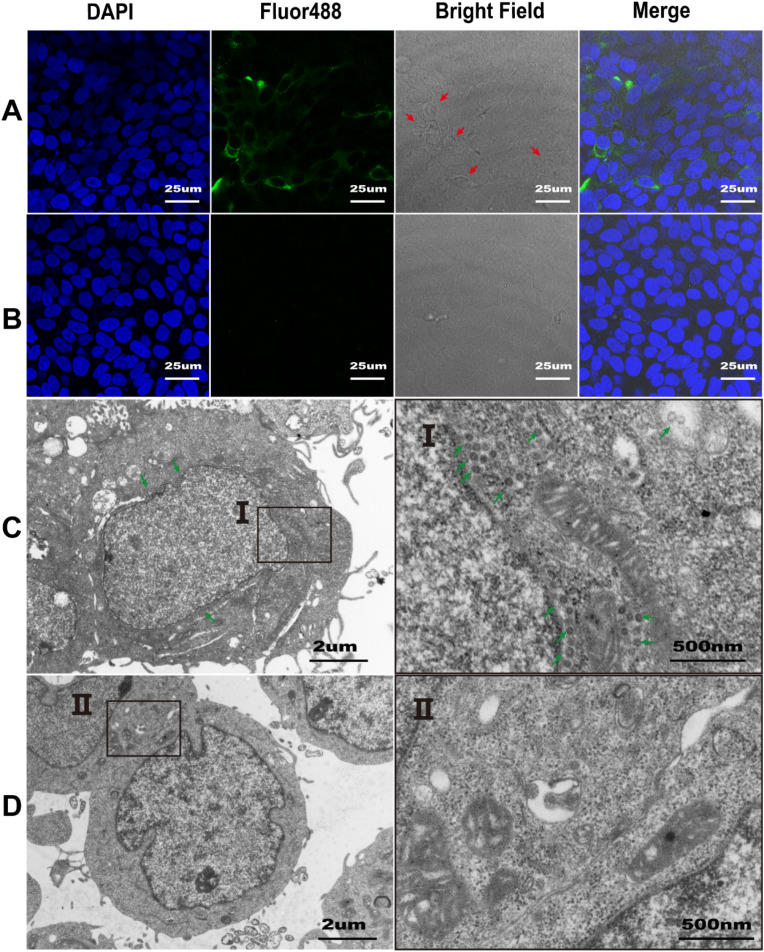
To further verify the antiviral activity of betulinic acid (1), a certain concentration (4 μM) of betulinic acid (1) was used to treat HCoV-OC43 infected cells, then IF analysis for the HCoV-OC43 nucleocapsid in these cells was conducted. The results showed that strong green fluorescence particles appeared prominently around the hornet's nest-like lesion of the control cells infected with HCoV-OC43, indicating that the hornet's nest-like lesion (red arrows) was caused by HCoV-OC43 infection (Fig. 4 A). Betulinic acid (1) substantially reduced the fluorescence signal of virus particles and attenuated the effect of cytopathic lesions (Fig. 4B). Subsequently, we performed TEM analysis of infected cells treated by betulinic acid (1) or DMSO. As demonstrated in Fig. 4C, DMSO-treated infected cells contained a large number of viral particles (green arrows) that were mainly distributed in the double-membrane vesicle (DMV) around the nucleus. It has been shown that the DMV acts as a viral factory that protects the coronavirus from immune attack by the host cells (Cortese et al., 2020). In contrast, no viral particles were detected in the infected cells treated with betulinic acid (1) at the concentration of 4 μM (Fig. 4D).
Fig. 4.
