Abstract
The microbial community structure and activity dynamics of a phosphate-removing biofilm from a sequencing batch biofilm reactor were investigated with special focus on the nitrifying community. O2, NO2−, and NO3− profiles in the biofilm were measured with microsensors at various times during the nonaerated-aerated reactor cycle. In the aeration period, nitrification was oxygen limited and restricted to the first 200 μm at the biofilm surface. Additionally, a delayed onset of nitrification after the start of the aeration was observed. Nitrate accumulating in the biofilm in this period was denitrified during the nonaeration period of the next reactor cycle. Fluorescence in situ hybridization (FISH) revealed three distinct ammonia-oxidizing populations, related to the Nitrosomonas europaea, Nitrosomonas oligotropha, and Nitrosomonas communis lineages. This was confirmed by analysis of the genes coding for 16S rRNA and for ammonia monooxygenase (amoA). Based upon these results, a new 16S rRNA-targeted oligonucleotide probe specific for the Nitrosomonas oligotropha lineage was designed. FISH analysis revealed that the first 100 μm at the biofilm surface was dominated by members of the N. europaea and the N. oligotropha lineages, with a minor fraction related to N. communis. In deeper biofilm layers, exclusively members of the N. oligotropha lineage were found. This separation in space and a potential separation of activities in time are suggested as mechanisms that allow coexistence of the different ammonia-oxidizing populations. Nitrite-oxidizing bacteria belonged exclusively to the genus Nitrospira and could be assigned to a 16S rRNA sequence cluster also found in other sequencing batch systems.
Modern biological treatment of wastewater involves not only C removal, but also elimination of the nutrients P and N (5, 20). This requires the combined or sequential actions of various groups of microorganisms, such as heterotrophic bacteria, phosphate-accumulating organisms (PAO), and nitrifying and denitrifying bacteria. Consequently, purification plants and processes have become increasingly complex to satisfy the needs of the different microorganisms, usually in several reactor stages (5, 27). The integration of different functions in a single reactor would save reaction space and time and therefore is desirable from an economical point of view. However, difficulties often arise in establishing stable nitrification in such complex systems. Nitrifying bacteria (i.e., ammonia-oxidizing bacteria [AOB] and nitrite-oxidizing bacteria [NOB]) usually show low maximum growth rates, relatively low substrate affinities, and high sensitivity to toxic shocks or sudden pH changes (17, 25, 41). In the presence of organic matter, they can be easily outcompeted by heterotrophs for oxygen (56) and ammonia (19). Other problems to be solved are the inhibition of denitrification by the presence of oxygen (5) and the need for cyclic changes of oxic and anoxic (i.e., free of oxygen and nitrate) conditions for biological phosphate removal (34). Biofilm systems are an obvious option for such multifunctional reactors. Slow-growing organisms remain in the reactor by their attached growth; the biofilm matrix might protect bacteria from grazing, harmful substances, or sudden pH shifts; and biofilms can be stratified and therefore provide oxic and anoxic reaction zones (11). During the last 5 years, several studies have addressed nitrifying biofilms through a combination of microsensor measurements and 16S rRNA-based methods, such as fluorescence in situ hybridization (FISH) (39, 48, 50). This approach revealed, e.g., the identity and spatial arrangement of AOB and NOB in various nitrifying systems (39, 48–50) and provided a first estimate of their in situ reaction rates and substrate affinities (48). However, very little is known about how and which nitrifying bacteria are adapted to competition with heterotrophs in more complex systems and how they interact with other processes.
Recently, a biofilm system was proposed that integrates enhanced biological phosphate removal (EBPR) and nitrification and denitrification in a single reactor (3, 18). The biofilm is subjected to a sequencing batch mode, in which an anoxic treatment period is followed by an oxic period to allow for net accumulation of polyphosphate in the biomass, which is removed from the system by backwashing at regular intervals. Substrate balances revealed that the removal of organic carbon and EBPR were successfully combined with nitrogen removal via nitrification and denitrification (3).
In the present study, the microbial ecology of this combined nitrification-EBPR biofilm process was investigated by using microsensor analysis and various molecular techniques, i.e., FISH and analysis of 16S ribosomal DNA (rDNA) and amoA gene sequences. The objectives were to reveal which populations contribute to which part of the process, which AOB and NOB persist under these highly competitive and transient conditions, and how nitrifying activity overlaps, in time and space, with heterotrophic activity, especially EBPR.
MATERIALS AND METHODS
Process description.
A 20-liter sequencing batch biofilm reactor (SBBR) was established as described previously (18). The artificial wastewater was composed of Na(CH3COO) x 3H2O (103 mg liter−1), peptone (200 mg liter−1), (NH4)2SO4 (63 mg liter−1), KH2PO4 (44 mg liter−1), KCl (14 mg liter−1), and yeast extract (3 mg liter−1), leading to influent concentrations of 12 mg liter−1 for P and 38 mg liter−1 for N and a chemical oxygen demand (COD) of about 270 mg liter−1. Oxygen and phosphate concentrations in the bulk water were regularly monitored by online measurements with an oxygen electrode (Oxy 196; WTW, Weilheim, Germany) and with a P analyzer (Phosphax Inter; Dr. Lange, Düsseldorf, Germany). Ammonium, nitrate, and COD were determined photometrically with standard test kits (LCK 303, 339, and 314; digital photometer ISIS 6000; Dr. Lange, Düsseldorf, Germany). The lengths of the operation periods were as follows: 20 min of filling (min 0 to 20), 160 min of nonaerated recirculation (min 20 to 180), 260 min of recirculation with aeration (min 180 to 440), and 40 min of draining (min 440 to 480). The process temperature was kept at 20°C. Biofilm was grown on substratum, Kaldnes elements (Purac, Merseburg, Germany): i.e., plastic rings (diameter, 8 mm, height, 8 mm) designed so biofilm could adhere to both the outer surface and the central spaces within the ring. To remove biofilm material with incorporated polyphosphate, the system was backwashed once a week with pressurized air and water. Removal of biomass from the central spaces of the Kaldnes elements was not efficient, leading to complete clogging of these spaces. The preceding FISH analysis of the biofilm structure as described below revealed no visual difference in the composition and spatial organization of the main microbial populations in the upper parts of biofilms originating from the external substratum surface and from the biofilm of clogged central spaces. Therefore, elements filled completely with biogenic material were chosen for microsensor measurements and FISH.
Microsensor measurements.
To allow measurements during reactor operation, Kaldnes elements with biofilm were transferred during the initial filling period from the reactor to a separate 750-cm3 flow chamber coupled to the recirculation of the reactor. Microsensors were inserted through small holes in the top lid that were closed with stoppers during the nonaeration period. Vertical concentration microprofiles in the biofilm were measured for oxygen with Clark-type microsensors (45) and for ammonium, nitrite, and nitrate with potentiometric ion-selective microelectrodes of the LIX type (15) as described previously. At least 20 profiles were measured for each parameter at different times covering the whole course of a treatment cycle. Measurements were distributed over four cycles to check for the similarity of conditions on different days of operation.
Rate calculations.
Due to the periodic changes in the process conditions, the microprofiles measured in situ do not represent a steady-state situation. Therefore, in the case of nitrate, profiles were corrected to allow the application of a one-dimensional diffusion-reaction model for calculation of specific volumetric rates of net consumption or production. In each nitrate profile, the first value measured in the bulk water was set to t = 0. Then for each depth, the concentration change over time was calculated from two subsequent profiles. The resulting slope was used to correct the time-dependent change of concentration points at t > 0 in each profile by linear interpolation. In that way, every profile was corrected for the dynamics of concentration changes leading to the elimination of transience. The correction procedure just described influenced the absolute values of concentration in the deeper biofilm, but did not change the shapes of the profiles. The correction-of-profile data were not applied to oxygen profiles, because their appearance and the stability of bulk values throughout the oxic period of the process indicated a clear steady-state situation with respect to oxygen.
Volumetric nitrification rates were calculated by applying a one-dimensional diffusion-reaction model to the corrected data. Each profile results from a combination of production (P), consumption (C), and diffusive transport, as described by Fick's second law of one-dimensional diffusion, which under steady-state conditions [δc(z, t)δt = 0)] can be written as P − C = −Ds · δ2c(z,t)/δz2, where DS is the effective diffusion coefficient, c is the solute concentration, z is depth, and t is time. Assuming a zero order reaction, volumetric net production was subsequently calculated by quadratic regression for each depth (38).
DNA extraction.
DNA was extracted from native biofilm samples stored at −70°C with the FastDNA-Extraction kit for soil (Bio 101, Carlsbad, Calif.), as described in the manufacturer's instructions. The quality of DNA was checked by agarose (1% [wt/vol]) gel electrophoresis.
