Abstract
The bacterial loads of air, surfaces, and personnel in clean rooms are routinely monitored using a set of standard media. Bacteria that can grow on these media are a tiny fraction of the total numbers in any environment. A substantial proportion of bacteria long thought to be unculturable were recently shown to be oligophilic. Oligophile counts in clean rooms in our studies exceeded the standard plate counts by up to 2 orders of magnitude. They responded to disinfection routines in ways similar to the responses of conventional bacteria. We suggest that oligophiles are better tools than conventional bacteria for environmental monitoring in aseptic pharmaceutical production units.
Clean rooms are essential in aseptic pharmaceutical production. Monitoring microbial and particle counts is part of good manufacturing practices (2, 4, 12, 16, 19, 23). The prescribed protocols for monitoring involve the use of media such as soybean casein digest agar (SCDA) and incubation at 30 to 35°C for 4 days (3). It is well documented that the counts can vary by 10 to 30% depending upon the choice of medium and incubation conditions (6, 15, 24). The standards suggested are often too limited to allow a statistical test of significance (10). These problems make the bacteriological standards for clean rooms a matter for debate (2, 12).
A variety of molecular techniques have made it clear during the last decade that the bacteria present in any environment far outnumber those which grow on commonly used media and are far more diverse. The difference has been estimated to be up to 2 orders of magnitude (7, 11, 18, 21). While the diverse bacteria that fail to grow on conventional media have been termed “unculturable,” it has been shown recently (21) that a substantial portion, if not all, of them can be cultured by using a dilute but diverse nutrient medium. The recovery of organisms such as Escherichia coli from a viable nonculturable state is also known to be better in dilute media (8). Oligophile colonies on dilute media appear slowly and are often microscopic. Oligophilic bacteria have been shown to be abundant in a wide variety of habitats (1, 8, 9, 13, 14, 21, 22), and the initial impression that they are restricted to oligotrophic habitats is no longer seen as correct.
Oligophilic bacteria have also been isolated from clinical materials, although their role is not known (17, 20). A number of oligophilic bacteria exhibit antibiotic resistance, most of which is plasmid borne (25). Oligophiles can therefore be a potential pool of antibiotic resistance genes that can be acquired by pathogens through plasmid transfer. Because of their potential clinical importance, oligophilic bacteria merit attention. We report here the presence of oligophilic bacteria in considerable numbers in clean rooms where the counts on conventional media were zero. The results indicate that counts of oligophilic bacteria should be a part of the environmental-monitoring schedules for clean rooms. Since oligophiles are present in greater numbers than other bacteria, counting them would result in a reduced coefficient of variation that can make statistical comparisons more meaningful.
For comparison of conventional bacterial and oligophile counts, we used SCDA (casein enzymic hydrolysate, 1.5 g; papaic digest of soybean meal, 0.5 g; sodium chloride, 0.5 g; agar-agar, 1.5 g; and distilled water to make 100 ml [pH 7.3 ± 0.2]; sterilized by autoclaving at 121°C for 20 min) (3) for conventional bacterial counts and the Ravan medium (glucose, 5 mg; peptone, 5 mg; sodium acetate, 5 mg; sodium citrate, 5 mg; yeast extract, 5 mg; sodium pyruvate, 2 mg; agarose, 1%; and distilled water to make 100 ml [pH 7.0 ± 0.2]; sterilized by autoclaving at 121°C for 20 min) (21) for oligophile counts.
The clean rooms of a pharmaceutical production unit manufacturing bacterial vaccines were sampled for the study, using settle plate counts for air flora, swabs from a variety of surfaces in the working area, and finger dabs of the working personnel before and after routine disinfection procedures. For settle plates, SCDA and Ravan medium plates were exposed for 4 h simultaneously in triplicate in the clean room, on the laminar bench, in the corridor, and in the medium preparation room during working hours. Sampling in each place was repeated twice. The clean rooms were sampled before and after fumigation with formalin. For surface sampling, a 25-cm2 area of each surface was swabbed (5). The surfaces sampled included a laminar bench, a writing desk, a beef-cutting table, the medium preparation room floor, and the walls of the clean room during working hours. All the surfaces within the clean room were sampled before and after routine disinfection, which included Sterillium (ethyl-hexadecyl-dimethyammoniumethylsulfate in N and isopropanol; Bode Chemie, Hamburg, Germany) spray. All surfaces were sampled twice. The hands of two personnel were sampled before they entered the anteroom, thrice each before and after disinfection with Sterillium, Levermed (benzalkonium chloride, N, and isopropanol; Glaxo India Ltd.), or 70% isopropyl alcohol plus 1% benzalkonium chloride. The plantar surfaces of the feet of two persons were swabbed over a 25-cm2 area before and after they walked for two steps on the Dycem polymeric flooring, which retains particles by electrostatic attraction. The wheels of trolleys were swabbed over 25 cm2 of surface before and after they were rolled over the Dycem floor for four or five turns. The walls in the clean room were swabbed over 25 cm2 before and after they were rolled with a Dycem polymeric roller. These samplings were replicated five times independently. The Dycem surfaces used for the above-mentioned experiments were swabbed similarly before and after use and after being washed with 10% liquid soap containing 3.5% Aseptik (chlorhexidine gluconate, cetrimide, and isopropyl alcohol; ICI India Ltd.).