Fluorescence microscopy of infected HRT-18 cells with HCoV-OC43 (A) and betulinic acid (1) treated infected HRT-18 cells (B) at 48 hpi. Immunofluorescence staining of the nucleus with 4,6-diamidino-2-phenylindole (DAPI) (blue) and HCoV-OC43 nucleocapsid protein with Fluor488(green). The hornet's nest-like lesion (red arrows) was detected in bright field of infected HRT-18 cells with HCoV-OC43. Virions (green arrows) were detected in the ultrathin sections of HCoV-OC43 infected HRT-18 cells (C) and no viral particles were detected in betulinic acid (1) treated infected HRT-18 cells (D) at 48 hpi under electron microscope. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.6. Analysis of the anti-coronavirus mechanism of action of the compounds from E. chinensis leaves
3.6.1. Molecular docking studies for the 23 natural compounds
To decipher the potential mechanism of action underlining the anti-coronaviruses effects of E. chinensis leaves, we performed molecular docking studies for the 23 natural compounds from E. chinensis leaves with 9 binding pockets of SARS-CoV-2 Nsps (Mpro, PLpro, Nsp3AMP, Nsp3MES, RdRp RTP, Nsp14 N7-MTase, Nsp16GTA, Nsp16MGP, Nsp16SAM) via COVID-19 Docking Server. In Table 1, molecular docking scores were used to determine the strength of interactions between small molecules and proteins; the higher the score value, the stronger the interaction. The results revealed that the affinities of betulinic acid (1) for PLpro and NSP14N7-MTase was −9.6 kcal/mol and −11.2 kcal/mol respectively, which was significantly stronger than that of the 22 other compounds and the origin ligands, indicating that betulinic acid (1) was the main antiviral active component of E. chinensis leaves by targeting PLpro and Nsp14N7-MTase.
The flavonoid components also exhibited notable affinities for PLpro, Nsp3MES, Nsp14N7-MTase, Nsp16GTA and Nsp16SAM with docking scores below −8 kcal/mol. Thus, the flavonoids from E. chinensis also played an important role in antiviral responses. The docking scores of the other 10 compounds except for scopoletin (14) were basically lower than −7 kcal/mol, implying these compounds may not be working for anti-coronavirus. The results of molecular docking scoring were principally consistent with the results of CPE and EC50 assays previously performed in this study -- that triterpenoids and flavonoids showed antiviral activity, but other types of compounds had no antiviral activity.
3.6.2. Docking and visualizing of betulinic acid (1) toward target protein PLpro and Nsp14N7-MTase
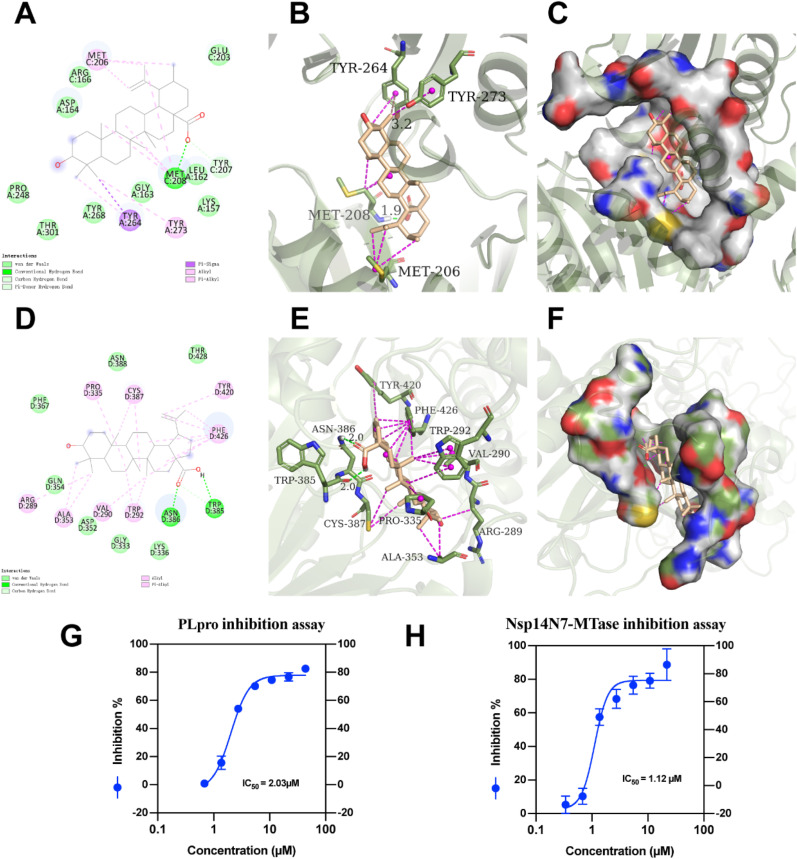
