Abstract
The spatial arrangements and associative behavior of Actinomyces naeslundii, Veillonella dispar, Fusobacterium nucleatum, Streptococcus sobrinus, and Streptococcus oralis strains in an in vitro model of supragingival plaque were determined. Using species-specific fluorescence-labeled antibodies in conjunction with confocal laser scanning microscopy, the volumes and distribution of the five strains were assessed during biofilm formation. The volume-derived cell numbers of each strain correlated well with respective culture data. Between 15 min and 64 h, populations of each strain increased in a manner reminiscent of batch growth. The microcolony morphologies of all members of the consortium and their distributions within the biofilm were characterized, as were interspecies associations. Biofilms formed 15 min after inoculation consisted principally of single nonaggregated cells. All five strains adhered strongly to the saliva-conditioned substratum, and therefore, coadhesion played no role during the initial phase of biofilm formation. This observation does not reflect the results of in vitro coaggregation of the five strains, which depended upon the nature of the suspension medium. While the possibility cannot be excluded that some interspecies associations observed at later stages of biofilm formation were initiated by coadhesion, increase in bacterial numbers appeared to be largely a growth phenomenon regulated by the prevailing cultivation conditions.
Polyspecies microbial consortia typically consist of cells and microcolonies embedded in exopolymer matrices perforated with channels through which contact with the milieu extérieur is maintained (50). Dental plaque is a clinically relevant example of such a consortium which mediates oral diseases of microbial etiology. The resistance or resilience of biofilms to antimicrobial agents appears to be related to their distinctive architectures (12, 17, 45), in which case an understanding of the fine structure of oral biofilms may lead to new or improved strategies for plaque control.
Efforts have been directed towards defining the temporal development and spatial organization of an in vitro model of supragingival plaque whose responses to various antimicrobial agents and proprietary oral hygiene products (15) mimic clinical observations. At the same time, information was sought on the importance of intraspecies aggregation, interspecies coaggregation, and coadhesion on surface attachment during the initial stages of biofilm formation.
MATERIALS AND METHODS
Experimental design.
Biofilms containing Actinomyces naeslundii OMZ 745, Veillonella dispar ATCC 17748T (OMZ 493), Fusobacterium nucleatum KP-F2 (OMZ 596), Streptococcus sobrinus OMZ 176, and Streptococcus oralis SK248 (OMZ 607) were formed on hydroxyapatite disks as previously described (15). Three independent trials were run, in each of which six or seven biofilms were recovered per time point (Fig. 1). At every time point in each trial, three disks were dip-washed to remove loosely adherent cells and vortexed, and the eluted cells were sonified, while the remaining disks were labeled with dye-conjugated antibodies (Abs) and examined by confocal laser scanning microscopy (CLSM).
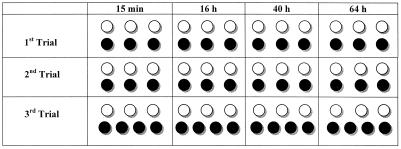
FIG. 1.
Experimental design for analysis of hydroxyapatite disks. The first, second, and third trials represent experiments done on different occasions as checks for repeatability. Solid circles, disks used for CLSM; open circles, disks from which biofilms were eluted and analyzed by conventional microscopy and plate counting. Tetrads rather than trios of disks were used in the third trial in order to obtain the total of 10 disks per time point needed for all NRPCs in CLSM analysis.
Quantification of eluted cells.
Suspensions (25 μl) of eluted cells were incubated on microscope slides in the dark with LIVE/DEAD BacLight Bacterial Viability Kit solutions (0.25 μl each; Molecular Probes B. V., Leiden, The Netherlands) for 15 min at room temperature. Three counts of 100 bacteria at different sites on the slides were made using a Leitz Dialux 22 fluorescence microscope (Leica Mikroskopie Systeme AG, Glattbrugg, Switzerland) equipped with an Osram 50-W high-pressure Hg lamp and an I2/3 filter block for fluorescein isothiocyanate fluorescence. Serial dilutions of eluted cells were prepared in physiological saline, and aliquots (50 μl) were plated onto Columbia blood agar, mitis salivarius agar, and a selective medium for fusobacteria based on Fastidious Anaerobe Agar (15). After anaerobic incubation for 72 h, differentiation of the five species was achieved by observation of colony gross morphology in conjunction with microscopic examination of cells from selected colonies. Nine independent CFU values per species per time point were averaged.
Influence of medium on in vitro coaggregation.
Tubes (9 ml) of each species were grown overnight in mFUM (15) at 37°C. Following examination of each liquid culture by phase-contrast microscopy, the cells were pelleted and washed twice in cold physiological saline, and each pellet was resuspended either in a 1:1 (vol/vol) mixture of mFUM plus saliva (five tubes) or in buffered KCl (7) (five tubes) such that the final optical density at 550 nm of each cell suspension was 0.5 ± 0.05. All 10 nonredundant pairwise combinations (NRPCs) were generated for each resuspension medium by mixing 1 ml of one species with 1 ml of another in a 15-ml polystyrene tube, vortexing the pairs (5 s), and incubating them at room temperature. After standing for 1 h, the tubes were viewed by eye for the presence or absence of a sediment and then vortexed and examined for macroscopic coaggregates by using a loupe and for minute coaggregates by using a microscope. Two hours later, the tubes were vortexed and viewed with a loupe. Macroscopic and microscopic coaggregation were scored semiquantitatively on a scale ranging from no coaggregation to profuse coaggregation.
Labeling, embedment, and viewing of biofilms.
A. naeslundii was detected with immunoglobulin M (IgM) monoclonal Ab (MAb) 396AN1 (51), V. dispar was detected with IgG3 MAb 349VP1.1 (14), F. nucleatum was detected with IgG3 MAb 395FN1 (52), and S. oralis was detected with IgM MAb 493SO1 (R. Gmür and T. Thurnheer, unpublished work). Culture supernatants with high MAb concentrations were produced in MiniPerm cell culture vessels (Heraeus Instruments GmbH & Co. KG, Hanau, Germany) using serum-free HP-1 medium (Cell Culture Technologies, Zürich, Switzerland). S. sobrinus was labeled with polyclonal rabbit anti-OMZ 176 Abs. Immunoglobulins were purified by protein A affinity chromatography (Affi-Gel protein A gel; Bio-Rad Laboratories AG, Glattbrugg, Switzerland) and coupled with Alexa 594 or Oregon Green 488 according to the manufacturer's guidelines (Molecular Probes B. V.).
