Abstract
Previous work with Pseudomonas aeruginosa showed that catalase activity in biofilms was significantly reduced relative to that in planktonic cells. To better understand biofilm physiology, we examined possible explanations for the differential expression of catalase in cells cultured in these two different conditions. For maximal catalase activity, biofilm cells required significantly more iron (25 μM as FeCl3) in the medium, whereas planktonic cultures required no addition of iron. However, iron-stimulated catalase activity in biofilms was still only about one-third that in planktonic cells. Oxygen effects on catalase activity were also investigated. Nitrate-respiring planktonic cultures produced approximately twice as much catalase activity as aerobic cultures grown in the presence of nitrate; the nitrate stimulation effect could also be demonstrated in biofilms. Cultures fermenting arginine had reduced catalase levels; however, catalase repression was also observed in aerobic cultures grown in the presence of arginine. It was concluded that iron availability, but not oxygen availability, is a major factor affecting catalase expression in biofilms.
Microbial biofilms have been the subject of significant interest because of accumulating evidence suggesting that surface-associated growth and proliferation are normal and preferable for microorganisms. The implications of this simple observation are beginning to emerge, with the most important being that bacteria growing in biofilms are much more resistant to antimicrobial agents than are bacteria growing as planktonic cultures. Antibiotic killing models thus far developed have used planktonic cells (i.e., batch cultures) extensively but are likely invalid for biofilms. It has been suggested that the physiology of bacteria in a biofilm is different from that in planktonic cells (3), yet there are few defined examples and even fewer explanations for described differences.
We have initiated a series of studies focused on examining biofilm cell physiology and response to environmental stimuli; we are using elements of the oxidative stress response in this work. The oxidative stress response is well suited for these experiments because (i) the genes and enzymes involved are well understood (9, 10, 11, 12, 14), (ii) it is an important defense mechanism of recalcitrant bacterial biofilms in infections against the oxidative burst of phagocytic cells (16), and (iii) oxidative biocides are also used in industry to remove problematic biofilms from piping systems. Further, in Pseudomonas aeruginosa, key enzymes of the oxidative stress response, such as the major catalase, KatA, and a manganese-cofactored superoxide dismutase (Mn-SOD), are controlled by quorum sensing (16, 17); in addition, the activities of these enzymes are also known to be very sensitive to iron and phosphate availability (9, 12, 13, 14). Therefore, these enzymes and the genes that encode them represent effective models for assessing certain important aspects of bacterial physiology in the biofilm environment.
In recent work, we observed that catalase activity in P. aeruginosa biofilm cells was considerably lower than that in planktonic cells (16). The most obvious environmental factors that could conceivably influence catalase levels in biofilms and potentially account for this difference include iron and oxygen availability and H2O2 production. Iron limitation significantly reduces KatA activity (13), and it is possible that movement of a cationic micronutrient into a biofilm is limited by anionic carboxyl or hydroxyl groups on bacterial surface polysaccharides. We also considered oxygen availability as a possible explanation for the depressed levels of catalase in biofilms. Studies have shown that oxygen tension is reduced in bacterial biofilms (3, 21). We postulated that if oxygen respiration is reduced, endogenous generation of H2O2 also will be reduced, thus reducing the requirement for elevated catalase levels. In an attempt to better understand the basis for the differential expression of catalase in these two growth modes, we conducted a series of experiments to determine the extent to which catalase levels can be manipulated. In so doing, we also hoped to gain a better understanding of the physiological differences between biofilm and planktonic bacteria.
Iron effects on catalase activity.
Planktonic cultures and biofilms of P. aeruginosa wild-type strain PAO1 (18) were grown in tryptic soy broth (TSB) as previously described (16). Briefly, batch (planktonic) cultures were grown to stationary phase in 1/10-strength TSB and then assayed for catalase activity (which is comprised of only KatA under these conditions [2]) using previously described methods (15). Biofilms were cultured on stainless steel slides (1.3 by 7.6 cm) in a drip-flow reactor (5) using 1/100-strength TSB at a constant flow of 50 ml per h. To assess and compare the response of biofilm catalase activity to iron availability, the TSB medium for both planktonic cultures and biofilms was amended with various amounts of FeCl3. To impose iron starvation conditions (12, 13, 14), the iron-specific chelator 2,2-dipyridyl (500 μM for planktonic cultures and 50 μM for biofilm cultures) was added.