To further reveal the outstanding affinities of betulinic acid (1) toward target protein PLpro and Nsp14N7-MTase of SARS-CoV-2, detailed docking and visualizing was performed by using AutoDock Tools. The interactions between betulinic acid (1) and PLpro were shown in Fig. 5 A and C, the carbonyl oxygen of betulinic acid (1) engages in a direct hydrogen-bonding interaction with Met208. Meanwhile, betulinic acid (1) has pi-sigma interactions with Tyr264 and engages in a big network of van der Waals interactions toward Pro248, Thr301, Thr268, Gly163, Leu162 and Lys157 in the very deep and broad S4 pocket of PLpro. Pro248 and Tyr268 are key residues to PLpro activity of SARS-CoV and SARS-CoV-2, which may be the reason for the good scores of betulinic acid (1) binding to PLpro (Ibrahim et al., 2020; Rut et al., 2020).
Fig. 5.
Interactions of betulinic acid (1) with target protein PLpro (A-C, G) and Nsp14N7-MTase (D-F, H). A. 2D interaction of betulinic acid (1) with target protein PLpro; B. 3D-box of betulinic acid (1) with target protein PLpro; C. 3D-osurf of betulinic acid (1) with target protein PLpro; D. 2D interaction of betulinic acid (1) with target protein Nsp14N7-MTase; E. 3D-box of betulinic acid (1) with target protein Nsp14N7-MTase; F. 3D-osurf of betulinic acid (1) with target protein Nsp14N7-MTase. G-H, the inhibitory activities of betulinic acid (1) against PLpro(G) and Nsp14N7-MTase(H).
Fig. 5D and F showed the interaction of betulinic acid (1) and Nsp14N7-MTase, the carbonyl oxygen of betulinic acid (1) engages in two direct hydrogen-bonding interactions with Asn386 and Trp385. Meanwhile, betulinic acid (1) engages in alkyl or pi-alkyl interactions with Pro335, Cys387, Try420, Phe426, Trp292, Val290, Ala353, Asn386, Phe426 and Trp292 are critical sites for N7-MTase and responsible for more than 50% of the N7-MTase activity of SARS-CoV (Y. Ma et al., 2015). Asn386 also holds a strong lead in the inhibition of SARS-CoV-2 N7-MTase (Selvaraj et al., 2021).
3.6.3. PLpro inhibition assay
A fluorogenic enzymatic assay was conducted to corroborate the inhibitory potency of betulinic acid (1) against PLpro. The 7- amino-4-methylcoumarin (AMC)-labeled pentapeptide Z-RLRGG-AMC was used in the assay, which represents the consensus cleavage sequence of PLpro. When covalent conjugated, the AMC motif is not fluorescent, however, it becomes dramatically fluorescent upon cleavage by PLpro, enabling efficient anti-proteolytic activity measurement (Ratia et al., 2008). Posoralidin, a known natural inhibitor of SARS-CoV PLpro, was used as the positive control (D. W. Kim et al., 2014). We found that betulinic acid (1) has significant anti-proteolytic activity against PLpro with IC50 values as low as 2.03 μM by this method (Fig. 5G).