Disks destined for CLSM were dipped three times in sterile physiological saline (room temperature) and then incubated in an opaque box at room temperature with appropriately diluted Abs. The box was agitated gently for 30 min (15-min biofilms) or 90 min (16-, 40-, and 64-h biofilms). Thereafter, Ab solutions were aspirated, and the disks were washed by immersion (5 min for 15-min biofilms; 10 min for 16-, 40-, and 64-h biofilms) in three changes of physiological saline (2 ml). Since Abs conjugated with either Alexa 594 or Oregon Green 488 were available for each species, two species at a time were viewed by red and green fluorescence. All species pairings in the biofilms were visualized using 10 biofilms (Fig. 1) with the 10 NRPCs of Abs, producing four sets of data per species per time point. The bottom of each stained disk was pressed firmly onto a small wad of plasticine affixed to a glass microscope slide, and the upper surfaces of the disks were covered immediately with Mowiol (8 μl) and topped with glass coverslips. Mowiol, a semipermeable mounting medium compatible with immunostaining (46), was prepared by mixing Mowiol 4-88 (2.4 g; Calbiochem-Novabiochem Corp., San Diego, Calif.) with 50% (vol/vol) aqueous glycerol (12 ml); Tris-HCl (12 ml; 0.2 M; pH 8.5) was added, and the mixture was stirred for 5 h and then left undisturbed for 2 h. 1,4-Diazabicyclo[2.2.2]octane (24 mg; Fluka Chemie AG, Buchs, Switzerland) was added to retard fading, and the suspension was incubated for 10 min at 50°C. The Mowiol suspension was clarified by centrifugation (15 min; room temperature; 5,000 × g), and the supernatant was removed and frozen at −20°C; aliquots were thawed as needed. Mowiol-mounted discs were incubated in opaque containers at room temperature for 6 h and then stored in the dark at 4°C. The stained biofilms were examined using a DM IRB E inverted microscope (Leica Mikroskopie und Systeme GmbH, Wetzlar, Germany) fitted with an Ar-Kr laser (model 543; Omnichrome, Inc., Chino, Calif.) and a TCS 4D computer-operated confocal scanning system (Leica Lasertechnik GmbH, Heidelberg, Germany). Confocal images were obtained with oil immersion objectives (×100 for 15-min biofilms; ×40 for 16-, 40-, and 64-h biofilms). Cells labeled with Oregon Green 488 were viewed with a 522-nm bandpass filter, whereas cells labeled with Alexa 594 were viewed with a 590-nm longpass filter. The microscope pinhole size was set to 100/255, with resulting radii of 636 nm (×40 objective) or 254 nm (×100 objective). Image acquisition was done in 8× line average mode.
Image analysis.
Nine randomly selected square spots were examined per biofilm disk. Z-series were generated by vertical optical sectioning of each spot into 20 equispaced xy focal planes (lateral strata) spanning the height (z) of immunolabeled biofilms, and each optodigital thin section was scanned once for green fluorescence and once for red fluorescence. The data were processed on an Indigo 2ex workstation (Silicon Graphics, Inc., Mountain View, Calif.). The area of each spot was transformed into a digital image containing 512 by 512 pixels; the side of each pixel represented 0.49 μm (×40 objective) or 0.2 μm (×100 objective). The ratio of total disk surface area to spot surface area was 1,299 for the ×40 objective and 7,781 for the ×100 objective.
The 40 scans per 15-min spot were recombined using Imaris 3.0 software (Bitplane AG, Zürich, Switzerland), and the bacterial cells in each reconstructed spot were counted manually. For 16-, 40-, and 64-h biofilms, where the extent of disk colonization precluded manual counting, VoxelShop Pro software (Bitplane AG) was used to calculate the volume occupied per species per reconstructed spot. The volume values for each species at 16, 40, and 64 h were averaged and scaled by the appropriate proportionality factor to obtain average volumes occupied per species per time point per disk. Dividing the average species volume per disk by the average volume per cell (Table 1) yielded the number of bacterial cells per species per disk at these time points. To study microcolony development within the biofilm and to characterize the associative behavior of the different species, the 40 scans per NRPC per time point were recombined and analyzed using Imaris 3.0, resulting in thousands of CLSM images, the analysis of which provided a description of the development of the consortium as a function of time.
TABLE 1.
Volumes of bacterial cells
| Species | Idealized shape | Calculated cell vol (μm3) (avg ± SD)a |
|---|---|---|
| A. naeslundii | Cylinder | 1.31 ± 0.39 |
| V. dispar | Sphere | 0.41 ± 0.35 |
| F. nucleatum | Cylinder | 0.86 ± 0.19 |
| S. sobrinus | Sphere | 0.82 ± 1.23 |
| S. oralis | Sphere | 0.74 ± 0.46 |
Lengths (L) and cross-sectional diameters (D) were measured for 10 cells of each species, and volumes were calculated as follows: spherical cells, V = πD3/6; cylindrical cells, V = πD2L/4. Approximately normal distributions of volume values per species were confirmed using the Ryan-Joiner test (47). SD, standard deviation at the 0.05 significance level.
RESULTS
Quantification of species within biofilms by culture and image analysis.
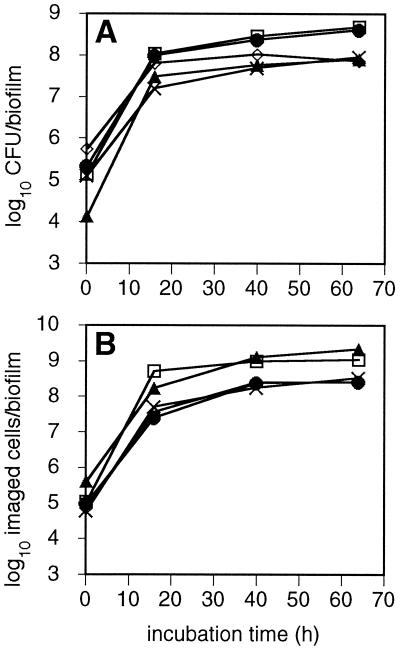
High numbers (104 to 105 CFU) of each species were adherent to the substratum within 15 min of exposure to the inoculum, achieving attachment densities of ≥102 CFU per mm2 of disk surface per species. Average population profiles per biofilm as a function of time for each species as determined from CFU counts are shown in Fig. 2A. Fifteen minutes after inoculation, the number of F. nucleatum KP-F2 CFU adherent to saliva-coated disks was at least an order of magnitude lower than that of any other species, though the subsequent growth of this strain was such that by 16 h its population slightly exceeded that of A. naeslundii OMZ 745. Only modest changes in CFU counts occurred between 16 and 64 h. Comparison of time-dependent population profiles from CFU counts (Fig. 2A) with those calculated from images of immunolabeled cells (Fig. 2B) demonstrated a reasonable correspondence between cell numbers inferred from plating and cell numbers calculated from image analysis, though F. nucleatum plate counts were consistently lower than CLSM cell counts. Live/dead staining of cells eluted from biofilms between 15 min and 64 h showed a slight decrease in percentage of viability as the biofilm aged, but the mean proportion of live cells never fell below 80%.