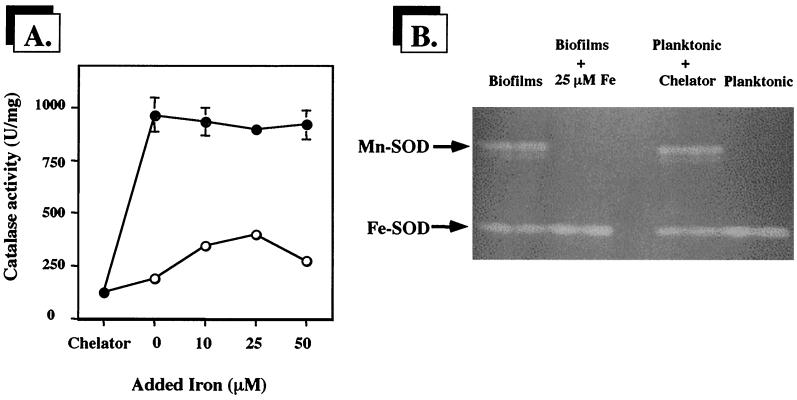
Under iron starvation conditions, catalase activities in planktonic cells and biofilm cells were essentially the same and were very low (approximately 125 U per mg of protein) (Fig. 1A). In contrast, when iron limitation was removed, catalase levels in planktonic cells were about 10-fold higher than those observed in biofilms, consistent with our previous observations (16). The inclusion of additional iron had no effect on catalase activity in planktonic cells. However, supplemental iron significantly and reproducibly increased catalase activity in biofilm cells. Catalase activity in biofilm cells increased approximately threefold in response to added iron but was still significantly lower than that in planktonic cells.
FIG. 1.
P. aeruginosa biofilms require more iron for maximal catalase activity. (A) Comparison of catalase activities in biofilms (○) and stationary-phase planktonic cultures (●) grown in the presence of the iron-specific chelator 2,2-dipyridyl, without added iron, or with increasing iron concentrations. Catalase activity is shown as units of activity per milligram of protein, where 1 U is defined as the amount which decomposes 1 μmol of H2O2 min−1. Error bars, where visible, represent ±1 standard error of the mean for three separate cultures. (B) SOD activity stain of planktonic and biofilm whole-cell proteins (50 μg per lane) resolved in a native gel. The locations of the isozymes Mn-SOD and Fe-SOD are designated to the left of the gel image. Total protein concentration was determined using a Bio-Rad protein assay kit with bovine serum albumin fraction V as a standard.
To confirm the iron-limited status of the planktonic and biofilm cells, we used SOD activity staining (15) of whole-cell proteins separated by native gel electrophoresis to assess the expression of Mn-SOD, which is strongly induced by iron starvation (12, 13, 14), and Fe-SOD, which is expressed constitutively but which is predictably not enhanced by iron starvation (12, 13, 14). As shown in Fig. 1B, biofilm cells expressed Mn-SOD when grown without added iron but not when the medium was amended with iron. Planktonic cells did not contain Mn-SOD unless 2,2-dipyridyl was included, suggesting they were not iron limited under normal growth conditions. Chemical analysis of 1/10-strength TSB medium showed an iron concentration of 1.5 μM. The iron content in the 1/100-strength TSB used for the biofilm experiments would be proportionately lower; however, the constant flow over the biofilm cells would continuously deliver fresh medium over the developing biofilm. Over the course of such experiments, the total iron made available to the biofilm would exceed that in batch conditions by at least eightfold. Regardless, in all subsequent experiments, 25 μM FeCl3 was included in the medium.
Effect of oxygen availability on catalase activity.