3.6.4. N7-MTase inhibition assay
The inhibitory activity of betulinic acid (1) against Nsp14N7-MTase was also conducted by a fluorescence screening assay. The Nsp14 C-terminal domain functions as a guanine-N7 methyl transferase (N7-MTase) for mRNA capping, which is methylated at N7 position of the guanosine in the presence of methyl donor S-adenosyl-L-methionine (SAM) yielding to a cap-0 (7mGpppN) and S-adenosyl-L-homocysteine (SAH) by-product (Bouvet et al., 2010). SAH is rapidly converted to highly fluorescent compound resorufin via a commercially available kit. Sinefungin, a known SARS-CoV N7-MTase inhibitor, was used as a positive control (Aouadi et al., 2017). In this work we identified that betulinic acid (1) exhibited superior inhibition of the N7-MTase activity of SARS-CoV-2 with an IC50 of 1.12 μM (Fig. 5H).
4. Discussion
In recent years, interest in medicinal plants has increased significantly because of the fewer side effects they produce. The leaves of Eurya chinensis(Chinese Dagang Tea)have been consumed as herbal tea for centuries and have also been used to prevent influenza and treat colds and fevers in traditional Chinese medicine, which suggests that there might be safe antiviral compounds in the leaves of Eurya chinensis. However, as far as we know there are no reports on the chemical profile and efficacy of its leaves for the treatment of fever and viral infections because of ignored antiviral research on medicinal plants until the COVID-19 outbreak.
In this study, we found for the first time that E. chinensis leaf extracts including water extract exhibited inhibitory effects against HCoV-OC43 without significant cytotoxicity in vitro. Antibody-based EC50 assay demonstrated that EtOAc-soluble fraction showed the strongest antiviral activity, which may be explained by the presence of more active compounds to be uncovered in this fraction that needs to be further clarified. Further, 23 monomer compounds were isolated from EtOAc fraction: the triterpenoids (betulinic acid, α-amyrin) and the flavonoids (naringenin, eriodictyol and quercetin) are the main ingredients responsible for the antiviral activity of E. chinensis leaves and were enriched in EtOAc fraction demonstrated by chemical fingerpringting.
Among 23 compounds betulinic acid (1) is the most prominent compound with EC50 value of 2.41 μM against HCoV-OC43 and high content of 0.64 (%w/w) in EtOAc fraction. Betulinic acid, a lupane-type pentacyclic triterpene, has been found to have inhibitory effects on HCoV-229E GFP, HCoV-OC43 and SARS-CoV (Murer et al., 2022; Wen et al., 2007). Betulinic acid was found to inhibit Mpro activity by competitive inhibition at 10 μmol/L in SARS-CoV model (Wen et al., 2007) and at 14.55 μmol/L in SARS-CoV-2 model (Alhadrami et al., 2021). Betulinic acid demonstrated the ability to bind to Nsp15 protein, and thereby could be useful for inhibiting the viral replication of SARS-CoV-2 (Kumar et al., 2021). Structure-activity relationship exploration of betulinic acid derivatives yielded a strong inhibitor of HCoV-229E replication via the nsp15 endoribonuclease (Stevaert et al., 2021). Betulinic acid was reported to interact strongly with the PLpro from docking and MD simulation studies (Rudrapal et al., 2022), but the inhibition activity and structural modification of betulinic acid against PLpro have not been further studied as far as we know. The affinity and the inhibition activity of betulinic acid (1) binding to N7-MTase have not been studied as far as we know.
In this study, docking analysis and enzyme inhibition experiments demonstrated betulinic acid (1) exhibited significant inhibition activity against PLpro and N7-MTase of SARS-CoV-2 mainly due to forming single and double hydrogen bonding with key residues located at the active site pocket of the enzyme, as described earlier in this study. Betulinic acid (1) could be considered a promising anti-coronaviruses lead that can suppress PLpro and N7-MTase catalytic activity, and we believe that the structure and mechanistic insights presented here would support the development of potent and specific anti-coronaviruses inhibitors.