FIG. 2.
Population profiles for each species, determined by culture or image analysis. (A) Average number of CFU recovered per species per time point per disk. (B) Average number of cells imaged per species per time point per disk. ×, A. naeslundii; ●, V. dispar; ▴, F. nucleatum; ◊, S. sobrinus; □, S. oralis.
Interspecies coaggregation of planktonic cells.
Intraspecies aggregation and interspecies coaggregation of planktonic bacteria are strongly influenced by environmental conditions, especially the growth or resuspension medium (6, 33). The effects of the medium on coaggregation among the bacteria comprising the biofilm model were gauged by resuspending cells in either the substrate used for biofilm formation (mFUM plus saliva) or buffered KCl (Table 2). In buffered KCl medium but not in mFUM plus saliva, coaggregation between F. nucleatum KP-F2 and all partners was seen; in mFUM plus saliva, however, it was S. sobrinus OMZ 176 which coaggregated with all partners. Other pairings in either medium did not lead to coaggregation. These results were independent of the time point and mode of observation.
TABLE 2.
In vitro coaggregation of NRPCs in mFUM plus saliva and in buffered KCl
| Species | Coaggregationa
|
|||||||
|---|---|---|---|---|---|---|---|---|
|
S. oralis
|
V. dispar
|
F. nucleatum
|
A. naeslundii
|
|||||
| mFUM + saliva | Buffered KCl | mFUM + saliva | Buffered KCl | mFUM + saliva | Buffered KCl | mFUM + saliva | Buffered KCl | |
| S. sobrinus | ++ | − | + | − | +++ | ++ | +++ | − |
| S. oralis | − | − | − | +++ | − | − | ||
| V. dispar | − | + | − | − | ||||
| F. nucleatum | − | ++ | ||||||
−, none; +, minor; ++, moderate; +++, profuse.
Structural features of species within 15-min biofilms.
At 15 min, very few F. nucleatum cells participated in intraspecies aggregates. While F. nucleatum formed interspecies coaggregates (particularly with streptococci) more frequently than any other species in the biofilm (Fig. 3A), aggregation and coaggregation clearly were not dominant modes of bacterial adhesion to the salivary pellicle of any of the five species, since at 15 min, single nonaggregated cells (constituting ≥96% of the total cell population) attached directly to the salivary pellicle.
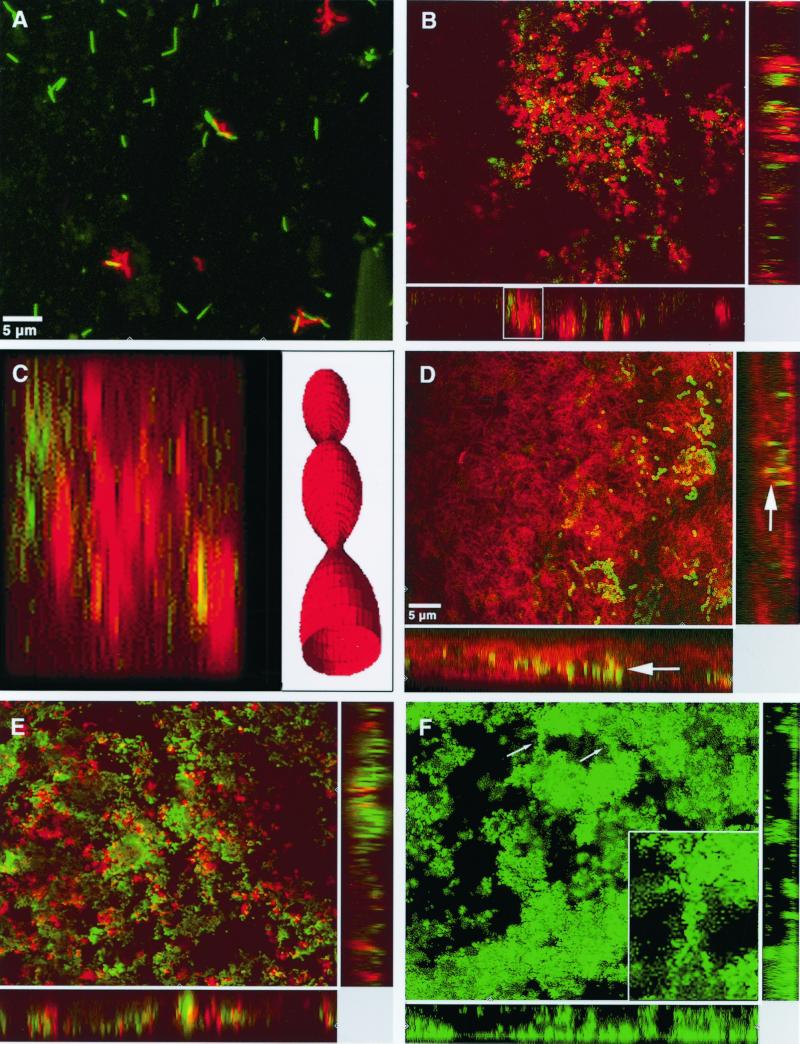
FIG. 3.
CLSM images of biofilms stained with pairs of species-specific Abs. (A) F. nucleatum (green) plus A. naeslundii (red); 15 min. (B) V. dispar (green) plus A. naeslundii (red); 16 h. (C) Sagittal section (the boxed area in panel B) showing stacked microcolonies of A. naeslundii; the image has been stretched by a factor of 1.5 along the xz axis. The drawing to the right is an idealized representation of the stacked microcolonies' unduloid appearance. (D) S. sobrinus (green) plus F. nucleatum (red); 64 h; the arrows indicate the z-plane of the main image. (E) V. dispar (green) plus S. sobrinus (red); 40 h. (F) S. oralis; 40 h. The arrows indicate cellular bridges linking microcolonies; the box on the lower right is an enlargement of an intermicrocolony bridge (the accompanying labeled partner species is not shown).