16S rDNA analysis.
A 1-kb fragment of the 16S rDNA gene was amplified with the complement of probe NOLI191 (43) as the forward primer and the unlabeled probe Nso1225 (35) as the reverse primer. The following reaction mixture was used: 50 pmol of each primer, 2.5 μmol of each deoxynucleoside triphosphate (dNTP), 1× PCR buffer, 1 U of SuperTaq DNA polymerase (HT Biotechnology, Cambridge, United Kingdom), and 50 to 100 ng of template DNA. The mixture was adjusted to 100 μl with sterile water. PCR was performed with an Eppendorf Mastercycler (Eppendorf, Hamburg, Germany) with 35 cycles with hot start. The annealing temperature of 56°C for the primer set was determined in previous PCRs run at different temperatures. After checking an aliquot of the PCR product by agarose gel electrophoresis, the DNA was directly ligated into the pGEM-T vector (Promega, Mannheim, Germany) according to the manufacturer's instructions and subsequently transformed to competent high-efficiency Escherichia coli cells (strain JM109; Promega, Mannheim, Germany). White and blue screening was used to screen for recombinant transformants. Inserts of positive clones were analyzed for redundancy by amplified rDNA restriction analysis (ARDRA) (44). One representative clone was sequenced for each distinct fragmentation pattern by Taq Cycle sequencing with the PCR primers or primers M13uni and M13rev on a model ABI 377 sequencer (PE Corporation, Norwalk, Conn.). The sequences were checked for chimera formation with the CHECK_CHIMERA software of the Ribosomal Database Project (32). Sequences were aligned and analyzed by use of the ARB software package (Technical University Munich, Munich, Germany) according to the last release of the 16S rDNA sequence database of December 1998 as well as individually added sequences of recent publications. Trees were calculated for the clones and selected related sequences following the suggestions of Ludwig et al. (31). For tree calculation, maximum-parsimony, distance matrix, and maximum-likelihood methods were used, and the results were combined in a consensus tree.
Probe design.
Based on the newly retrieved and published sequences of the Nitrosomonas oligotropha lineage (40), a probe was designed by using the PROBE_DESIGN tool of ARB. The dissociation temperature of the probe was determined by hybridization with a pure culture of Nitrosomonas ureae isolate Nm10 at formamide concentrations ranging from 0 to 60%. Analysis was done by confocal laser-scanning microscopy (CLSM) and image analysis as described elsewhere (13). The specificity of the probe was checked against the ARB database and experimentally tested under the optimal hybridization conditions with the strains listed in Table 1.
TABLE 1.
Organisms used for determination of Td, their sequence at the target region, and hybridization results with probe Nmo218
| Organism or probe | Sequence | FISH result with probe Nmo218 |
|---|---|---|
| Probe sequence Nmo218 (3′→5′) | TACGAAAACCTCGCCGGC | |
| Target sequence (5′→3′) | AUGCUUUUGGAGCGGCCG | |
| Nitrosomonas ureae Nm10a | .................. | + |
| Nitrosomonas cryotolerans Nm55a | G................. | + |
| Nitrosomonas aestuarii Nm36a | G................. | + |
| Pseudomonas lemoignei DSM 7445b | .....A............ | − |
| Nitrosomonas communis Nm2a | G.......A......... | − |
| Nitrosococcus mobilis Nc2a | GC................ | − |
Donated as fixed pure cultures from G. Timmermann, University of Hamburg, Hamburg, Germany.
Active culture received from the German Culture Collection (DSMZ) and fixed as described in the text.
FISH.
Complete substratum elements with adhering biofilm were fixed with fresh 4% paraformaldehyde solution and alternatively with ethanol, washed with phosphate-buffered saline (PBS), and stored in PBS-ethanol (1:1) at −20°C until further processing (1, 33). After freezing and removal of the plastic material, radial biofilm sections with a thickness of 14 μm were prepared at −18°C, immobilized on gelatin-coated microscope slides, and dehydrated in an ethanol series (50). In situ hybridizations of cells in the biofilm were performed with fluorescently labeled, rRNA-targeted oligonucleotide probes according to the method of Manz et al. (33). The probes and conditions used are listed in Table 2. Probes labeled with the sulfoindocyanine dyes Cy3 and Cy5 were obtained from Interactiva (Ulm, Germany) and Biometra (Göttingen, Germany). In cases in which stringency conditions did not allow simultaneous hybridization with several probes, multiple probe hybridization was performed in subsequent steps by first hybridizing with the probe of higher stringency (58). The biofilm was additionally stained with 4′, 6′-diamidino-2-phenylindole (DAPI) after the hybridization step with a solution of 1 mg liter−1 for 10 min or 100 mg liter−1 for 10 s for the purpose of polyphosphate staining (23). Samples were analyzed by standard epifluorescence microscopy on a Zeiss Axioplan II microscope and by CLSM on a Zeiss LSM 510 microscope (Carl Zeiss, Jena, Germany).
TABLE 2.
Oligonucleotide probes and hybridization conditions applied in this study
| Probe | Probe sequence (5′→3′) | Target sitea | Target organism(s) | % of formamideb | NaCl concn (mM)c | Reference |
|---|---|---|---|---|---|---|
| EUB338 | GCTGCCTCCCGTAGGAGT | 16S (338–355) | Domain Bacteria | 20 | 225 | 1 |
| NON338 | ACTCCTACGGGAGGCAGC | 16S (338–355) | Complementary to EUB338 | 20 | 225 | 33 |
| ALF968 | GGTAAGGTTCTGCGCGTT | 16S (968–985) | Most α-proteobacteria and other bacteria | 35 | 80 | 37 |
| ALF1b | CGTTCGYTCTGAGCCAG | 16S (19–35) | Most α-proteobacteria and other bacteria | 20 | 225 | 33 |
| ALF4-1322 | TCCGCCTTCATGCTCTCG | 16S (1322–1339) | Subgroup 4 of α-proteobacteria | 40 | 56 | 37 |
| RRP1088 | CGTTGCCGGACTTAACC | 16S (1088–1104) | Genera Rhodobacter, Rhodovulum, Roseobacter, and Paracoccus and other bacteria | 0 | 900 | 37 |
| BET42ad | GCCTTCCCACTTCGTTT | 23S (1027–1043) | β-Proteobacteria | 35 | 80 | 33 |
| HGC69a | TATAGTTACCACCGCCGT | 23S (1901–1918) | Gram-positive bacteria with high GC content | 20 | 225 | 46 |
| HGC1351 | CAGCGTTGCTGATCTGCG | 16S (1351–1368) | Gram-positive bacteria with high GC content | 30 | 112 | 16 |
| GAM42ad | GCCTTCCCACATCGTTT | 23S (1027–1043) | γ-Proteobacteria | 35 | 80 | 33 |
| Nso1225 | CGCCATTGTATTACGTGTGA | 16S (1225–1244) | Ammonia-oxidizing β-proteobacteria | 35 | 80 | 35 |
| Nso190 | CGATCCCCTGCTTTTCTCC | 16S (190–208) | Ammonia-oxidizing β-proteobacteria | 55 | 20 | 35 |
| Nsm156 | TATTAGCACATCTTTCGAT | 16S (156–174) | Various Nitrosomonas spp. | 5 | 636 | 35 |
| Nsv443 | CCGTGACCGTTTCGTTCCG | 16S (444–462) | Nitrosospira spp. | 30 | 112 | 35 |
| NmII | TTAAGACACGTTCCGATGTA | 16S (120–139) | Nitrosomonas communis lineage | 25 | 159 | 40 |
| NmIV | TCTCACCTCTCAGCGAGCT | 16S (1004–1023) | Nitrosomonas cryotolerans lineage | 35 | 80 | 40 |
| NEUd | CCCCTCTGCTGCACTCTA | 16S (653–670) | Halophilic and halotolerant members of the genus Nitrosomonas | 40 | 56 | 60 |
| Nse1472e | ACCCCAGTCATGACCCCC | 16S (1472–1489) | N. europaea | 50 | 28 | 22 |
| NmV | TCCTCAGAGACTACGCGG | 16S (174–191) | Nitrosococcus mobilis lineage | 35 | 80 | 40 |
| NOLI191 | CGATCCCCCACTTTCCTC | 16S (191–208) | Various members of the Nitrosomonas oligotropha lineage | 30 | 112 | 43 |
| cNOLIf | CGATCCCCCACTTTCCCC | 16S (191–208) | This study | |||
| Nmo218 | CGGCCGCTCCAAAAGCAT | 16S (218–235) | Nitrosomonas oligotropha lineage | 35 | 80 | This study |
| NIT3d | CCTGTGCTCCATGCTCCG | 16S (1035–1048) | Nitrobacter spp. | 40 | 56 | 61 |
| NSR826 | GTAACCCGCCGACACTTA | 16S (826–843) | Various Nitrospira spp. | 20 | 225 | 49 |
| NSR1156 | CCCGTTCTCCTGGGCAGT | 16S (1156–1173) | Various Nitrospira spp.g | 30 | 112 | 49 |
| Ntspa712dh | CGCCTTCGCCACCGGCCTTCC | 16S (712–732) | Phylum Nitrospira | 35 | 80 | 14 |
| Ntspa662dh | GGAATTCCGCGCTCCTCT | 16S (662–679) | Genus Nitrospira | 35 | 80 | 14 |
rRNA position according to E. coli numbering (9).