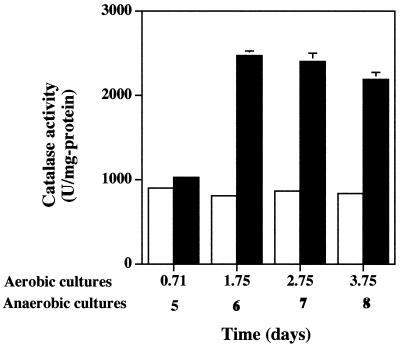
To test whether oxygen limitation might also partially account for the reduced catalase levels in P. aeruginosa biofilms, planktonic cells were grown anaerobically. P. aeruginosa is capable of anaerobic growth using nitrate as an electron acceptor or arginine for substrate-level phosphorylation (8). Strain PAO1 was cultured in sealed serum bottles containing medium that had been amended with 100 mM potassium nitrate and sparged with N2 gas prior to autoclaving. The medium also contained the oxygen indicator 0.1% resazurin to verify anaerobiosis. Cell growth was appreciably slower under these conditions, requiring 5 days to reach stationary phase (results not shown). However, when cells reached stationary phase, catalase activity was about twofold higher than that in aerobic cultures amended with nitrate and when measured at various times after stationary phase was reached (Fig. 2). The addition of nitrate had no effect on catalase activity in aerobic cultures (compare to Fig. 1A). Also, as with the iron effects shown in Fig. 1A, the increase in catalase activity was due to increased levels of KatA. In native gel analysis, the oxidative-stress-inducible catalases KatB and KatC were not evident (results not shown). The presence of nitrate had no effect on Mn-SOD expression, which was absent in the cells grown in these experiments (results not shown).
FIG. 2.
Catalase activity is increased in nitrate-respiring planktonic cultures of P. aeruginosa. Potassium nitrate (100 mM) was included in cultures incubated aerobically (open bars) and anaerobically (filled bars). Culture age is shown on the x axis; anaerobic cultures grew more slowly. Error bars, where visible, represent ±1 standard error of the mean for three separate cultures. Results are from one representative experiment of three independent experiments for each type of culture. Catalase activity is shown as units of activity per milligram of protein, where 1 U is defined as the amount which decomposes 1 μmol of H2O2 min−1.
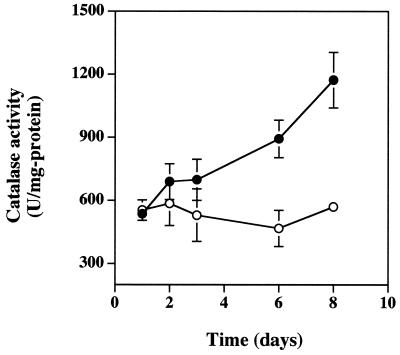
The effect of nitrate was also tested with P. aeruginosa biofilms. Catalase activity was significantly increased in mature biofilms that had been grown with 10 mM potassium nitrate (Fig. 3). The nitrate effect was not apparent in young biofilms, but as with the anaerobic NO3-respiring planktonic culture comparisons (Fig. 2), the presence of nitrate caused a doubling of catalase activity as the biofilms aged (Fig. 3).
FIG. 3.
Catalase activity in P. aeruginosa biofilms grown in the presence (●) or absence (○) of 10 mM nitrate. Catalase activity is shown as units of activity per milligram of protein, where 1 U is defined as the amount which decomposes 1 μmol of H2O2 min−1. Error bars, where visible, represent ±1 standard error of the mean for at least two biofilms at each time point.
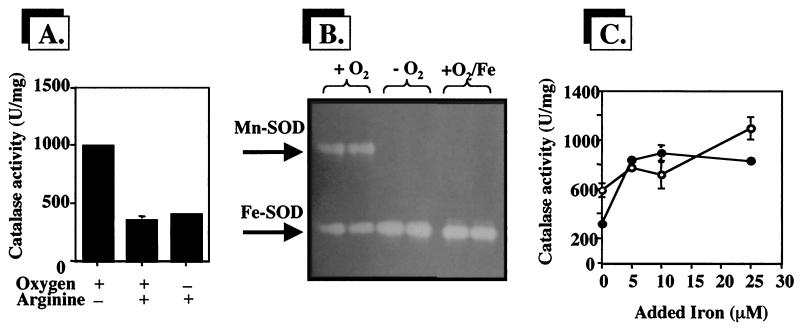
To assess the effects of anoxic substrate-level phosphorylation on catalase levels, planktonic cells were cultured anaerobically in 100 mM phosphate-buffered 1/10-strength TSB containing 40 mM arginine. Aerobic cultures were included as controls. Surprisingly, catalase levels were significantly depressed in both anaerobic and aerobic planktonic cultures (Fig. 4A). As the anaerobic metabolism of arginine results in increased culture pH (through the release of NH3), we tested catalase levels in aerobic cultures grown in 1/10-strength TSB buffered to pH 6 to pH 9 but observed no effect on catalase activity (results not shown). The possibility that arginine acts as an iron chelator and thus limits catalase activity was also tested by staining for SOD activity in native gels (Fig. 4B). Iron-starvation-inducible Mn-SOD was strongly expressed in cells growing aerobically in medium containing arginine, but the addition of 10 μM FeCl3 repressed the expression of this isozyme. Surprisingly, Mn-SOD was absent in anaerobic arginine-fermenting planktonic cultures (Fig. 4B). When iron was added to cells growing aerobically in TSB-arginine medium, catalase activity was restored to nearly normal levels (Fig. 4C).