We reported that the flavonoids such as naringenin(5), eriodictyol(8) and quercetin(11) exhibited antiviral activity against HCoV-OC43 and notable affinities for PLpro, Nsp3MES, Nsp14N7-MTase, Nsp16GTA and Nsp16SAM of SARS-CoV-2 by virtual screening in this study. Naringenin and quercetin are well-known flavonoids whose antiviral properties have been investigated in numerous studies exhibiting anti-SARS-CoV-2 activity (Clementi et al., 2020).The flavonoids such as quercetin, quercetin-3-O-glucoside, kaempferol and quercitrin has been reported to exhibited excellent binding affinity towards with PLpro of SARS-CoV-2 (Derosa, Maffioli, D'Angelo and Di Pierro, 2021; Mouffouk et al., 2021). The best docking score was observed in quercetin-3-O-glucoside, which binds to His74, Arg83, Tyr155, Asn157, His176 amino acid residues of pocket of PLpro with eight hydrogen bonds (Hiremath et al., 2021). However, the affinities of flavonoids binding to SARS-CoV-2 Nsp3, Nsp14 and Nsp16 were rarely reported, the multifaceted aspect of flavonoids should be taken into consideration.
5. Conclusion
In conclusion, triterpenoids and flavonoids are the main ingredients responsible for the antiviral activity of E. chinensis leaves via multi-target mechanisms against coronaviruses, and thus hold great promise for the development of functional dietary supplements and health products for the prevention and treatment of coronaviruses. Betulinic acid (1) was highlighted as a promising structural motif for developing anti-coronaviruses inhibitors via suppression PLpro and N7-MTase catalytic activity.
Funding
This work was supported by the National Natural Science Foundation of China (NSFC) (No.82073733) and the Science and Technology Service Network Initiative of the Chinese Academy of Sciences (CAS) (KFJ-STS-QYZD-2021-03-002).
CRediT authorship contribution statement
Liyun Zhao: performed antiviral activities assays, Writing – original draft, IF, TEM, virtual screening and manuscript drafting. Xubing Qin: Formal analysis, performed extraction, purification and chemical structure analysis. Tingting Lin: performed CCK-8 and antiviral activities assays. Fuda Xie: performed virtual screening and docking. Liyuan Yao: performed chemical structure identified. Yulin Li: performed chemical structure identified. Binhong Xiong: critically discussed and confirmed data. Zhifang Xu: critically discussed and confirmed data. Yongchang Ye: performed plant identified and revised the article for important intellectual content, S.Q. critically revised the article for important intellectual content and final approval of the article. Hongfeng Chen: performed plant identified and revised the article for important intellectual content, S.Q. critically revised the article for important intellectual content and final approval of the article.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We are grateful to Prof. Jincun Zhao and Dr. Fang Li of Guangzhou Medical University for their guidance in culturing HCoV-OC43, and to Dr. Hanxiang Li, Dr. Guangyi Dai and Rufang Deng of South China Botanical Garden Public Laboratory for their help in using the LC/MS and electron microscope.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jep.2022.115528.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
The data that has been used is confidential.