Percentage of formamide in the hybridization buffer.
Concentration of sodium chloride in the washing buffer.
Used together with an equimolar amount of unlabeled competitor oligonucleotides as indicated in the reference.
Referred to as S-*-Nse-1472-a-A-18 in the reference.
Unlabeled competitor oligonucleotide applied in equimolar amount to discriminate against weak mismatches in Thiobacillus thioparus and two sequences of Azoarcus spp.
Excluding clones described by Burrell et al. (10).
Referred to as S-*-Ntspa-0712-a-A-21 and S-G-Ntspa-0662-a-A-18, respectively, in the reference.
Quantification of AOB and NOB.
Total AOB abundance was quantified by microscopical counting of cells hybridized with probes Nso1225 and Nso190 in several fields of view along a 50-μm-thick horizontal layer up to a biofilm depth of 600 μm. Distinct populations of AOB were counted after hybridization with probes NEU and NOLI191. Quantification of NOB was done by CLSM as previously described (48); i.e., optical sections with a defined thickness of 0.6 μm were scanned, and by determining the signal area and subsequent multiplication with a volume-specific cell abundance, the cell number for a given volume was calculated. Thorough calibration for NOB quantification was performed by counting DAPI-stained cells in various scanned fields of view (n = 30), leading to volumetric indices with a 95% confidence interval of ±7%. Results with all probes used for quantification were corrected for nonspecific binding according to results with probe NON338 as a negative control.
For quantitative population analysis with FISH, means and medians were calculated to describe the distribution of the AOB and NOB populations.
Analysis of amoA sequences.
To supplement results from FISH and the specific 16S rDNA library, comparative sequence analysis of biofilm-derived 491-bp fragments of the ammonia monooxygenase gene (amoA) was performed as previously described (42). Amplificates of amoA were separated according to their GC content by agarose gel retardation as described by Schmid et al. (47).
Nucleotide sequence accession number.
The 16S rDNA partial sequences obtained in this study are available from the EMBL nucleotide sequence database under accession no. AJ297415 to AJ297419. The amoA partial sequences appear under accession no. AF293065 to AF293075 and AY007575.
RESULTS
Functional analysis.
Concentrations of the solutes measured by microsensors in the bulk liquid of the measuring setup closely reflected the bulk liquid composition in the reactor as determined independently by online monitoring (data not shown).
During the nonaeration period, oxygen could be detected in neither the bulk water nor the biofilm at any time. During the aeration period of the process, the biofilm was supplied with oxygen at 227.8 ± 5.4 μM (mean ± standard deviation [SD], n = 16), corresponding to 80.2% ± 1.9% air saturation. Oxygen penetration into the biofilm was limited to a depth of 200 μm (Fig. 1), leaving substantial parts of the biofilm anoxic during the whole treatment. The average areal oxygen uptake remained stable throughout the oxic period at 0.84 ± 0.05 μmol cm−2 h−1 (mean ± SD, n = 16). Neither the penetration depth nor the slope through the diffusive boundary layer was significantly altered during the oxic period (Fig. 1).
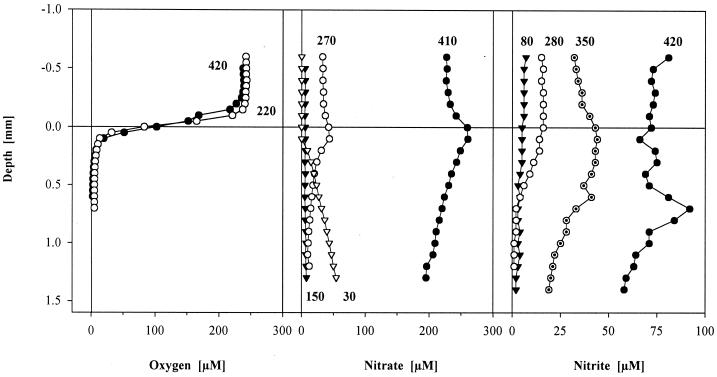
FIG. 1.
Representative examples of vertical concentration microprofiles of oxygen, nitrate, and nitrite measured in biofilm at different times of the reactor cycle. Numbers refer to the time in minutes after start of the treatment cycle (start of aeration, t = 180 min).
With ammonium, nitrate, and nitrite, absolute concentrations showed a certain variability, but the shapes and time courses of profiles between different cycles were similar. A strong initial decrease of ammonium directly after filling from 2,300 μM (t = 25 min) to 950 μM (t = 60 min) could be observed in the biofilm, analogous to the decline within the bulk water in the first 60 min (profiles not shown). Later, the concentration of ammonium decreased less but continuously up to the end of the treatment cycle to 800 μM. The concentration in the biofilm followed this pattern. Because no gradients of ammonium were observed above or within the biofilm, ammonium uptake of the whole biofilm or of any specific layer could not be quantified.
Production of nitrate during oxic conditions was restricted to a narrow surface layer of about 200 μm, causing an accumulation of nitrate in the bulk water of up to 230 μM in the final period of the treatment (Fig. 1). The highest concentration of nitrate measured in the productive layer was 260 μM. During the following nonaeration period (after draining and refilling the system), the remaining nitrate was detected up to a concentration of 200 μM in the deeper biofilm zones, but it continuously decreased during the anoxic period. At the beginning of the aeration (t = 180 min), no nitrate from the previous treatment cycle was left in the biofilm (Fig. 1). The shape of nitrate profiles supported a denitrifying activity in the deeper biofilm layer. Nitrite accumulated during aeration up to 75 μM. In the second half of the aeration treatment period, an even stronger local production of nitrite occurred in the deeper biofilm layers at a depth of 600 to 700 μm, which was probably due to denitrification, with concentrations of more than 90 μM (Fig. 1).
During the aeration period, there was a conspicuous delay in the first occurrence of both nitrite and nitrate compared to the onset of aeration. Detectable amounts of both solutes (>1 μM) were measured first at t = 270 min, i.e., 90 min after onset of aeration.
A nitrogen balance based on nitrification products (300 μM), remaining ammonium (800 μM), and an assumed stripping and incorporation into bacterial biomass of 25% (61) indicates an unresolved N loss of about one-third.
Calculation of nitrate production.
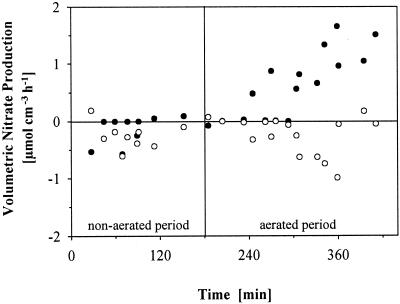
Volumetric nitrate production rates were separately estimated for the productive surface (depth of 0 to 200 μm) and the deeper biofilm (300 to 600 μm) from profile data measured in four different batch runs. The deeper layer showed some denitrifying activity up to about 1.0 μmol cm−3 h−1 at t = 360 min, evolving together with nitrifying activity in the upper layer. The nitrate production in the surface layer during aeration reached maximum estimated rates of 1.7 μmol cm−3 h−1 (corresponding to 0.03 μmol cm−2 h−1) and thus accounted for about 7% of the oxygen uptake at the end of the process (Fig. 2).
FIG. 2.
Volumetric net production and consumption of nitrate at the biofilm surface (0 to 200 μm [●]) and in the deeper biofilm (300 to 600 μm [○]) during the reactor cycle. Data points originate from measurements of nitrate microprofiles of four different batch runs. For details of calculations, see text.
Broad-scale community structure.
Among all phylogenetic groups tested, four groups dominated the biofilm community, as shown by FISH: (i) members of the gram-positive bacteria with high DNA G+C content (GPBHGC), which were mainly coccoid cells forming loose aggregates; (ii) members of the β-proteobacteria, forming dense layers at the very surface and dense globular aggregates mostly located within the upper 200 μm; (iii) a population morphologically similar to the GPBHGC, with cells typically arranged in tetrads, that hybridized with probes ALF968, ALF1b, and GAM42a, but not with probes for subgroups of the α-proteobacteria (Fig. 3A), leaving their phylogenetic affiliation as yet unresolved; and (iv) members of the phylum Nitrospira (see below). Other phylogenetic groups together did not account for more than 20% of the microbial community in the biofilm (data not shown). The vast majority of all cells were located in the upper 300 μm of the biofilm, whereas in the deeper, permanently anoxic layers, only a few cell aggregates could be detected occasionally.