Structural features of species in biofilms between 16 and 64 h.
Live/dead stains of entire biofilms revealed that at 16 h these consisted mostly of discrete microcolonies with minimal intercolony association. Images at 40 and 64 h showed more densely populated biofilms containing large numbers of microcolonies with few unstained regions between them.
(i) A. naeslundii.
Microcolonies of A. naeslundii OMZ 745 consisted mainly of small spheroid structures interspersed with columnar microcolonies, few of which spanned the height of the biofilm. By 16 h (Fig. 3B), spheroid microcolonies were scattered throughout the biofilm, with a slight preference for the lower and central strata; individual cells were noted near the disk surface. By 40 h, there had been a slight shift in microcolony distribution towards the central strata, and lone cells were absent. Microcolonies in the upper strata often contacted the roof of the biofilm. Loose associations of small spheroid microcolonies stacked so as to resemble upright Delaunay unduloids were seen in sagittal (xz; yz) views (Fig. 3C). By 64 h, these unduloids were plentiful in the central strata.
(ii) V. dispar.
Of the five strains comprising the biofilm, V. dispar ATCC 17748T displayed the greatest morphologic transition between 16 and 40 h. By 16 h, single cocci and mostly small spheroid microcolonies were scattered sparsely throughout the biofilm (Fig. 3B) and were slightly concentrated within the central strata; few columnar or mushroom-shaped microcolonies were observed, and the few large colonies seen were oriented horizontally. Some microcolonies in the upper strata extended to the roof of the biofilm. By 40 h (Fig. 3E), single cells were absent, and most microcolonies were large oblate structures linked by slender cellular bridges.
(iii) F. nucleatum.
By 16 h, F. nucleatum KP-F2 existed mostly as small oblate microcolonies in the lower strata, though some single cells were seen in the central strata. A few columnar microcolonies spanned the height of the biofilm. By 40 h (Fig. 3D), only oblate microcolonies were present, equidistributed throughout the biofilm and often stretching from the disk surface to the roof of the biofilm.
(iv) S. sobrinus.
S. sobrinus OMZ 176 formed mainly spheroid microcolonies residing in the lower and middle strata; the few microcolonies stretching from the disk surface to the roof of the biofilm were columnar or mushroom shaped. Microcolonies bordering the biofilm roof rarely extended below the central strata. By 16 h, spheroid microcolonies consisted of three or four short, usually intertwined chains localized to the upper half of the biofilm, while independent cells and chains clustered near the surface of the disk. By 40 h (Fig. 3E), a few large, elongate microcolonies lying parallel to the disk surface had formed, though some lone cells and chains were seen principally in the central strata. By 64 h (Fig. 3D), the proportion of large horizontal microcolonies had increased.
(v) S. oralis.
S. oralis SK248 formed spheroid or ovoid microcolonies which expanded laterally. A greater proportion of microcolonies extended from the disk surface to the roof of the biofilm than had been observed for S. sobrinus. By 16 h, microcolonies were equidistributed in size and dispersed evenly throughout the biofilm; some small columnar microcolonies and a few very large amorphous microcolonies were seen. A mass of single cells and short chains occurred near the surfaces of the disks. By 40 h, microcolonies, usually large oblate structures linked by narrow cellular bridges (Fig. 3F) and anchored to the disk surface by slender podia, had collected in the central strata of the biofilm. Between 40 and 64 h, the microcolonies appeared to have drifted apart.
Associative behavior.
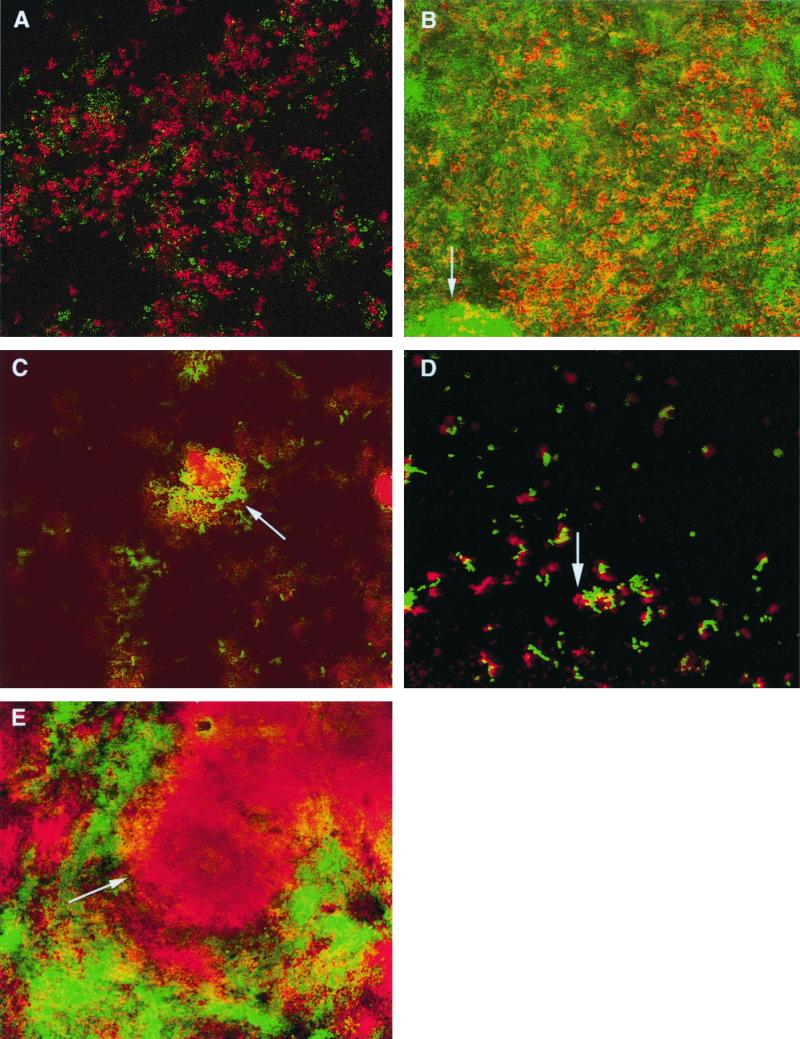
The associative behavior of biofilm strains was classified into the five general spatial types illustrated in Fig. 4. At 16 h, the frequency of the modes of association of the different species followed the order interlaced > interosculated > coated or embedded; by 40 h, however, there had been a shift away from interlacing or interosculation towards embedment, possibly a consequence of growth obscuring the other, more discrete forms of interspecies associations (Table 3). S. oralis and F. nucleatum dominated the images they shared with other species (Fig. 3D and 4B). No special associations between the two streptococcal species were noted. A. naeslundii, and especially F. nucleatum, displayed a pronounced propensity to combine with all species except V. dispar; indeed, V. dispar usually collected into autonomous microcolonies, displaying minimal association with other strains (Fig. 3E and 4A). At 40 and 64 h, microcolonies of S. oralis and F. nucleatum formed a mosaic pattern devoid of extensive intermingling (Fig. 4E).