References
- Alhadrami H.A., Sayed A.M., Sharif A.M., Azhar E.I., Rateb M.E. Olive-Derived triterpenes suppress sars COV-2 main protease: a promising scaffold for future therapeutics. Molecules. 2021;26(9) doi: 10.3390/molecules26092654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aouadi W., Eydoux C., Coutard B., Martin B., Debart F., Vasseur J.J.…Decroly E. Toward the identification of viral cap-methyltransferase inhibitors by fluorescence screening assay. Antivir. Res. 2017;144:330–339. doi: 10.1016/j.antiviral.2017.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet M., Debarnot C., Imbert I., Selisko B., Snijder E.J., Canard B., Decroly E. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6(4) doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A.…Xie X.S. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2021 doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H.…Sigal A. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2021 doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Wu Y., Chen C., Gu Y., Zhu C., Wang S.…Wu C. Identifying potential anti-COVID-19 pharmacological components of traditional Chinese medicine Lianhuaqingwen capsule based on human exposure and ACE2 biochromatography screening. Acta Pharm. Sin. B. 2021;11(1):222–236. doi: 10.1016/j.apsb.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementi N., Scagnolari C., D'Amore A., Palombi F., Criscuolo E., Frasca F., Pierangeli A., Mancini N., Antonelli N., Clementi M., Carpaneto A., Filippini A. Naringenin is a powerful inhibitor of SARS-CoV-2 infection in vitro. Pharmacol. Res. 2021;163:105255. doi: 10.1016/j.phrs.2020.105255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese M., Lee J.Y., Cerikan B., Neufeldt C.J., Oorschot V.M.J., Kohrer S.…Bartenschlager R. Integrative imaging reveals SARS-CoV-2-induced reshaping of subcellular morphologies. Cell Host Microbe. 2020;28(6):853–866 e855. doi: 10.1016/j.chom.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7 doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J.P., Almasy K.M., McDonald E.F., Plate L. Comparative multiplexed interactomics of SARS-CoV-2 and homologous coronavirus nonstructural proteins identifies unique and shared host-cell dependencies. ACS Infect. Dis. 2020;6(12):3174–3189. doi: 10.1021/acsinfecdis.0c00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derosa G., Maffioli P., D'Angelo A., Di Pierro F. A role for quercetin in coronavirus disease 2019 (COVID-19) Phytother Res. 2021;35(3):1230–1236. doi: 10.1002/ptr.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay M., Thomas D.W., Craighead J.L., Economides C., Rosenthal J. Clinical development success rates for investigational drugs. Nat. Biotechnol. 2014;32(1):40–51. doi: 10.1038/nbt.2786. [DOI] [PubMed] [Google Scholar]
- Hiremath S., Kumar H.D.V., Nandan M., Mantesh M., Shankarappa K.S., Venkataravanappa V.…Reddy C.N.L. In silico docking analysis revealed the potential of phytochemicals present in Phyllanthus amarus and Andrographis paniculata, used in Ayurveda medicine in inhibiting SARS-CoV-2. 3 Biotech. 2021;11(2):44. doi: 10.1007/s13205-020-02578-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim T.M., Ismail M.I., Bauer M.R., Bekhit A.A., Boeckler F.M. Supporting SARS-CoV-2 papain-like protease drug discovery: in silico methods and benchmarking. Front. Chem. 2020;8 doi: 10.3389/fchem.2020.592289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasereka M.C., Hawkes M.T. Neuroinvasive potential of human coronavirus OC43: case report of fatal encephalitis in an immunocompromised host. J. Neurovirol. 2021;27(2):340–344. doi: 10.1007/s13365-020-00926-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Min, Jang Lee, Biomolecules K.J. Natural bis-benzylisoquinoline alkaloids-tetrandrine, fangchinoline, and cepharanthine. Inbit Human Coronavirus Infect MRC-5 Human Lung Cell. 2019;9(11):696. doi: 10.3390/biom9110696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.W., Seo K.H., Curtis-Long M.J., Oh K.Y., Oh J.W., Cho J.K.…Park K.H. Phenolic phytochemical displaying SARS-CoV papain-like protease inhibition from the seeds of Psoralea corylifolia. J. Enzym. Inhib. Med. Chem. 2014;29(1):59–63. doi: 10.3109/14756366.2012.753591. [DOI] [PubMed] [Google Scholar]