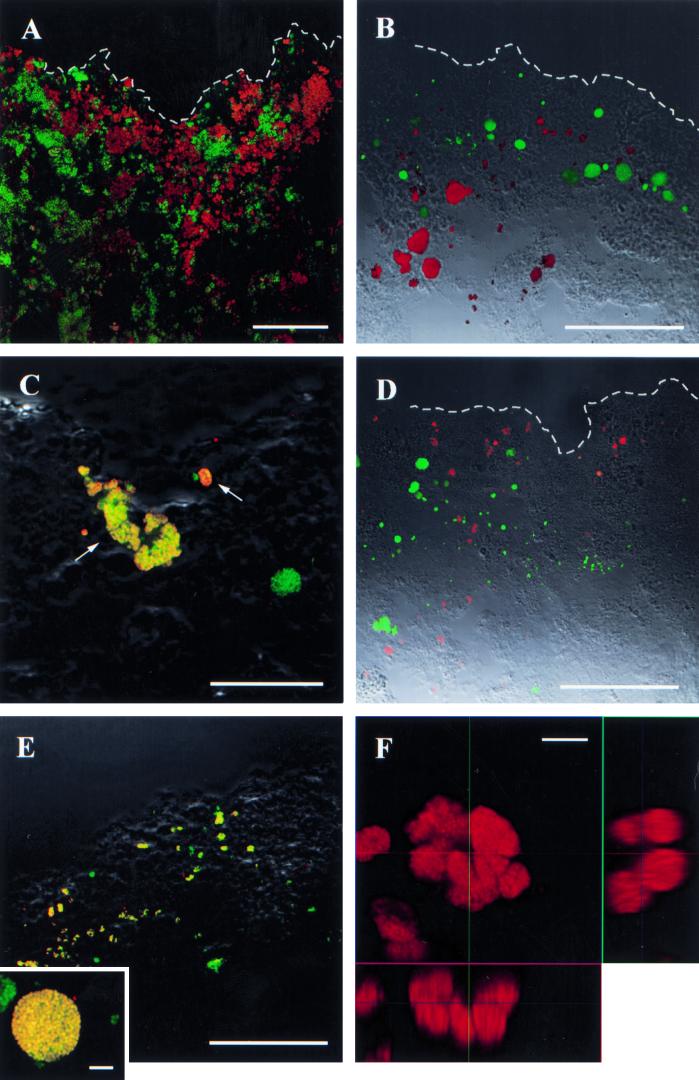
FIG. 3.
Confocal laser-scanning micrographs of vertical thin sections of biofilm as hybridized with different fluorescent oligonucleotide probes. (A) Overview of the biofilm surface hybridized with ALF968 and HGC1351 (in green and red, respectively). (B) Aggregates of β-subclass AOB as hybridized with Nso1225 (green) and NOB of the genus Nitrospira (Ntspa662, red). (C) Clustered aggregates (arrows) of members of the N. communis lineage (NmII, red) among the population β-subclass AOB (Nso1225, green). Colocalization of the two probes results in a yellow color. (D) Aggregates of members of the N. europaea lineage as hybridized with probe NEU (green) and of the N. oligotropha lineage as hybridized with probe NOLI191 (red). (E) Colocalization of signals after hybridization with probes specific for the β-subclass AOB (Nso1225, green) and the N. oligotropha lineage (Nmo218, red); the insert shows a big aggregate as hybridized with probes Nso1225 (green) and NOLI191 (red). (F) Orthogonal representation of a dense assemblage of Nitrospira sp. as hybridized with probe Ntspa662. Scale bars are 50 μm (A, B, D, and E), 25 μm (C), and 5 μm (insert in panels E and F), respectively. Dashed lines indicate the surface of the biofilm exposed to the wastewater.
Additional staining with DAPI in high concentrations, indicating polyanionic inclusions by yellow fluorescence (23), was exclusively colocalized with hybridization signals for GPBHGC. However, only a fraction of the GPBHGC population was stained yellow with DAPI.
AOB community structure.
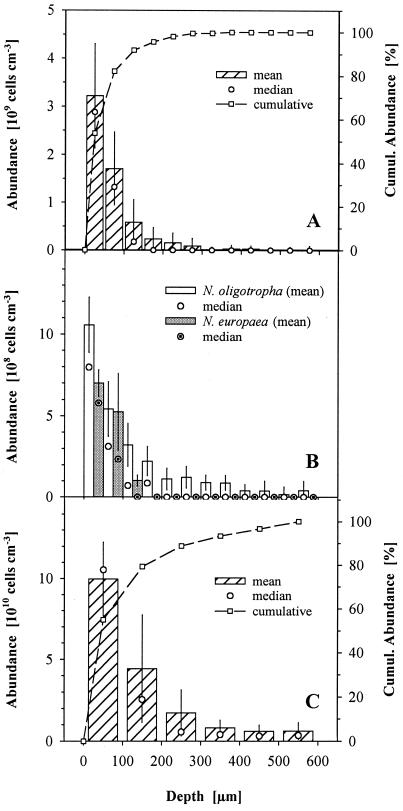
A common problem for the quantification of nitrifying bacteria is the formation of dense aggregates resulting in a typical patchy distribution of AOB and NOB (Fig. 3). Therefore, neither normal distribution of values nor homogeneity of variances is given throughout the biofilm. For that reason, the median may be a better representative of cell densities than the mean and will be given in the following sections. To allow comparison with other studies, however, both means and medians are displayed in Fig. 4.
FIG. 4.
Depth distribution of β-subclass AOB (A), of populations affiliated with different lineages of the genus Nitrosomonas (B), and of the NOB Nitrospira spp. (C) in the biofilm investigated. Numbers are given as volume-specific abundances. Cumul, cumulative. Note the different scales of the ordinates.
The globular aggregates hybridizing with probe BET42a were shown to belong to the AOB of β-proteobacteria by FISH with probes Nso1225 and Nso190 (Fig. 3B). The abundance of AOB was highest at the biofilm surface (Nso1225, 2.9 × 109 cm−3; Nso190, 2.0 × 109 cm−3) and declined below 1 × 108 cm−3 within the first 200 μm. A cumulative mean of 95% of all AOB could be detected within the first 200 μm (Fig. 4A). A few single aggregates of β-ammonia oxidizers occurred in the deeper biofilm as well, but the abundance in these layers on average was very low. Hybridization with probes Nsm156 and Nsv443 revealed that the complete AOB community consisted of members of the genus Nitrosomonas. Within this genus, three different subgroups of AOB were detected. The smallest fraction (which was not further quantified) belonged to the Nitrosomonas communis lineage (40) of β-subclass AOB, as identified by hybridization with probes Nso1225 and NmII (Fig. 3C). They formed small aggregates and were restricted to the upper 100 μm. The two dominant subpopulations showed distinct distribution patterns. One population belonged to the Nitrosomonas europaea lineage of β-subclass AOB (40), as identified by hybridization with probes Nso1225, Nsm156, and NEU (Fig. 3D). Hybridization with Nse1472 or NmV resulted in no signals, indicating that the population is not identical to N. europaea or Nitrosococcus mobilis. The second population hybridized with Nso1225 and NOLI191 (Fig. 3D and E), a probe that had been designed for the N. oligotropha lineage (40) based solely on the sequence of N. ureae (43). Both groups together accounted for from 55 to 100% of β-subclass AOB hybridizing with probe Nso1225 in the upper 200 μm of the biofilm. At the surface, 22 and 33% of Nso1225-positive cells hybridized with probes NEU and NOLI191, respectively. At a depth of 200 μm, about 90% of Nso1225-positive cells hybridized with probe NOLI191, whereas the abundance of the N. europaea lineage declined to less than 10% (Fig. 4B). No signals were detected after hybridizations with probe NmIV specific for Nitrosomonas cryotolerans.
AOB-specific PCR.