FIG. 4.
Categories of spatial arrangements between different species in the biofilm. (A) Code 0 (unassociated): V. dispar (green) plus A. naeslundii (red); 16 h. (B) Code 1 (embedded): F. nucleatum (green) plus A. naeslundii (red); 64 h. (C) Code 2 (coated): S. sobrinus (green) plus F. nucleatum (red); 16 h. (D) Code 3 (interlaced): S. sobrinus (green) plus A. naeslundii (red); 16 h. (E) Code 4 (interosculated): S. oralis (green) plus F. nucleatum (red); 40 h.
TABLE 3.
Spatial arrangements of pairs of species in the biofilm
| Species | Arrangementa
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 16 h
|
40 and 64 h
|
|||||||
| V. dispar | F. nucleatum | S. sobrinus | S. oralis | V. dispar | F. nucleatum | S. sobrinus | S. oralis | |
| A. naeslundii | 0 | 3, 4 | 4 > 2, 3b | 0 > 3, 4 | 1 (An > Vd) | 1 (Fn > An) | 3, 4 | 1 (So > An) |
| V. dispar | 3, 4 (Fn > Vd) | 0 ≫ 4 | 1 (So > Vd) | 1 (Fn > Vd) | 1, 3 | 1 (So > Vd) | ||
| F. nucleatum | 2,c 4 | 3, 4 | 2, 4 (Fn > Ss) | 4d | ||||
| S. sobrinus | 1 (So > Ss) | 1 (So > Ss) | ||||||
Code numbers are defined as follows: 0, unassociated (no conspicuous association between species [Fig. 4A]); 1, embedded (discrete microcolonies of one species dispersed or sprinkled within a much denser background of microcolonies of a different species [Fig. 4B]); 2, coated (surface of intraspecies aggregates encased or coated in whole or in part by a mono- or bilayer of cells of a different species [Fig. 4C]); 3, interlaced (discrete, contacting intraspecies aggregates, in which contact between aggregates may be tangential or extensive but usually without a filigreed appearance [Fig. 4D]); 4, interosculated (interspecies complexes or, more commonly, aggregates of various shapes and sizes [Fig. 4E]). While the relative spatial arrangements of pairs of species were similar at 40 and 64 h, the 64-h images typically showed higher cell densities than the 40-h images. Where particular species or arrangements predominate, these are indicated (>, predominates over; ≫, greatly predominates over). An, A. naeslundii; Fn, F. nucleatum; Vd, V. dispar; So, S. oralis; Ss, S. sobrinus.
S. sobrinus cells coating A. naeslundii microcolonies.
S. sobrinus cells coating F. nucleatum microcolonies.
Each species nestled within microcolony surface pockets of the other species, contacting only at microcolony boundaries without extensive intermingling.
DISCUSSION
Tagged antibodies are useful tools for probing the fine structure of microbial ecosystems (3), though their use for this purpose in conjunction with CLSM remains underexploited (44, 49, 50). Since methodologies for simultaneously visualizing all five species in the biofilm are not yet generally available (5), recourse was made to pairwise application of fluorescence-labeled Abs followed by CLSM. Though no single image taken through a single plane of confocality can capture the complex spatial characteristics of a species during its growth in the consortium, careful examination of the thousands of images recorded showed enough interdisk consistencies per species per time point to permit identification of the most salient morphological features and spatial arrangements for each member of the biofilm.
After a 15-min exposure to the five-species inoculum, saliva-covered disks were coated with a shallow layer of cells, most of which bound directly to the pellicle. Given the presumed importance of intraspecies aggregation and interspecies coaggregation in establishing dental plaque (20, 24, 25, 29), 15-min biofilms were scrutinized for the presence of these cell complexes, but they were seldom encountered at this time. By 16 h, the adherent cells had given rise to microcolonies of various sizes and shapes (predominantly spheroid or ovoid), while single cells occurred preferentially near the disk surface (A. naeslundii, S. sobrinus, and S. oralis) or the central strata (V. dispar and F. nucleatum). By 40 h, single cells either had vanished (A. naeslundii, V. dispar, F. nucleatum, and S. oralis) or were confined to the central strata in reduced numbers (S. sobrinus). There was an increase in the diversity of microcolony form (columnar or mushroom shaped), including some large, quite distinctive morphologies (stacked spheroid microcolonies of A. naeslundii; oblate interbridged microcolonies of S. oralis). The population structures of only a few polymicrobial communities are known in sufficient detail for comparison with that of our oral biofilm model. Cook et al. (8) described the sequential deposition of Streptococcus gordonii and Porphyromonas gingivalis onto saliva-coated glass surfaces; after 2 h, this accretive biofilm consisted of obconical porphyromonad microcolonies anchored to a confluent streptococcal monolayer. Most bacterial consortia whose microanatomies have been described were recovered from environments unrelated to the oral cavity (1, 30, 34, 39, 48, 54), and therefore they might be expected to differ substantially from our plaque model. Nonetheless, these divergent biofilms share numerous structural features.
Dosani (10) reported that biofilms begin as an assemblage of discrete microcolonies which eventually grow together and over one another to form a continuous mass; our live/dead-stained biofilms revealed such a coalescence to have taken place between 16 and 40 h. James et al. (G. A. James, D. E. Caldwell, and J. W. Costerton, Abstr. Can. Soc. Microbiol./Soc. Ind. Microbiol. Annu. Meet., abstr. P138, 1993), Amann et al. (2), and Møller et al. (35) each observed species-specific spatial heterogeneity within biofilms (also see reference 31). Formation of palisades and other vertical structures from initially adherent cells and the presence of free-floating grapelike clusters have been noted (23, 28, 38, 54). Zhang et al. (56) suggested that mushroom-shaped microcolonies might be gravitational artifacts of biofilms cultivated in an inverted position, though mushroom-shaped microcolonies of V. dispar and S. sobrinus occurred in biofilms cultivated and stained in upright positions, concordant with predictions by three-dimensional models of substrate-limited biofilms (41, 42). Neu and Lawrence (38) demonstrated that some structural features in biofilms are artifacts of fluid flow. Our plaque model was not subject to continuous drag forces, and lift forces were exerted only during brief episodes of disk dip-washing, punctuated by long periods of stagnant incubation. Shear-induced perturbation in our biofilms, therefore, will have been slight, so that the microcolony morphologies and associative (or partitive) behaviors adopted by the cells will have arisen principally from physiological and physicochemical interactions among themselves.