- Kong R., Yang G., Xue R., Liu M., Chang S. COVID-19 Docking Server: a meta server for docking small molecules, peptides and antibodies against potential targets of COVID19. Bioinformatics. 2020;36(20):5109–5111. doi: 10.1093/bioinformatics/btaa645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Kashyap P., Chowdhury S., Kumar S., Panwar A., Kumar A. Identification of phytochemicals as potential therapeutic agents that binds to Nsp15 protein target of coronavirus (SARS-CoV-2) that are capable of inhibiting virus replication. Phytomedicine. 2021;85 doi: 10.1016/j.phymed.2020.153317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Leung K.S., Wong M.H., Ballester P.J. Correcting the impact of docking pose generation error on binding affinity prediction. BMC Bioinf. 2016;17(Suppl. 11):308. doi: 10.1186/s12859-016-1169-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Ma L., Hu Y., Li X., Yu M., Shang H., Zou Z. Natural biflavones are potent inhibitors against SARS-CoV-2 papain-like protease. Phytochemistry. 2022;193 doi: 10.1016/j.phytochem.2021.112984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H.…Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/s0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Chen X., Mei F., Xiong Q., Liu Q., Dong L.…Lan K. Drastic decline in sera neutralization against SARS-CoV-2 Omicron variant in Wuhan COVID-19 convalescents. Emerg. Microb. Infect. 2022:1–17. doi: 10.1080/22221751.2022.2031311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Wu L., Shaw N., Gao Y., Wang J., Sun Y.…Rao Z. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc. Natl. Acad. Sci. U. S. A. 2015;112(30):9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani J.S., Johnson J.B., Steel J.C., Broszczak D.A., Neilsen P.M., Walsh K.B., Naiker M. Natural product-derived phytochemicals as potential agents against coronaviruses: a review. Virus Res. 2020;284 doi: 10.1016/j.virusres.2020.197989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J.M., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S.N.…Chan S.Y. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Milani M., Donalisio M., Bonotto R.M., Schneider E., Arduino I., Boni F.…Mastrangelo E. Combined in silico and in vitro approaches identified the antipsychotic drug lurasidone and the antiviral drug elbasvir as SARS-CoV2 and HCoV-OC43 inhibitors. Antivir. Res. 2021;189 doi: 10.1016/j.antiviral.2021.105055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfopoulou S., Brown J.R., Davies E.G., Anderson G., Virasami A., Qasim W.…Breuer J. Human coronavirus OC43 associated with fatal encephalitis. N. Engl. J. Med. 2016;375(5):497–498. doi: 10.1056/NEJMc1509458. [DOI] [PubMed] [Google Scholar]
- Mouffouk C., Mouffouk S., Mouffouk S., Hambaba L., Haba H. Flavonols as potential antiviral drugs targeting SARS-CoV-2 proteases (3CL(pro) and PL(pro)), spike protein, RNA-dependent RNA polymerase (RdRp) and angiotensin-converting enzyme II receptor (ACE2) Eur. J. Pharmacol. 2021;891 doi: 10.1016/j.ejphar.2020.173759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murer L., Volle R., Andriasyan V., Petkidis A., Gomez-Gonzalez A., Yang L.…Greber U.F. Identification of broad anti-coronavirus chemical agents for repurposing against SARS-CoV-2 and variants of concern. Curr. Res. Virol. Sci. 2022;3 doi: 10.1016/j.crviro.2022.100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myint S.H. Springer US; 1995. Human Coronavirus Infections. [Google Scholar]
