To the Editor—During the coronavirus-disease-2019 (COVID-19) pandemic, in-hospital surveillance via serial severe acute respiratory coronavirus virus 2 (SARS-CoV-2) testing has repeatedly demonstrated its utility in early detection, isolation, and interruption of nosocomial transmission. 1 However, analytical sensitivity of real-time, reverse-transcriptase, polymerase chain reaction (rRT-PCR) testing for SARS-CoV-2 is crucial in ensuring accurate results and early detection of COVID-19 cases and, thus, to mitigate nosocomial COVID-19 outbreaks. The emergence of SARS-CoV-2 variants has been demonstrated to negatively affect analytical sensitivity of rRT-PCR assays. For instance, failure of the N-gene assay has occurred with certain SARS-CoV-2 variants. 2 N-gene point mutations have been reported in the literature, resulting in cases of diagnostic escapes. 3–5 To date, however, no confirmed case of nosocomial transmission arising from N-gene dropout has been reported in the literature. We highlight here a case of nosocomial SARS-CoV-2 transmission arising from a case of N-gene dropout, which illustrates the importance of proper interpretation when discrepant results are encountered on different gene-targets utilized in SARS-CoV-2 diagnostic testing. This study was conducted as part of an outbreak investigation, and ethics approval was not required under our institutional review board guidelines.
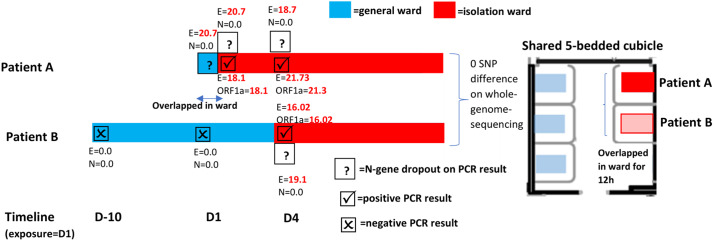
At our institution, a large tertiary-care hospital in Singapore, from June 21, 2021, onward, all inpatients were routinely tested for SARS-CoV-2 on admission via both rapid antigen detection (RAD) testing as well as PCR, as part of enhanced infection-prevention measures. 6,7 Routine SARS-CoV-2 PCR testing was conducted using the Cepheid GeneXpert Xpert Xpress assay (Cepheid, Sunnyvale, CA), a proprietary FDA-approved assay targeting both the E-gene and N-gene; while the BD-Veritor SARS-CoV-2 antigen rapid test kit (Becton Dickinson, Franklin Lakes, NJ) was used for SARS-CoV-2 RAD testing. Inpatients were subsequently tested at weekly intervals for SARS-CoV-2 infection via PCR. 7 Up to January 2022, 49,933 admissions were screened for SARS-CoV-2, with 3,155 (6.3%) of 49,933 testing positive. In January 2022, an asymptomatic female aged 74 years (patient A), doubly vaccinated with mRNA vaccines, was admitted to our institution. SARS-CoV-2 RAD testing on admission was negative; SARS-CoV-2 PCR on admission screening via GeneXpert-Xpert-Xpress returned a positive result on the E-gene gene target (cycle-threshold [Ct], 20.7) but a negative result on the N2 gene target. Because our institution had not encountered N-gene dropout cases among hospitalized inpatients prior to January 2022, the practice at that time was to await results of repeated SARS-CoV-2 PCR testing using a different assay (Roche cobas-6800, Roche Diagnostics, Indianapolis, IN) that utilized a separate set of gene targets (ORF-1a and E-gene regions). As such, the patient was not initially isolated until repreated testing revealed persistent positive results at a low Ct value for the E-gene target and repeated SARS-CoV-2 PCR using a different assay (ie, Roche cobas-6800) returned positive. The index patient was subsequently transferred to isolation. Also, 5 other patients in the shared cubicle were exposed. The fully vaccinated patient in the adjacent bed (patient B), with an exposure time of 12 hours, subsequently tested positive on day 4 after exposure (ie, day 14 of admission), with a similar pattern of N-gene dropout on testing (Fig. 1). Previously, our institution reported 2 cases of N-gene dropout arising from a point mutation in G29195T; however, these cases were submitted from an outpatient ambulatory clinic in October 2021 and did not overlap with this cluster of cases in space or time. 5
Fig. 1.
Nosocomial transmission arising from a case of N-gene diagnostic escape.