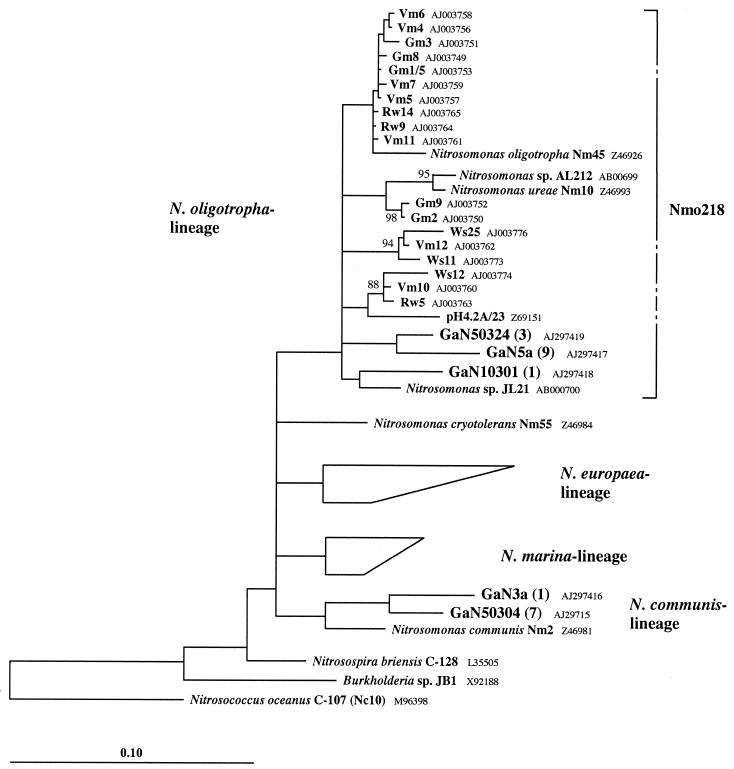
Because hybridization with a single probe is sometimes not sufficient to prove the identity of a given cell (2), a specific PCR and cloning strategy was applied to support the occurrence of populations affiliated with the N. oligotropha lineage within the biofilm. From the 26 clones analyzed by ARDRA, 10 different restriction patterns were obtained. Three of the patterns, representing a total of 13 clones, were indicative of sequences most similar to Nitrosomonas isolate JL21 (55), a member of the N. oligotropha lineage (Fig. 5). Two patterns, representing a total of eight clones, belonged to sequences most similar to Nitrosomonas communis Nm2. The remaining five clones with different ARDRA patterns represented single sequences not related to the genus Nitrosomonas. The amplification of these sequences and of the ones similar to Nitrosomonas communis was due to insufficient primer discrimination under the conditions chosen.
FIG. 5.
Phylogenetic tree of the genus Nitrosomonas inferred from comparative analysis of 16S rDNA sequence data. The accession numbers of the published sequences used are given in the tree. Sequences with accession no. AB000701, AB000702, AJ005546, M96399, M96402, M96403, and Z46987 represent the N. europaea lineage, and sequences with accession no. AJ003777, M96400, Z46990, Z69091, and Z69097 represent the N. marina lineage. The numbers of clones with identical ARDRA patterns for each sequence are given in parentheses. Phylogenetic reconstruction is based on a maximum-likelihood tree calculated from 950 informative positions with a genus-specific 50% positional variability filter. Tree topology was tested by distance matrix and maximum-parsimony methods, and a consensus tree was drawn. Multifurcations connect branches for which a relative order could not unambiguously be determined by the different treeing methods used. Bootstrap values (100 cycles) refer to the maximum-parsimony tree. Values smaller than 80% were omitted. The sequence of N. oligotropha isolate Nm45 was added to the consensus tree by the ARB maximum-parsimony method without changing the tree topology. The bar represents 10% estimated sequence divergence.
Design and application of probe Nmo218.
The sequence of probe NOLI191 differs from the target sites of most of the recently reported 16S rRNA sequences of the N. oligotropha lineage (42, 52, 55). Furthermore, the probe hybridizes to organisms (e.g., Pseudomonas lemoignei) not belonging to the intended target group. Therefore, we designed probe Nmo218 encompassing the whole N. oligotropha lineage (Fig. 5). The probe sequence and binding sites of target and nontarget organisms are displayed in Table 1. The dissociation temperature of the probe was 40.1 ± 1.7°C, and the optimal formamide concentration in the hybridization buffer was 35%. When applied under these conditions, probe Nmo218 did not hybridize with any negative controls, except for Nitrosomonas cryotolerans and Nitrosomonas aestuarii (Table 1). (The sequences of five clones with identical binding sites to N. cryotolerans or N. aestuarii might also not be discriminated.) The occurrence of N. cryotolerans can, however, be ruled out by parallel use of probe NmIV. Hybridization of probe Nmo218 to the biofilm resulted in a picture similar to that with probe NOLI191 (Fig. 3D and E). However, simultaneous hybridization with both probes revealed a certain number of organisms only hybridizing with either of the two probes. Cells of N. aestuarii hybridized with probe Nmo218 (Table 1), but not with probe NOLI191. Consequently, cells showing this hybridization pattern might be related to N. aestuarii.
AOB diversity assessed by comparative amoA sequence analysis.
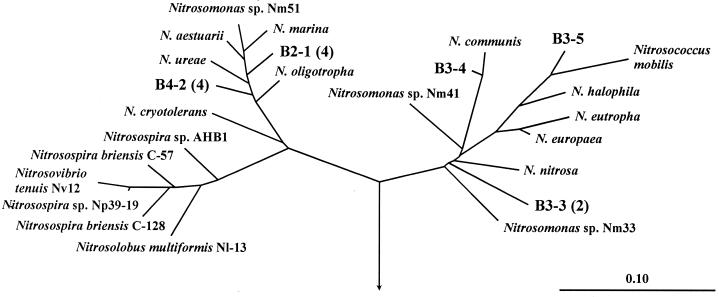
Specific amplification of the amoA gene fragments from extracted biofilm DNA and subsequent separation by gel retardation resulted in three clearly visible amoA bands. All bands were excised and separately cloned and sequenced. A total of 12 amoA clones (4 from each band) were analyzed, which represented five phylogenetically distinct β-subclass AOB (Fig. 6). Two clusters (representing two different bands), each containing four amoA sequences, were found within the Nitrosomonas marina-N. oligotropha lineages (which cannot clearly be distinguished by using amoA sequences) (42). The third band contained a higher diversity of amoA sequences. One amoA clone was closely related to Nitrosomonas communis. Another clone was affiliated with Nitrosococcus mobilis, which could not be detected in the biofilm by FISH. The two remaining amoA clones obtained from the third band clustered together and formed an independent lineage not closely related to any described AOB.
FIG. 6.
Phylogenetic FITCH-Margoliash amoA tree (using global rearrangement and randomized input order [7 jumbles]) showing the position of the 12 recovered biofilm sequences in relation to described β-subclass AOB (42). The bar indicates 10% estimated sequence divergence. The root was determined with the amoA sequences of the γ-subclass ammonia-oxidizing bacteria (42). Cloned amoA sequences with amino acid similarities >99% are represented by a single clone: the number in parentheses indicates the number of clones for each representative. Clones labeled with the prefixes B2, B3, and B4 were obtained from different gel retardation bands.
NOB community structure.
The only NOB detected in the system were of the genus Nitrospira, as identified by hybridization with probes Ntspa712, Ntspa662, and NSR826 (Fig. 3B and F). Low numbers of cells also hybridized with probe NSR1156. The abundance of Nitrospira spp. in the upper 100 μm of the biofilm was 1.1 × 1011 cm−3 and therefore was about 30 times higher than the abundance of β-subclass AOB, as quantified with probe Nso1225. In comparison to the AOB, the vertical distribution was broadened towards depth. Within the section investigated, a 95% limit in cumulative (mean) abundance was reached, although not before a depth of 400 μm, and the numbers were high even in deeper layers of the biofilm (Fig. 4C).
Cell-specific nitrification rates.
The average cell density of NOB in the upper 200 μm of the biofilm where nitrification was mainly performed was 6.6 × 1010 cm−3. Because nitrate production in this layer during aeration on average was 0.7 μmol cm−3 h−1, conversion rates were about 0.01 fmol of nitrite cell−1 h−1 for the final aeration period. Maximum rates were 0.025 fmol of nitrite cell−1 h−1. Cell-specific ammonia oxidation rates can only be roughly estimated based on nitrate production rates. With the average abundance of AOB of 1.09 × 109 cm−3 in the upper 200 μm, mean conversion rates were at least 0.65 fmol of ammonium cell−1 h−1 (maximum of 1.5 fmol of ammonium cell−1 h−1). However, because of the accumulation of nitrite within the nitrification zone, this number is clearly an underestimate.
DISCUSSION
Biofilm activity and community structure.
In the biofilm studied, biological phosphorus removal is combined with nitrogen removal via nitrification and denitrification. Nitrification is typically performed by distinct phylogenetic groups and will be discussed later. For biological P removal, involvement of various groups has been suggested: e.g., GPBHGC (12, 59), members of the genus Rhodocyclus (7, 8, 21), or a mixture of multiple species (34). The high abundance of GPBHGC and the detection of DAPI-stainable inclusions indicative of polyphosphate (23) in part of this population strongly promote the first hypothesis for our system. However, the fixation procedure used might have led to the disappearance of polyphosphate granules (21), and therefore PAO, especially among gram-negative bacteria, might have been overlooked.