The precise manner in which surface-adherent cells at 15 min gave rise to free-floating microcolonies in the biofilm was not determined. They may have formed de novo by growth of detached single cells or small clusters, by exfoliation of upper portions of microcolonies, or by detachment of whole microcolonies from the substratum. A shift away from free-floating single cells and towards free-floating microcolonies between 16 and 40 h is consistent with the formation of suspended microcolonies by the growth of detached cells, while the large oblate microcolonies of S. oralis hovering over the substratum but still attached to it by slender cellular threads might represent transitional forms in the process of detachment. Such transitional structures may have been observed by de Beer et al. (9), who found that as biofilms aged the substratal base of cell clusters became increasingly lacunose.
Besides CLSM, cultivation on Columbia blood agar and two selective media for enumeration of fusobacteria and streptococci was used for species quantification. For four of the five species, image analysis counts were in agreement with CFU counts of eluted cells; however, microscopic cell counts of F. nucleatum KP-F2 often exceeded CFU by a factor of ≥20. That much of the adherent population of F. nucleatum is in a viable but noncultivable state is unlikely, since the F. nucleatum population increases by some 3.5 orders of magnitude between 15 min and 16 h, more than any other species in the biofilm, though this strain has a somewhat longer doubling time than any other species comprising the biofilm (unpublished observations). More likely, the cell numbers of F. nucleatum obtained from CFU data are underestimated. Discrepancies between microscopic cell counts and CFU counts are encountered commonly for aggregatory microorganisms or for those species whose cells are not dispersed readily following mitosis (16, 19, 21, 53). In addition, suboptimal recovery of F. nucleatum on the selective Fastidious Anaerobe Agar medium may have contributed to the low CFU counts of this species.
The predominance of F. nucleatum in our biofilm model—by 64 h this species still represented >50% of the total cell load—is consistent with the report by Moore and Moore (36) that F. nucleatum figures among the taxa most frequently isolated from gingival-crevice plaque. It has been claimed that F. nucleatum binds poorly to the acquired pellicle (22), though in fact the species has an affinity for salivary components, such as statherin (55) and proline-rich glycoproteins (13); indeed, the adherence strength of F. nucleatum to saliva is comparable to that of Streptococcus sanguis (43).
A. naeslundii and S. oralis are regarded as pioneer colonizers of tooth surfaces (26, 37, 40), the coaggregation of which with the reputed secondary colonizers Fusobacterium and Veillonella are said to facilitate formation of dental plaque (18, 32). However, neither A. naeslundii nor S. oralis formed interspecies coaggregates with one another or with F. nucleatum or V. dispar, even after 3 h in a saliva-based medium promoting the formation and growth of oral biofilms (Table 2). The only bacterium to coaggregate with the other four species in mFUM plus saliva was S. sobrinus (27). It would appear that, at least in our in vitro model of dental plaque, intraspecies aggregation and interspecies coaggregation are not crucial for biofilm formation (11). Coadhesion of planktonic cells to sessile cells has been postulated to play a role in the formation of dental plaque (4); however, careful examination of CLSM images of 15-min polyspecies biofilms failed to identify any of the five species acting as conspicuous anchors or nuclei for attachment of any of the other species. Increase in bacterial numbers in this biofilm appears to be largely a growth phenomenon regulated by the prevailing cultivation conditions rather than the result of specific or primary aggregation or coadhesion.
ACKNOWLEDGMENTS
We thank André Meier and Yvonne Helweg for excellent technical assistance. The help of Matthias Höchli (Elektronenmikroskopisches Zentrallaboratorium der Universität Zürich) and René Fischer (Institut für Biochemie, ETH Zentrum, Zürich, Switzerland) with CLSM and hybridoma supernatant production, respectively, is gratefully acknowledged.
REFERENCES
- 1.Abella C A, Cristina X P, Martinez A, Pibernat I, Vila X. Two new motile phototrophic consortia: “Chlorochromatium lunatum” and “Pelochromatium selenoides.”. Arch Microbiol. 1998;169:452–459. doi: 10.1007/s002030050596. [DOI] [PubMed] [Google Scholar]
- 2.Amann R I, Stromley J, Devereux R, Key R, Stahl D A. Molecular and microscopic identification of sulfate-reducing bacteria in multispecies biofilms. Appl Environ Microbiol. 1992;58:614–623. doi: 10.1128/aem.58.2.614-623.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bohlool B, Schmidt E. The immunofluorescence approach to microbial ecology. Adv Microb Ecol. 1980;4:203–236. [Google Scholar]
- 4.Bos R, van der Mei H C, Busscher H J. Co-adhesion of oral microbial pairs under flow in the presence of saliva and lactose. J Dent Res. 1996;75:809–815. doi: 10.1177/00220345960750021201. [DOI] [PubMed] [Google Scholar]
- 5.Castro, S. 1999. Fluorescent staining advances. Genet. Eng. News vol. 19(17), 1 October.