The nasopharyngeal swab samples of patient A and patient B were subjected to whole-genome sequence analysis. A point mutation, C29200T, was detected in the consensus sequences of both samples. G29200T is a synonymous mutation that was previously reported to cause N-gene detection failure. 4 Whole-genome similarity analysis was performed using previously published workflows utilizing the ARTIC protocol on Oxford Nanopore minION sequencers. 8 The SARS-CoV-2 genomes from patient A and patient B were identical (0 SNP difference). The identical genomes and epidemiological link support the hypothesis of nosocomial transmission. No additional cases of N-gene dropout were detected amongst inpatients or healthcare workers (HCWs) despite intensive surveillance including weekly, rostered, routine testing. 7 This result is likely due to enhanced infection prevention measures in-place during the study period, including universal N95 usage by HCWs and high vaccination-uptake rates (≥80%) among inpatients and HCWs. 9
The potential of nosocomial SARS-CoV-2 transmission from cases of N-gene dropout is not purely theoretical. Clinicians and diagnostic laboratories need to be vigilant for such cases because emergence of novel SARS-CoV-2 variants with the potential to escape existing diagnostic tests remains inevitable. Test interpretation is critical. According to the manufacturer instructions for the Cepheid GeneXpert-Xpert-Xpress test kit, cases with discrepant results (negative on the N2-gene gene target but positive on the E-gene gene-target) should have been reported as presumptive positive with a recommendation for repeated testing. 10 Had this interpretation been used, the patient would have been isolated upon admission and the additional period of transmission risk would not have occurred, despite the N-gene dropout. SARS-CoV-2 diagnostic assays should ideally be based on ≥2 gene targets, and healthcare institutions need to maintain a robust molecular surveillance system to detect emergent diagnostic escapes. Reliable diagnostic detection of SARS-CoV-2 is crucial for containment of COVID-19 and prevention of spread within healthcare facilities.
Acknowledgments
Financial support
This work was not grant funded. Funding for consumables was provided by the Singapore General Hospital. K.K.K.K. is supported by the Singapore National Medical Research Council Research Training Fellowship.
Conflicts of interest
The authors report no conflicts of interest relevant to this report.
References
- 1. Prabhu RM, Firestone MJ, Bergman K, et al. Use of serial testing to interrupt a SARS-CoV-2 outbreak on a hospital medical floor—Minnesota, October–December 2020. Infect Control Hosp Epidemiol 2022. doi: 10.1017/ice.2022.40. [DOI] [PMC free article] [PubMed]
- 2. Chen Y, Han Y, Yang J, Ma Y, Li J, Zhang R. Impact of SARS-CoV-2 variants on the analytical sensitivity of rRT-PCR assays. J Clin Microbiol 2022;60:e0237421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hasan MR, Sundararaju S, Manickam C, et al. A novel point mutation in the N gene of SARS-CoV-2 may affect the detection of the virus by reverse transcription-quantitative PCR. J Clin Microbiol 2021;59:e03278–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ziegler K, Steininger P, Ziegler R, Steinmann J, Korn K, Ensser A. SARS-CoV-2 samples may escape detection because of a single point mutation in the N gene. Euro Surveill 2020;25:2001650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ko KKK, Abdul Rahman NB, et al. SARS-CoV-2 N gene G29195T point mutation may affect diagnostic reverse transcription-PCR detection. Microbiol Spectr 2022;10:e0222321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wee LE, Conceicao EP, Sim JX, et al. Utilization of rapid antigen assays for detection of severe acute respiratory coronavirus virus 2 (SARS-CoV-2) in a low-incidence setting in emergency department triage: does risk-stratification still matter? Infect Control Hosp Epidemiol 2021. doi: 10.1017/ice.2021.407. [DOI] [PubMed]
- 7. Wee LEI, Conceicao EP, Aung MK, et al. Rostered routine testing for healthcare workers and universal inpatient screening: the role of expanded hospital surveillance during an outbreak of coronavirus disease 2019 (COVID-19) in the surrounding community. Infect Control Hosp Epidemiol 2021. doi: 10.1017/ice.2021.366. [DOI] [PMC free article] [PubMed]
- 8. Wee LE, Ko KK, Conceicao EP, et al. Linking sporadic hospital clusters during a community surge of the severe acute respiratory coronavirus virus 2 (SARS-CoV-2) B.1.617.2 delta variant: the utility of whole-genome sequencing. Infect Control Hosp Epidemiol 2022. doi: 10.1017/ice.2022.106. [DOI] [PMC free article] [PubMed]
- 9. Liang En W, Ko KK, Conceicao EP, et al. Enhanced infection-prevention measures including universal N95 usage and daily testing: the impact on SARS-CoV-2 transmission in cohorted hospital cubicles through successive delta and omicron waves. Clin Infect Dis 2022:ciac320. [DOI] [PMC free article] [PubMed]
- 10. Xpert Xpress SARS-CoV-2 test: package insert. Cepheid website. https://www.cepheid.com/Package%20Insert%20Files/Xpert%20Xpress%20SARS-CoV-2%20Assay%20ENGLISH%20Package%20Insert%20302-3787%20Rev.%20B.pdf. Published 2021. Accessed June 6, 2022.