- Oleg T., O. A J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Comput. Chem. 2009;31(2) doi: 10.1002/jcc.21334. NA-NA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordonez A.A., Bullen C.K., Villabona-Rueda A.F., Thompson E.A., Turner M.L., Merino V.F.…Jones-Brando L. Sulforaphane exhibits antiviral activity against pandemic SARS-CoV-2 and seasonal HCoV-OC43 coronaviruses in vitro and in mice. Commun Biol. 2022;5(1):242. doi: 10.1038/s42003-022-03189-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park R., Jang M., Park Y.I., Park Y., Jung W., Park J., Park J. Epigallocatechin gallate (EGCG), a green tea polyphenol, reduces coronavirus replication in a mouse model. Viruses. 2021;13(12) doi: 10.3390/v13122533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J.…Schwartz O. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2021 doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- Ratia K., Pegan S., Takayama J., Sleeman K., Coughlin M., Baliji S.…Johnson M.E. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. U.S.A. 2008;105:16119–16124. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudrapal M., Gogoi N., Chetia D., Khan J., Banwas S., Alshehri B.…Walode S.G. Repurposing of phytomedicine-derived bioactive compounds with promising anti-SARS-CoV-2 potential: molecular docking, MD simulation and drug-likeness/ADMET studies. Saudi J. Biol. Sci. 2022;29(4):2432–2446. doi: 10.1016/j.sjbs.2021.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runfeng L., Yunlong H., Jicheng H., Weiqi P., Qinhai M., Yongxia S.…Zifeng Y. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2) Pharmacol. Res. 2020;156 doi: 10.1016/j.phrs.2020.104761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rut W., Lv Z., Zmudzinski M., Patchett S., Olsen S.K. Activity profiling and crystal structures of inhibitor-bound sars-cov-2 papain-like protease: a framework for anti-covid-19 drug design. Sci. Adv. 2020;6(42) doi: 10.1126/sciadv.abd4596. 6(42), eabd4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirtzinger E.E., Kim Y., Davis A.S. Improving human coronavirus OC43 (HCoV-OC43) research comparability in studies using HCoV-OC43 as a surrogate for SARS-CoV-2. J. Virol. Methods. 2022;299 doi: 10.1016/j.jviromet.2021.114317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj C., Dinesh D.C., Panwar U., Abhirami R., Boura E., Singh S.K. Structure-based virtual screening and molecular dynamics simulation of SARS-CoV-2 Guanine-N7 methyltransferase (nsp14) for identifying antiviral inhibitors against COVID-19. J. Biomol. Struct. Dyn. 2021;39(13):4582–4593. doi: 10.1080/07391102.2020.1778535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Yuan Y., Nie L., Li B., Qin X., Li Y.…Qiu S. A new ent-kaurane diterpene derivative from the stems of Eurya chinensis R. Br. Nat Prod Res. 2018;32(2):182–188. doi: 10.1080/14786419.2017.1343327. [DOI] [PubMed] [Google Scholar]
- Song J., Yuan Y., Nie L., Li B., Qin X., Li Y.…Qiu S. Two new ent-kaurane diterpenes from the stems of Eurya chinensis. J. Asian Nat. Prod. Res. 2018;20(10):962–968. doi: 10.1080/10286020.2017.1373099. [DOI] [PubMed] [Google Scholar]
- Song J., Yuan Y., Tan H., Wu J., Huang R., Li H.…Qiu S. Euryachins A and B, a new type of diterpenoids from Eurya chinensis with potent NO production inhibitory activity. RSC Adv. 2016;6(89):85958–85961. doi: 10.1039/c6ra11994h. [DOI] [Google Scholar]
- Song J.L., Yuan Y., Tan H.B., Huang R.M., Liu H.X., Xu Z.F., Qiu S.X. Anti-inflammatory and antimicrobial coumarins from the stems of Eurya chinensis. J. Asian Nat. Prod. Res. 2017;19(3):222–228. doi: 10.1080/10286020.2016.1191474. [DOI] [PubMed] [Google Scholar]
- Stevaert A., Krasniqi B., Van Loy B., Nguyen T., Thomas J., Vandeput J.…Naesens L. Betulonic acid derivatives interfering with human coronavirus 229E replication via the nsp15 endoribonuclease. J. Med. Chem. 2021;64(9):5632–5644. doi: 10.1021/acs.jmedchem.0c02124. [DOI] [PubMed] [Google Scholar]
- Wen C.C., Kuo Y.H., Jan J.T., Liang P.H., Wang S.Y., Liu H.G., Y N.S. Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus. J. Med. Chem. 2007;(50):4087–4095. doi: 10.1021/jm070295s. [DOI] [PubMed] [Google Scholar]
- Zhang R., Wang K., Ping X., Yu W., Qian Z., Xiong S., Sun B. The ns12.9 accessory protein of human coronavirus OC43 is a viroporin involved in virion morphogenesis and pathogenesis. J. Virol. 2015;89(22):11383–11395. doi: 10.1128/JVI.01986-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J.…Research T. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that has been used is confidential.