- 6.Chisari G, Gismondo M R. Coaggregation between Actinomyces viscosus with Streptococcus pyogenes and Streptococcus agalactiae. Microbiologica. 1986;9:393–398. [PubMed] [Google Scholar]
- 7.Clark W B, Bammann L L, Gibbons R J. Comparative estimates of bacterial affinities and adsorption sites on hydroxyapatite surfaces. Infect Immun. 1978;19:846–853. doi: 10.1128/iai.19.3.846-853.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cook G S, Costerton J W, Lamont R J. Biofilm formation by Porphyromonas gingivalis and Streptococcus gordonii. J Periodont Res. 1998;33:323–327. doi: 10.1111/j.1600-0765.1998.tb02206.x. [DOI] [PubMed] [Google Scholar]
- 9.de Beer D, Stoodley P, Roe F, Lewandowski Z. Effects of biofilm structures on oxygen distribution and mass transport. Biotechnol Bioeng. 1994;43:1131–1138. doi: 10.1002/bit.260431118. [DOI] [PubMed] [Google Scholar]
- 10.Dosani R. An electron microscopic study of wastewater biofilm formation. M.S. thesis. Cincinnati, Ohio: The University of Cincinnati; 1991. [Google Scholar]
- 11.Ganeshkumar N, Hughes C V, Weiss E I. Co-aggregation in dental plaque formation. In: Busscher H J, Evans L V, editors. Oral biofilms and plaque control. Amsterdam, The Netherlands: Harwood Academic Publishers; 1998. pp. 125–143. [Google Scholar]
- 12.Gilbert P, Allison D G. Biofilms and their resistance towards antimicrobial agents. In: Newman H N, Wilson M, editors. Dental plaque revisited: oral biofilms in health and disease. Cardiff, United Kingdom: Bioline; 1999. pp. 125–143. [Google Scholar]
- 13.Gillece-Castro B L, Prakobphol A, Burlingame A L, Leffler H, Fisher S J. Structure and bacterial receptor activity of a human salivary proline-rich glycoprotein. J Biol Chem. 1991;266:17358–17368. [PubMed] [Google Scholar]
- 14.Gmür R, Guggenheim B, Giertsen E, Thurnheer T. Automated immunofluorescence for enumeration of selected taxa in supragingival dental plaque. Eur J Oral Sci. 2000;108:393–402. doi: 10.1034/j.1600-0722.2000.108005393.x. [DOI] [PubMed] [Google Scholar]
- 15.Guggenheim B, Giertsen E, Schüpbach P, Shapiro S. Validation of an in vitro biofilm model of supragingival plaque. J Dent Res. 2001;80:363–370. doi: 10.1177/00220345010800011201. [DOI] [PubMed] [Google Scholar]
- 16.Harmsen H J M, Gibson G R, Elfferich P, Raangs G C, Wildeboer-Veloo A C M, Argaiz A, Roberfroid M B, Welling G W. Comparison of viable cell counts and fluorescence in situ hybridization using specific rRNA-based probes for the quantification of human fecal bacteria. FEMS Microbiol Lett. 2000;183:125–129. doi: 10.1111/j.1574-6968.2000.tb08945.x. [DOI] [PubMed] [Google Scholar]
- 17.Helmerhorst E J, Hodgson R, van't Hof W, Veerman E C I, Allison C, Nieuw Amerongen A V. The effects of histatin-derived basic antimicrobial peptides on oral biofilms. J Dent Res. 1999;78:1245–1250. doi: 10.1177/00220345990780060801. [DOI] [PubMed] [Google Scholar]
- 18.Jacquelin L F, Brisset L, Le Magrex E, Carquin J, Gelle M P, Choisy C. Prévention de la plaque dentaire cariogène. Étude des structures impliquées dans l'adhésion et la coagrégation chez Streptococcus mutans et Streptococcus sobrinus. Pathol Biol. 1995;43:371–379. [PubMed] [Google Scholar]
- 19.Jannasch H W, Jones G E. Bacterial populations in seawater as determined by different methods of enumeration. Limnol Oceanogr. 1959;4:128–139. [Google Scholar]
- 20.Jones S J. A special relationship between spherical and filamentous microorganisms in mature human dental plaque. Arch Oral Biol. 1972;17:613–616. doi: 10.1016/0003-9969(72)90081-7. [DOI] [PubMed] [Google Scholar]
- 21.Karlsson K, Malmberg P. Characterization of exposure to molds and actinomycetes in agricultural dusts by scanning electron microscopy, fluorescence microscopy and the culture method. Scand J Work Environ Health. 1989;15:353–359. doi: 10.5271/sjweh.1847. [DOI] [PubMed] [Google Scholar]
- 22.Kaufman J, DiRienzo J M. Isolation of a corncob (coaggregation) receptor polypeptide from Fusobacterium nucleatum. Infect Immun. 1989;57:331–337. doi: 10.1128/iai.57.2.331-337.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keevil C W, Walker J T. Nomarksi DIC microscopy and image analysis of biofilms. Binary-Comput Microbiol. 1992;4:93–95. [Google Scholar]
- 24.Kohlenbrander P E, London J. Ecological significance of coaggregation among oral bacteria. Adv Microb Ecol. 1992;12:183–217. [Google Scholar]
- 25.Kohlenbrander P E, London J. Adhere today, here tomorrow: oral bacterial adherence. J Bacteriol. 1993;175:3247–3252. doi: 10.1128/jb.175.11.3247-3252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohlenbrander P E, Andersen R N, Clemans D L, Whittaker C J, Klier C M. Potential role of functionally similar coaggregation mediators in bacterial succession. In: Newman H N, Wilson M, editors. Dental plaque revisited: oral biofilms in health and disease. Cardiff, United Kingdom: Bioline; 1999. pp. 171–186. [Google Scholar]
- 27.Lamont R J, Rosan B. Adherence of mutans streptococci to other oral bacteria. Infect Immun. 1990;58:1738–1743. doi: 10.1128/iai.58.6.1738-1743.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawrence J R, Korber D R, Wolfaardt G M, Caldwell D E. Behavioral strategies of surface-colonizing bacteria. Adv Microb Ecol. 1995;14:1–75. [Google Scholar]
- 29.Listgarten M A, Mayo H E, Tremblay R. Development of dental plaque on epoxy resin crowns in man. A light and electron microscopic study. J Periodontol. 1975;46:10–26. doi: 10.1902/jop.1975.46.1.10. [DOI] [PubMed] [Google Scholar]
- 30.Lünsdorf H, Brümmer I, Timmis K N, Wagner-Döbler I. Metal selectivity of in situ microcolonies in biofilms of the Elbe River. J Bacteriol. 1997;179:31–40. doi: 10.1128/jb.179.1.31-40.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manz W, Arp G, Schumann-Kindel G, Szewzyk U, Reitner J. Widefield deconvolution epifluorescence microscopy combined with fluorescence in situ hybridization reveals the spatial arrangement of bacteria in sponge tissue. J Microbiol Methods. 2000;40:125–134. doi: 10.1016/s0167-7012(99)00103-7. [DOI] [PubMed] [Google Scholar]