The SCDA plates were incubated at 30 to 35°C, and the visual colony counts were taken after 96 h. The Ravan medium plates were incubated at 20 to 22°C. Visual and microscopic colony counts were taken after 4, 7, 14, 21, and 28 days. As a large proportion of colonies developing on oligotrophic media were microscopic, colony counts were done microscopically. Under a stereoscopic microscope with a 4× objective, parallel strips of the field width were scanned for microscopic colonies. If the counts were more than 1,000 per plate, colonies in 15 fields in each plate were counted. The mean counts were multiplied by the ratio of the area of the plate to the area of the field to get the number of colonies per plate. In order to avoid edge effect, only the colonies whose centers were in the field were counted.
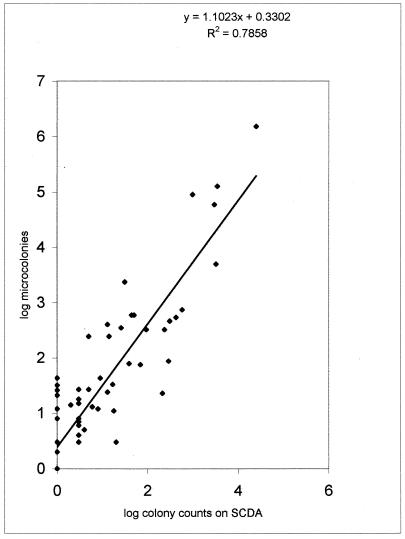
A comparison of plate counts on SCDA and dilute Ravan medium revealed that in surface sampling the oligophilic counts were greater than the conventional counts in 46 out of 66 pairs of plate counts (Fig. 1). In four pairs, the oligophile counts were less. This difference is highly significant (P < 0.001) in a table-wide Dixon and Mood sign test. The differences were individually significant by t test in 30 pairs, in 28 of which oligophile counts were significantly greater and in 2 of which copiophile counts were significantly greater. The difference was most marked for samples from surfaces that normally gather dust and are more likely to harbor surface growers. The soles of feet and the wheels of trolleys showed the maximum differences. Correlation between SCDA and Ravan medium counts has a significantly positive y intercept (Fig. 1), indicating that a negative report on SCDA is often accompanied by a positive count on oligotrophic medium.
FIG. 1.
Correlation between colony counts of copiophilic and oligophilic organisms on a double log plot. The slope is >1, and a positive intercept demonstrates the consistently higher counts of oligophiles.
The microcolony counts on Ravan medium increased with prolonged incubation, but the slopes declined after 21 days. Microcolonies are likely to go undetected, particularly if they are few. There is a possible negative bias, therefore, in the oligophile counts. In spite of the bias, microcolony counts on oligotrophic medium were consistently greater.
The differences between conventional and oligophilic bacterial counts were not significant for settle plates used for air flora. However, fungal colonies were observed only on Ravan medium, where they constituted 15 to 20% of the colonies, whereas no fungal colonies were observed on the SCDA medium after 96 h of incubation. Among bacterial colonies, the majority on SCDA comprised micrococci, staphylococci, and spore-forming gram-positive rods, whereas those on Ravan medium comprised gram-negative rods, gram-positive and -negative coccobacilli, and gram-positive non-spore-forming rods in addition to the bacteria mentioned above.
After the use of a disinfectant or a routine fumigation procedure, the counts of oligophilic organisms dropped rapidly, often reaching zero, as in the case of copiophiles (Table 1). Oligophilic organisms seemed to respond well to the disinfection procedures. However, on a number of occasions, a zero count on SCDA was accompanied by a positive count on the Ravan medium. Out of the 25 samples for which the counts on SCDA were zero, 12 showed growth on oligotrophic medium. A maximum count of 42 was recorded, and between 20 and 30 colonies were commonly seen. This is important, since generally a zero count on conventional medium is taken as satisfactory. The count differences between conventional and oligotrophic media indicate that a substantial number of viable organisms can still be present when the conventional plate counts fail to detect any.