Denitrification cannot at all be assigned to a certain microbial population, nor is it restricted to a certain zone of the biofilm. During the anoxic period, the surface layers denitrify nitrate that has been accumulated during the oxic period of the preceding cycle (Fig. 1 and 2) and by that contribute about 20% to the total N loss. During the oxic period, the deeper parts of the biofilm remain anoxic and are provided with nitrate via nitrification (Fig. 1). The contribution of denitrification in the 300- to 600-μm-deep biofilm during the oxic period to total N loss is about 15%. Because there are reports of denitrifying PAO (28, 57) and because GPBHCG as potential PAO were found in close proximity to nitrifying bacteria, the source of nitrate, involvement of PAO in denitrification appears to be possible. However, this cannot be proven based upon our data.
Competition for oxygen.
Both nitrification and phosphate uptake require oxygen. Therefore, competition for oxygen between the respective microbial populations during the aeration period is to be expected. Microsensor data indicate that the nitrifying bacteria were oxygen limited. Oxygen penetration was low, whereas ammonium was present in excess during the whole process. The distribution of AOB and NOB in general corresponds to the oxygen penetration depth, supporting the findings of earlier studies (39, 50). The delayed onset of nitrification after the start of the aeration and the accumulation of nitrite despite the high abundance of NOB very likely reflect the limited supply of oxygen. Due to their high Km for oxygen, AOB and NOB are poor competitors compared to heterotrophic bacteria (17, 41, 56). Thus, during the initial aeration period, oxygen is taken up preferentially by heterotrophs as the PAO. As the phosphate uptake rate declines with the ongoing aeration period (data not shown), the stoichiometric oxygen demand of PAO metabolism decreases as well (51). Consequently, during the course of the aeration period, phosphate accumulation is most likely replaced by nitrification in terms of oxygen consumption.
Nitrification rates.
The handling of concentration data to estimate volumetric nitrate production rates (see Materials and Methods) is based on two assumptions: (i) low lateral heterogeneity of the concentration in the biofilm at a certain time and (ii) slow dynamics of the concentration changes due to activity compared to those due to diffusion (i.e., a pseudo-steady-state situation). Structural analysis showed a certain lateral inhomogeneity based mostly on the clustering of nitrifiers (Fig. 3B), but inhomogeneity is low in the horizontal direction, and the resolution of the measurements was not in the range of the cluster size. Therefore, the first assumption is not violated. For a similar system (36), it was shown that the time required to reach equilibrium was 1.8 min in a biofilm with a thickness of 500 μm. This is quite short compared to the overall cycle dynamics observed here. Nevertheless, the second assumption may introduce some error, especially in highly dynamic layers. The rates determined here should therefore be regarded as best estimates.
The rates of both estimated cell-specific in situ ammonia oxidation and nitrate production are on the order of magnitude of those calculated for aggregates of a nitrifying fluidized bed reactor based on microsensor measurements (48) and the ammonia oxidation rate estimated by a process mass balance of a sewage treatment plant (60). Unfortunately, the resolution of our measurements was too low to distinguish between the ammonia oxidation rate of the surface layer (with the mixed nitrifying community) and that of the deeper layer (with the N. oligotropha-like population). Also, the different activities of each of the individual AOB populations in the surface layer or of individual cells within a monospecies cluster cannot be resolved. Therefore, it has to be noted that the cell-specific ammonia oxidation rate calculated here represents an average value for a diverse assemblage of AOB consisting of at least three different populations.
Community structure of AOB.
The combined approach of FISH, 16S rDNA, and amoA analyses led to the unexpected but consistent finding that there were several different AOB populations occurring in close spatial vicinity. In contrast, previous studies in biofilms and activated sludge usually reported a single population dominating the system, e.g., populations related to N. europaea (39, 50), Nitrosococcus mobilis (22), or members of the genus Nitrosospira (39, 49). This raises the question of what mechanisms allow for the coexistence of the different AOB populations in the biofilm studied here.
N. communis-like AOB were found only within the first 100 μm at the biofilm surface and only in low abundance. Isolates of this lineage originally were obtained from soils (24) and were recently also detected in activated sludge and biofilm systems (42). Unfortunately, the lack of ecophysiological data about N. communis and its exclusive occurrence in the same zone with both members of the N. europaea- and N. oligotropha-like AOB leave the specific adaptation of N. communis-like AOB to our system unresolved.
The second population was identified as members of the N. europaea lineage, although these bacteria were not identical to N. europaea itself. The low number of amoA sequences analyzed allows for some hidden diversity, because several clones might be represented by one band in the gel retardation. The N. europaea-like population detected by FISH thus might be unrepresented by any amoA clone. Members of this phylogenetic cluster, like, e.g., N. europaea and N. eutropha, are typically isolated from activated sludge systems (24), and the maximum substrate conversion rates for N. europaea are high compared to those of other strains of AOB (41). Pure culture and chemostat experiments revealed low substrate affinities for N. europaea with Km (NH4+) values in the range of 0.4 to 7 mM (41) and 0.88 to 1.96 mM (29), respectively. The same is true for oxygen affinity, with Km (O2) values between 6.9 and 17.4 μM (29). While ammonium concentrations in the biofilm exceeded most of the reported Km values, oxygen limitation for N. europaea-like AOB is obvious. Between a depth of 100 and 200 μm, the oxygen concentration decreases from 33.8 ± 5.45 μM to 7.0 ± 1.01 μM (mean ± SD, n = 15). Consequently, N. europaea-like AOB virtually disappear within these layers (Fig. 4B). Here, the biofilm is dominated by N. oligotropha-like AOB with an abundance of 1 × 108 to 2 × 108 cells cm−3 down to a depth of 400 μm. Based on 16S rDNA sequence analysis, members of the N. oligotropha lineage (also referred to as Nitrosomonas cluster 6a [26, 54]) have recently been detected in freshwater and brackish environments (52, 53), terrestrial habitats (26, 54), and activated sludge (42, 55). This suggests high physiological versatility and ecological importance. Isolates of this lineage are sensitive to ammonium concentrations exceeding 10 to 60 mM (24, 53, 55), show low Km (NH4+ plus NH3) values, and possess urease activity (53). These features support adaptation of the N. oligotropha lineage to low substrate concentrations. Although no kinetic data with respect to oxygen are available, this might also imply high affinity towards oxygen. Lower Km (O2) values of the N. oligotropha-like AOB than the values reported for N. europaea-related AOB could be responsible for the outcompetition of the latter at the oxic-anoxic transition zone in the biofilm. At the biofilm surface, however, both populations were found to coexist in almost equal abundance. Assuming higher maximum substrate conversion rates for N. europaea-like AOB (41), they should be able to outcompete other AOB in this zone. For explanation of the co-occurrence, the dynamics of the system have to be taken into account (i.e., the metabolic activity of the populations might be separated in time). In the initial aeration period, N. oligotropha-like AOB in particular might be active, because, as suggested above, their higher oxygen affinity allows competition for oxygen with the highly active heterotrophic PAO. In the late oxic period, when oxygen uptake of the heterotrophic PAO decreases, N. europaea-like AOB could become more active. Their relatively high maximum substrate conversion rate might compensate for their time-limited activity and contribute to the successful establishment of this population in the biofilm. It has been shown for biofilm populations of AOB that the abundance is likely not to be affected by short starvation periods (i.e., a few days). Furthermore, recovery for both growth and activity of AOB from starvation is very rapid. This effect is likely to be coupled to high densities of AOB, as found in biofilms, and is probably due to cell-cell signaling (4). The high cell density and occurrence in dense clusters we found are in agreement with this hypothesis. Based on this strategy, it can be argued that even merely short-term activity would allow AOB to persist in the biofilm.
In addition, there are reports about an anoxic type of metabolism in N. europaea and N. eutropha when electron acceptors other than oxygen are used (6). The ability to survive or even thrive during anoxic conditions is, however, likely to differ among the three AOB populations. Therefore, the nonaeration period of the reactor cycle might be another factor to support a mixed community of AOB. However, alternative hypotheses explaining the coexistence are possible. Detection of AOB populations in situ does not prove their recent activity (60). It can be speculated that the ability to maintain the ribosome content during inactive periods might be stronger in the N. oligotropha lineage than in the N. europaea lineage. This would cause N. europaea-like AOB to become more rapidly undetectable by FISH in deeper layers of the biofilm compared to the results with N. oligotropha. Currently, we have no evidence in favor of this hypothesis, but future studies including the detection of local activity (e.g., by use of microautoradiography) (30) might help to support or reject this hypothesis. By amoA analysis, three more clones could be identified, related to Nitrosococcus mobilis (B3–5) and not closely affiliated with any isolated Nitrosomonas strain (B3–3). Because neither population could be identified by FISH, we assume the populations were either low in numbers or were in a dormant state.