- 32.Marsh P, Martin M. Oral microbiology. 3rd ed. London, United Kingdom: Chapman & Hall; 1992. pp. 109–110. [Google Scholar]
- 33.McIntire F C, Vatter A E, Baros J, Arnold J. Mechanism of coaggregation between Actinomyces viscosus T14V and Streptococcus sanguis 34. Infect Immun. 1978;21:978–988. doi: 10.1128/iai.21.3.978-988.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Møller S, Pedersen A R, Poulsen L K, Arvin E, Molin S. Activity and three-dimensional distribution of toluene-degrading Pseudomonas putida in a multispecies biofilm assessed by quantitative in situ hybridization and scanning confocal laser microscopy. Appl Environ Microbiol. 1996;62:4632–4640. doi: 10.1128/aem.62.12.4632-4640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Møller S, Sternberg C, Andersen J B, Christensen B B, Ramos J L, Givskov M, Molin S. In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members. Appl Environ Microbiol. 1998;64:721–732. doi: 10.1128/aem.64.2.721-732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore W E C, Moore L V H. The bacteria of periodontal diseases. Periodontol 2000. 1994;5:66–77. doi: 10.1111/j.1600-0757.1994.tb00019.x. [DOI] [PubMed] [Google Scholar]
- 37.Morou-Bermudez E, Burne R A. Genetic and physiologic characterization of urease of Actinomyces naeslundii. Infect Immun. 1999;67:504–512. doi: 10.1128/iai.67.2.504-512.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neu T, Lawrence J R. Development and structure of microbial biofilms in river water studied by confocal laser scanning microscopy. FEMS Microbiol Ecol. 1997;24:11–25. [Google Scholar]
- 39.Nielsen A T, Tolker-Nielsen T, Barken K B, Molin S. Role of commensal relationships on the spatial structure of a surface-attached microbial consortium. Environ Microbiol. 2000;2:59–68. doi: 10.1046/j.1462-2920.2000.00084.x. [DOI] [PubMed] [Google Scholar]
- 40.Pearce C, Bowden G H, Evans M, Fitzsimmons S P, Johnson J, Sheridan M J, Wientzen R, Cole M F. Identification of pioneer viridans streptococci in the oral cavity of human neonates. J Med Microbiol. 1995;42:67–72. doi: 10.1099/00222615-42-1-67. [DOI] [PubMed] [Google Scholar]
- 41.Picioreanu C, van Loosdrecht M C M, Heijnen J J. Mathematical modeling of biofilm structure with a hybrid differential-discrete cellular automaton approach. Biotechnol Bioeng. 1998;58:101–116. doi: 10.1002/(sici)1097-0290(19980405)58:1<101::aid-bit11>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 42.Picioreanu C, van Loosdrecht M C M, Heijnen J J. Discrete-differential modelling of biofilm structure. Water Sci Technol. 1999;39:115–122. [Google Scholar]
- 43.Prakobphol A, Burdsal C A, Fisher S J. Quantifying the strength of bacterial adhesive interactions with salivary glycoproteins. J Dent Res. 1995;74:1212–1218. doi: 10.1177/00220345950740051101. [DOI] [PubMed] [Google Scholar]
- 44.Prensier G, Dubourguier H C, Thomas I, Albagnac G, Buisson M N. Specific immunological probes for studying the bacterial associations in granules and biofilms. In: Lettinga G, Zehnder A J B, Grotenhuis J T C, Hulshoff Pol L W, editors. Granular anaerobic sludge: microbiology and technology. Wageningen, The Netherlands: Centre for Agricultural Publishing and Documentation; 1988. pp. 55–61. [Google Scholar]
- 45.Reid G. Biofilms in infectious disease and on medical devices. Int J Antimicrob Agents. 1999;11:223–226. doi: 10.1016/s0924-8579(99)00020-5. [DOI] [PubMed] [Google Scholar]
- 46.Rodriguez J, Deinhardt F. Preparation of a semipermanent mounting medium for fluorescent antibody studies. Virology. 1960;12:316–317. doi: 10.1016/0042-6822(60)90205-1. [DOI] [PubMed] [Google Scholar]
- 47.Ryan T A, Jr, Joiner B L. Normal probability plots and tests for normality. Technical report. Department of Statistics. The Pennsylvania State University, University Park; 1976. [Google Scholar]
- 48.Schwarzer C, Auer B, Klima J, Haselwandter K. Physiological and electron microscopical investigations on syntrophic dicyandiamide degradation by soil bacteria. Soil Biol Biochem. 1998;30:385–391. [Google Scholar]
- 49.Singleton S, Albiston L, Treloar R, Mahers E, Hodgson R, Watson K, Schilling K, Allison C. Optical imaging and characterisation of oral biofilm structures using viral stains and specific antibody probes. In: Wimpenny J, Handley P, Gilbert P, Lappin-Scott H, editors. The life and death of biofilm. Cardiff, United Kingdom: Bioline; 1995. pp. 33–36. [Google Scholar]
- 50.Singleton S, Treloar R, Warren P, Watson G K, Hodgson R, Allison C. Methods for microscopic characterization of oral biofilms: analysis of colonization, microstructure, and molecular transport phenomena. Adv Dent Res. 1997;11:133–149. doi: 10.1177/08959374970110010401. [DOI] [PubMed] [Google Scholar]
- 51.Thurnheer T, Guggenheim B, Gmür R. Characterization of monoclonal antibodies for rapid identification of Actinomyces naeslundii in clinical samples. FEMS Microbiol Lett. 1997;150:255–262. doi: 10.1111/j.1574-6968.1997.tb10378.x. [DOI] [PubMed] [Google Scholar]
- 52.Thurnheer T, Guggenheim B, Gruica B, Gmür R. Infinite serovar and ribotype heterogeneity among oral Fusobacterium nucleatum strains? Anaerobe. 1999;5:79–92. [Google Scholar]
- 53.Wagner M, Amann R, Lemmer H, Schleifer K-H. Probing activated sludge with oligonucleotides specific for proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure. Appl Environ Microbiol. 1993;59:1520–1525. doi: 10.1128/aem.59.5.1520-1525.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolfaardt G M, Lawrence J R, Robarts R D, Caldwell S J, Caldwell D E. Multicellular organization in a degradative biofilm community. Appl Environ Microbiol. 1994;60:434–446. doi: 10.1128/aem.60.2.434-446.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie H, Gibbons R J, Hay D I. Adhesive properties of strains of Fusobacterium nucleatum of the subspecies nucleatum, vincentii and polymorphum. Oral Microbiol Immunol. 1991;6:257–263. doi: 10.1111/j.1399-302x.1991.tb00488.x. [DOI] [PubMed] [Google Scholar]
- 56.Zhang T C, Fu Y-C, Bishop P L. Competition for substrate and space in biofilms. Water Environ Res. 1995;67:992–1003. [Google Scholar]