TABLE 1.
Responses of oligophilic and copiophilic organisms to routine disinfection proceduresa
| Sample | Oligophile count
|
Copiophile count
|
||
|---|---|---|---|---|
| Before disinfection | After disinfection | Before disinfection | After disinfection | |
| Hand | 42 | 0 | 8 | 0 |
| Hand | 22 | 10 | 211 | 17 |
| Hand | 541 | 2 | 422 | 19 |
| Hand | 78 | 0 | 38 | 0 |
| Hand | 32 | 1 | 16 | 0 |
| Hand | 87 | 4 | 289 | 3 |
| Settle plate | 20 | 2 | 38 | 1 |
| Settle plate | 11 | 2 | 4 | 0 |
| Settle plate | 6 | 0 | 4 | 0 |
| Settle plate | 8 | 0 | 6 | 0 |
| Laminar table | 26 | 0 | 2 | 0 |
| Laminar table | 12 | 0 | 5 | 0 |
| Floor | 463 | 0 | 306 | 0 |
| Wall | 1 | 1 | 0 | 0 |
| Wall | 7 | 0 | 0 | 0 |
| Wall | 3 | 1 | 2 | 0 |
| Wall | 6 | 1 | 2 | 0 |
| Wall | 2 | 0 | 2 | 0 |
| Foot | 243 | 11 | 4 | 0 |
| Foot | 594 | 5 | 49 | 2 |
| Foot | 324 | 2 | 92 | 0 |
| Foot | 594 | 25 | 43 | 0 |
| Wheel | 2,349 | 14 | 30 | 2 |
Copiophile counts were taken on SCDA medium after 4 days, and oligophile counts were taken on Ravan medium after 28 days. The numbers are means of four plates approximated to the nearest integer. The coefficients of variation ranged between 10 and 20% except when the counts were <10.
The failure to get significant differences in counts in settle plates for air flora despite consistent and substantial differences in surface sampling suggests that oligophiles are not more efficient dispersers than copiophiles but are more efficient colonizers of surfaces. Since these organisms can grow on exceedingly low nutrient concentrations, they may grow on surfaces which hardly support the growth of copiophilic organisms. Since oligophiles respond well to disinfectants, conventional surface cleaning and disinfection protocols need not be modified. The efficiency of disinfection, however, can be better judged by monitoring oligophiles rather than copiophiles, owing to their greater initial numbers and detectable presence in samples for which copiophile counts are not recorded.
The permissible limits of bacterial counts for clean room standards are often too low. For example, according to the European Union's good manufacturing practice directive, the permissible number of CFU for surface contact plates for grade A is 1 and that for grade B is 5; the number permitted by USP for class 100 is 3, and that for class 10000 is 5 (4). Since chance differences and errors in counting bacteria by colony counts are large, the small permissible numbers make statistical inferences difficult (10). If the same samples are subjected to oligophile counts, substantially higher counts could be obtained, which would make statistical tests more meaningful. With oligophiles, it is possible to use the same recommended levels of sanitation as for conventional bacteria but with a statistically sounder testing protocol. Oligophile counts, therefore, would serve a useful purpose in the environmental monitoring of aseptic pharmaceutical production units. The drawback of oligophile counts is the greater incubation time required. They are thus not suitable for quick appraisals. For long-term monitoring and maintenance of sanitation, on the other hand, using counts of oligophilic bacteria in addition to the conventional methods would certainly prove useful.