NOB population.
Using different probes for NOB, the existence of a certain population of Nitrospira spp. could be proven, whereas Nitrobacter spp. were not detected. This is in agreement with several other culture-independent studies of engineered systems (10, 22, 39, 48, 49, 61). Analysis of the probe match pattern in the current data set by the software package ARB (Technical University Munich, Munich, Germany) showed that it fits a cluster of closely related sequences in the genus Nitrospira (strains with accession no. Y14636 to Y14643) retrieved from a nitrite-oxidizing sequencing batch reactor by Burrell et al. (10). The NOB population in our system is thus likely to be affiliated with this particular cluster of the genus Nitrospira. It might be speculated whether this cluster possesses common functional features leading to a competitive advantage under the periodically changing conditions typical for sequencing batch reactors. The cell densities of Nitrospira spp. found in this study are on the order of magnitude of those reported from a purely nitrifying fluidized bed reactor (48). However, the abundance is 1 order of magnitude higher than that of the AOB. The question remains of how such a high abundance is supported, when the substrate turnover is speculated to be low (48).
Conclusions.
Through the combination of microsensor measurements and molecular methods, it was possible to resolve the structure and activity of the nitrifying community in a complex, P-removing biofilm. Some first insights were obtained into the mechanism that allows for the coexistence of different populations of AOB in the same biofilm, i.e., separation of their distributions in space and separation of their activities in time. The design of probe Nmo218 specific for the N. oligotropha lineage will enable in situ detection and quantification of N. oligotropha-like AOB in future studies to collect more information about their natural abundance and ecology.
ACKNOWLEDGMENTS
This study was supported by the German Research Foundation (SFB 411, Project A1—Research Center for Fundamental Studies of Aerobic Biological Wastewater Treatment, Munich, Germany) and by the Max-Planck-Society.
We are indebted to Patrik Arnz for the maintenance of the reactor and Jakob Pernthaler for image analysis for melting temperature determination. Gabriele Eickert, Anja Eggers, and Vera Hübner are acknowledged for the preparation of oxygen microeletrodes, and Dirk de Beer and Olivier Pringault are acknowledged for valuable comments on the handling of microsensor data. Harold L. Drake is acknowledged for support.
REFERENCES
- 1.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnz, P., E. Arnold, and P. A. Wilderer. Enhanced biological phosphorus removal in a semi full-scale SBBR. Water Sci. Technol., in press. [PubMed]
- 4.Batchelor S E, Cooper M, Chhabra S R, Glover L A, Stewart G S A B, Williams P, Prosser J I. Cell density-regulated recovery of starved biofilm populations of ammonia-oxidizing bacteria. Appl Environ Microbiol. 1997;63:2281–2286. doi: 10.1128/aem.63.6.2281-2286.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bitton G. Wastewater microbiology. 2nd ed. New York, N.Y: Wiley-Liss; 1999. [Google Scholar]
- 6.Bock E, Schmidt I, Stüven R, Zart D. Nitrogen loss caused by denitrifying Nitrosomonas cells using ammonium or hydrogen as electron donors and nitrite as electron acceptor. Arch Microbiol. 1995;163:16–20. [Google Scholar]
- 7.Bond P L, Hugenholtz P, Keller J, Blackall L L. Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Appl Environ Microbiol. 1995;61:1910–1916. doi: 10.1128/aem.61.5.1910-1916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bond P L, Erhart R, Wagner M, Keller J, Blackall L L. Identification of some of the major groups of bacteria in efficient and nonefficient biological phosphorus removal activated sludge systems. Appl Environ Microbiol. 1999;65:4077–4084. doi: 10.1128/aem.65.9.4077-4084.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 10.Burrell P C, Keller J, Blackall L L. Microbiology of a nitrite-oxidizing bioreactor. Appl Environ Microbiol. 1998;64:1878–1883. doi: 10.1128/aem.64.5.1878-1883.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Characklis W G, Wilderer P A. Structure and function of biofilms. Chichester, United Kingdom: John Wiley & Sons; 1989. [Google Scholar]
- 12.Christensson M, Blackall L L, Welander T. Metabolic transformations and characterisation of the sludge community in an enhanced biological phosphorus removal system. Appl Microbiol Biotechnol. 1998;49:226–234. [Google Scholar]
- 13.Daims H, Brühl A, Amann R, Schleifer K-H, Wagner M. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol. 1999;22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- 14.Daims H, Nielsen P, Nielsen J L, Juretschko S, Wagner M. Novel Nitrospira-like bacteria as dominant nitrite-oxidizers in biofilms from wastewater treatment plants: diversity and in situ physiology. Water Sci Technol. 2000;41:85–90. [Google Scholar]
- 15.de Beer D, Schramm A, Santegoeds C M, Kühl M. A nitrite microsensor for profiling environmental biofilms. Appl Environ Microbiol. 1997;63:973–977. doi: 10.1128/aem.63.3.973-977.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erhart R. In situ Analyse mikrobieller Biozönosen in Abwasserreinigungsanlagen. Ph.D. thesis. Munich, Germany: Technical University Munich; 1997. [Google Scholar]
- 17.Focht D D, Verstraete W. Biochemical ecology of nitrification and denitrification. Adv Microb Ecol. 1977;1:135–214. [Google Scholar]
- 18.González-Martinez S, Wilderer P A. Phosphate removal in a biofilm reactor. Water Sci Technol. 1991;23:1405–1415. [Google Scholar]
- 19.Hanaki K, Wanatwin C, Ohgaki S. Effects of the activity of heterotrophs on nitrification in a suspended-growth reactor. Water Res. 1990;24:289–296. [Google Scholar]
- 20.Hartmann L. Historical development of wastewater treatment process. In: Rehm H-J, Reed G, Pühler A, Stadler P, editors. Biotechnology. 2nd ed. 11a. Weinheim, N.Y: Wiley VCH; 1999. pp. 5–16. [Google Scholar]
- 21.Hesselmann R P X, Werlen C, Hahn D J, van der Meer R, Zehnder A J B. Enrichment, phylogenetic analysis and detection of a bacterium that performs enhanced biological phosphate removal in activated sludge. Syst Appl Microbiol. 1999;22:454–465. doi: 10.1016/S0723-2020(99)80055-1. [DOI] [PubMed] [Google Scholar]
- 22.Juretschko S, Timmermann G, Schmid M, Schleifer K-H, Pommerening-Röser A, Koops H P, Wagner M. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl Environ Microbiol. 1998;64:3042–3051. doi: 10.1128/aem.64.8.3042-3051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawaharasaki M, Tanaka H, Kanagawa T, Nakamura K. In situ identification of polyphosphate-accumulating bacteria in activated sludge by dual staining with rRNA-targeted oligonucleotide probes and 4′, 6′-diamidino-2-phenylindole (DAPI) at a polyphosphate-probing concentration. Water Res. 1999;33:257–265. [Google Scholar]
- 24.Koops H P, Böttcher B, Möller U C, Pommerening-Röser A, Stehr G. Classification of eight new species of ammonia-oxidizing bacteria: Nitrosomonas communis sp. nov., Nitrosomonas ureae sp. nov., Nitrosomonas aestuarii sp. nov., Nitrosomonas marina sp. nov., Nitrosomonas nitrosa sp. nov., Nitrosomonas eutropha sp. nov., Nitrosomonas oligotropha sp. nov., Nitrosomonas halophila sp. nov. J Gen Microbiol. 1991;137:1689–1699. [Google Scholar]
- 25.Koops H-P, Möller U C. The lithotrophic ammonia-oxidizing bacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. A handbook of the biology of bacteria: ecophysiology, isolation, identification, applications. 2nd ed. Vol. 3. New York, N.Y: Springer; 1992. pp. 2625–2637. [Google Scholar]
- 26.Kowalchuk G A, Stienstra A W, Heilig H J, Stephen J R, Woldendorp J W. Molecular analysis of ammonia-oxidising bacteria in soil of successional grasslands of the Drentsche A (The Netherlands) FEMS Microbiol Ecol. 2000;31:207–215. doi: 10.1111/j.1574-6941.2000.tb00685.x. [DOI] [PubMed] [Google Scholar]