REFERENCES
- 1.Akagi Y, Taga N, Simidu U. Isolation and distribution of oligotrophic marine bacteria. Can J Microbiol. 1977;23:981–987. doi: 10.1139/m77-146. [DOI] [PubMed] [Google Scholar]
- 2.Akers J E. Environmental monitoring and control: proposed standards, current practices, and future directions. J Pharm Sci Technol. 1997;51:36–47. [PubMed] [Google Scholar]
- 3.Akers M J. Parenteral quality control. 2nd ed. New York, N.Y: Marcel Dekker, Inc; 1994. [Google Scholar]
- 4.Anonymous. EU guide to good manufacturing practice; annex on the manufacture of sterile medicinal products. Association commentary. PDA J Pharm Sci Technol. 1996;50:138–143. [PubMed] [Google Scholar]
- 5.Anonymous. USP on microbiological evaluation of clean rooms and other controlled environments. Association commentary. PDA J Pharm Sci Technol. 1997;51:222–226. [PubMed] [Google Scholar]
- 6.Bathgate H, Lazzari D, Cameron H, McKay D. The incubation period in sterility testing. J Parenter Sci Technol. 1993;47:254–257. [PubMed] [Google Scholar]
- 7.Bintrim S B, Donohue T J, Handelsman J, Robert G P, Goodman R M. Molecular phylogeny of Archaea from soil. Proc Natl Acad Sci USA. 1997;94:277–282. doi: 10.1073/pnas.94.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bloomfield S F, Stewart G S A B, Dodd C E R, Booth I R, Power E G M. The viable but non-culturable phenomenon explained? Microbiology. 1998;144:1–3. doi: 10.1099/00221287-144-1-1. [DOI] [PubMed] [Google Scholar]
- 9.Button D K, Schut F, Quang P, Martin R, Robertson B R. Viability and isolation of marine bacteria by dilution culture: theory, procedures, and initial results. Appl Environ Microbiol. 1993;59:881–891. doi: 10.1128/aem.59.3.881-891.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cundell A M, Bean R, Massimore L, Maier C. Statistical analysis of environmental monitoring data: does a worst case time for monitoring clean rooms exist? J Pharm Sci Technol. 1998;52:326–330. [PubMed] [Google Scholar]
- 11.Fuhrman J A, Campbell L. Marine ecology: microbial microdiversity. Nature. 1998;393:410–411. [Google Scholar]
- 12.Hertroys R, Van Vught P A M, Van De Donk H J M. Moving towards a (microbiological) environmental monitoring programme that can be used to release aseptically produced pharmaceuticals: a hypothesis, a practical programme, and some results. J Pharm Sci Technol. 1997;51:52–59. [PubMed] [Google Scholar]
- 13.Kuznetsov S I, Dubinina G A, Lapteva N A. Biology of oligotrophic bacteria. Annu Rev Microbiol. 1979;33:377–387. doi: 10.1146/annurev.mi.33.100179.002113. [DOI] [PubMed] [Google Scholar]
- 14.Mallory L M, Austin B, Colwell R R. Numerical taxonomy and ecology of oligotrophic bacteria isolated from the estuarine environment. Can J Microbiol. 1977;23:733–750. doi: 10.1139/m77-110. [DOI] [PubMed] [Google Scholar]
- 15.Marshall V, Poulson-Cook S, Moldenhauer J. Comparative mold and yeast recovery analysis (the effect of differing incubation temperature ranges and growth media) J Pharm Sci Technol. 1998;52:165–169. [PubMed] [Google Scholar]
- 16.Schicht H H. New European GMP rules for manufacturing sterile medicinal products. Cleanrooms Int. 1998;2:17–28. [Google Scholar]
- 17.Tada Y, Ihmori M, Yamaguchi J. Oligotrophic bacteria isolated from clinical materials. J Clin Microbiol. 1995;33:493–494. doi: 10.1128/jcm.33.2.493-494.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torsvik V, Goksoyr J, Daae F L. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.U.S. General Services Administration. Federal standards 209E. Airborne particulate cleanliness classes in cleanrooms and clean zones. Washington, D.C.: General Services Administration; 1992. [Google Scholar]
- 20.Wainwright M, Barakah F, Turk I A, Ali T A. Oligotrophic micro-organisms in industry, medicine and the environment. Sci Prog. 1991;75:313–322. [PubMed] [Google Scholar]
- 21.Watve M G, Shejwal V, Sonawane C, Rahalkar M, Matapurkar A, Shouche Y, Patole M, Padnis N, Champhenkar A, Damle K, Karandikar S, Krhirsagar V, Jog M. The ‘K’ selected oligophilic bacteria: a key to uncultured diversity? Curr Sci. 2000;78:1535–1542. [Google Scholar]
- 22.Whang K, Hattori T. Oligotrophic bacteria from rendzina forest soil. Antonie Leeuwenhoek. 1988;54:19–36. doi: 10.1007/BF00393955. [DOI] [PubMed] [Google Scholar]
- 23.Whyte W. Cleanroom design. 2nd ed. Chichester, West Sussex, England: John Wiley & Sons Ltd.; 1999. pp. 35–39. [Google Scholar]
- 24.Wilson J D, Varney M. Sterility test incubation issue. J Parenter Sci Technol. 1995;49:157–159. [PubMed] [Google Scholar]
- 25.Zlatkin I V, Vishnevetskaia O, Nikitin D I. Some aspects of antibiotic resistance of oligotrophic bacteria. Antibiot Khimioter. 1991;36:34–37. [PubMed] [Google Scholar]