- 27.Kuba T, van Loosdrecht M C M, Heijnen J J. Phosphorus and nitrogen removal with minimal COD requirement by integration of denitrifying dephosphatation and nitrification in a two-sludge system. Water Res. 1996;30:1702–1710. [Google Scholar]
- 28.Kuba T, Wachtmeister A, van Loosdrecht M C M, Heijnen J J. Effect of nitrate on phosphorus release in biological phosphorus removal systems. Water Sci Technol. 1994;30:263–269. [Google Scholar]
- 29.Laanbroek H J, Bodelier P L E, Gerards S. Oxygen consumption kinetics of Nitrosomonas europaea and Nitrobacter hamburgensis grown in mixed continuous cultures at different oxygen concentrations. Arch Microbiol. 1994;161:156–162. [Google Scholar]
- 30.Lee N, Nielsen P H, Andreasen K H, Juretschko S, Nielsen J L, Schleifer K-H, Wagner M. Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl Environ Microbiol. 1999;65:1289–1297. doi: 10.1128/aem.65.3.1289-1297.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ludwig W, Strunk O, Klugbauer S, Klugbauer N, Weizenegger M, Neumaier J, Bachleitner M, Schleifer K-H. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis. 1998;19:554–568. doi: 10.1002/elps.1150190416. [DOI] [PubMed] [Google Scholar]
- 32.Maidak B L, Cole J R, Parker C T, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M T J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 34.Mino T, van Loosdrecht M C M, Heijnen J J. Microbiology and biochemistry of the enhanced biological phosphate removal process. Water Res. 1998;32:3193–3207. [Google Scholar]
- 35.Mobarry B K, Wagner M, Urbain V, Rittmann B E, Stahl D A. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl Environ Microbiol. 1996;62:2156–2162. doi: 10.1128/aem.62.6.2156-2162.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morgenroth E. Enhanced biological phosphorus removal in biofilm reactors. PhD thesis. Munich, Germany: Technical University Munich; 1998. [Google Scholar]
- 37.Neef A. Anwendung der in situ-Einzelzell-Identifizierung von Bakterien zur Populationsanalyse in komplexen mikrobiellen Biozönosen. Ph.D. thesis. Munich, Germany: Technical University Munich; 1997. [Google Scholar]
- 38.Nielsen L P, Christensen P B, Revsbech N P, Sørensen J. Denitrification and oxygen respiration in biofilms studied with a microsensor for nitrous oxide and oxygen. Microb Ecol. 1990;19:63–72. doi: 10.1007/BF02015054. [DOI] [PubMed] [Google Scholar]
- 39.Okabe S, Satoh H, Watanabe Y. In situ analysis of nitrifying biofilms as determined by in situ hybridization and the use of microelectrodes. Appl Environ Microbiol. 1999;65:3182–3191. doi: 10.1128/aem.65.7.3182-3191.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pommerening-Röser A, Rath G, Koops H P. Phylogenetic diversity within the genus Nitrosomonas. Syst Appl Microbiol. 1996;19:344–351. [Google Scholar]
- 41.Prosser J I. Autotrophic nitrification in bacteria. Adv Microb Physiol. 1989;30:125–181. doi: 10.1016/s0065-2911(08)60112-5. [DOI] [PubMed] [Google Scholar]
- 42.Purkhold U, Pommerening-Röser A, Juretschko S, Schmid M C, Koops H-P, Wagner M. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl Environ Microbiol. 2000;66:5368–5382. doi: 10.1128/aem.66.12.5368-5382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rath G. Entwicklung eines Nachweissystems zur in situ-Analyse nitrifizierender Bakterienpopulationen auf der Basis spezifischer 16S rRNA-Gensequenzen. Ph.D. thesis. Hamburg, Germany: University of Hamburg; 1996. [Google Scholar]
- 44.Ravenschlag K, Sahm K, Pernthaler J, Amann R. High bacterial diversity in permanently cold marine sediments. Appl Environ Microbiol. 1999;65:3982–3989. doi: 10.1128/aem.65.9.3982-3989.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Revsbech N P. An oxygen microelectrode with a guard cathode. Limnol Oceanogr. 1989;34:474–478. [Google Scholar]
- 46.Roller C, Wagner M, Amann R, Ludwig W, Schleifer K-H. In situ probing of Gram-positive bacteria with high DNA G+C content using 23S rRNA-targeted oligonucleotides. Microbiology. 1994;140:2849–2858. doi: 10.1099/00221287-140-10-2849. [DOI] [PubMed] [Google Scholar]
- 47.Schmid M, Twachtmann U, Klein M, Strous M, Juretschko S, Jetten M S M, Metzger J W, Schleifer K-H, Wagner M. Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst Appl Microbiol. 2000;23:93–106. doi: 10.1016/S0723-2020(00)80050-8. [DOI] [PubMed] [Google Scholar]
- 48.Schramm A, de Beer D, van den Heuvel J C, Ottengraf S, Amann R. Microscale distribution of populations and activities of Nitrosospira and Nitrospira spp. along a macroscale gradient in a nitrifying bioreactor: quantification by in situ hybridization and the use of microsensors. Appl Environ Microbiol. 1999;65:3690–3696. doi: 10.1128/aem.65.8.3690-3696.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schramm A, de Beer D, Wagner M, Amann R. Identification and activities in situ of Nitrosospira and Nitrospira spp. as dominant populations in a nitrifying fluidized bed reactor. Appl Environ Microbiol. 1998;64:3480–3485. doi: 10.1128/aem.64.9.3480-3485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schramm A, Larsen L H, Revsbech N P, Ramsing N B, Amann R, Schleifer K-H. Structure and function of a nitrifying biofilm as determined by in situ hybridization and the use of microelectrodes. Appl Environ Microbiol. 1996;62:4641–4647. doi: 10.1128/aem.62.12.4641-4647.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smolders G J F, van der Meij J, van Loosdrecht M C M, Heijnen J J. Stoichiometric model of the aerobic metabolism of the biological phosphorus removal process. Biotechnol Bioeng. 1994;44:837–848. doi: 10.1002/bit.260440709. [DOI] [PubMed] [Google Scholar]
- 52.Speksnijder A, Kowalchuk G, Roest K, Laanbroek H. Recovery of a Nitrosomonas-like 16S rDNA sequence group from freshwater habitats. Syst Appl Microbiol. 1998;21:321–330. doi: 10.1016/S0723-2020(98)80040-4. [DOI] [PubMed] [Google Scholar]
- 53.Stehr G, Böttcher B, Dittberner P, Rath G, Koops H P. The ammonia-oxidizing nitrifying population of the river Elbe estuary. FEMS Microbiol Ecol. 1995;17:177–186. [Google Scholar]
- 54.Stephen J R, McCaig A E, Smith Z, Prosser J I, Embley T M. Molecular diversity of soil and marine 16S rRNA gene sequences related to β-subgroup ammonia-oxidizing bacteria. Appl Environ Microbiol. 1996;62:4147–4154. doi: 10.1128/aem.62.11.4147-4154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suwa Y, Sumino T, Noto K. Phylogenetic relationships of activated sludge isolates of ammonia oxidizers with different sensitivities to ammonium sulfate. J Gen Appl Microbiol. 1997;43:373–379. doi: 10.2323/jgam.43.373. [DOI] [PubMed] [Google Scholar]
- 56.van Niel E W J, Robertson L A, Kuenen J G. A mathematical description of the behaviour of mixed chemostat cultures of an autotrophic nitrifier and a heterotrophic nitrifier/aerobic denitrifier; a comparison with experimental data. FEMS Microbiol Ecol. 1993;102:99–108. [Google Scholar]
- 57.Vlekke G J F M, Comeau Y, Oldham W K. Biological phosphate removal from wastewater with oxygen or nitrate in sequencing batch reactors. Environ Technol Lett. 1988;9:791–796. [Google Scholar]
- 58.Wagner M, Amann R, Kämpfer P, Assmus B, Hartmann A, Hutzler P, Springer N, Schleifer K-H. Identification and in situ detection of gram-negative filamentous bacteria in activated sludge. Syst Appl Microbiol. 1994;17:405–417. [Google Scholar]
- 59.Wagner M, Erhart R, Manz W, Amann R, Lemmer H, Wedi D, Schleifer K-H. Development of an rRNA-targeted oligonucleotide probe specific for the genus Acinetobacter and its application for in situ monitoring in activated sludge. Appl Environ Microbiol. 1994;60:792–800. doi: 10.1128/aem.60.3.792-800.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wagner M, Rath G, Amann R, Koops H P, Schleifer K-H. In situ identification of ammonia-oxidizing bacteria. Syst Appl Microbiol. 1995;18:251–264. [Google Scholar]
- 61.Wagner M, Rath G, Koops H P, Flood J, Amann R. In situ analysis of nitrifying bacteria in sewage treatment plants. Water Sci Technol. 1996;34:237–244. [Google Scholar]