FIG. 4.
Effect of arginine on catalase and SOD activities in P. aeruginosa. (A) Catalase activity in planktonic cultures grown aerobically with or without 40 mM arginine and in planktonic cultures fermenting arginine. Catalase activity is shown as units of activity per milligram of protein, where 1 U is defined as the amount which decomposes 1 μmol of H2O2 min−1. (B) SOD activity stain of a native gel used to resolve Mn-SOD and Fe-SOD activities (50 μg of total cell protein per lane). Oxygen and iron treatments are noted above each lane. (C) Catalase activity in aerobic (●) and anaerobic (○) planktonic cultures grown in the presence of 40 mM arginine and various concentrations of added iron. For all assays, the cell extract protein concentration was determined using a Bio-Rad protein assay kit with bovine serum albumin fraction V as a standard. In panels A and C, error bars, where visible, represent ±1 standard error of the mean for three replicate cultures. Results are from one representative experiment for each treatment. Each outcome was verified to be reproducible.
Conclusions.
By measuring two different oxidative stress enzymes that are affected in different ways by iron availability, we found that P. aeruginosa biofilms exhibit behavior that is consistent with an iron starvation response. Biofilms expressed high levels of Mn-SOD (Fig. 1B), an enzyme that is strongly stimulated by iron limitation (12, 13, 14). However, when iron was added to the medium, Mn-SOD was not expressed and biofilm catalase levels increased (Fig. 1A). These two observations are consistent with each other and suggest either that, relative to planktonic cells, biofilm cells have a higher iron requirement or that iron movement within the biofilm is somehow constricted, thus limiting iron bioavailability. Previous studies provided evidence that bacteria colonizing cystic fibrosis lung tissue grow under iron-limited conditions (1, 6, 7). Further experiments aimed at understanding in situ conditions and gene expression, particularly as they are related to P. aeruginosa infections, should offer significant opportunities to improve the understanding of diseases caused by this organism. Further, determining the physiological differences between biofilm cells and planktonic cells will likely be critical in the understanding of and the eventual development of treatments for P. aeruginosa biofilm infections like those found in cystic fibrosis lung tissue.
We found no evidence that depressed catalase activity in biofilms is due to oxygen limitation per se. Catalase levels in planktonic cells were actually significantly increased under anaerobic nitrate-respiring conditions (Fig. 2). The same effect was also apparent in biofilms grown with nitrate-amended medium. The latter observation is consistent with previous studies showing the oxygen-limited nature of the biofilm interior (3) and the use of nitrate as an alternative electron acceptor for cells in a biofilm. Interestingly, catalase activity was significantly depressed under either aerobic and anaerobic culture conditions when arginine was added to the medium (Fig. 4A). The exact role of arginine (or products of arginine metabolism, e.g., ornithine) in catalase control is not clear at this time, but it is likely linked somehow with iron availability, as the addition of iron to the medium reversed the arginine effect (Fig. 4C). This result is also in agreement with the presence of iron-starvation-inducible Mn-SOD in aerobic cultures (Fig. 4B) but not under anaerobic conditions, where Mn-SOD was absent. The latter observation is perhaps an important clue about how sodA (the gene coding for Mn-SOD) may be regulated while P. aeruginosa is involved in anaerobic arginine metabolism. Under anaerobic culture conditions, P. aeruginosa metabolizes arginine via the arginine deiminase pathway (20), with arginine being stoichiometrically converted to ornithine (4). Ornithine derivatives are common features of microbial iron chelates (19); thus, it is possible that subsequent metabolic alterations of the resulting ornithine contribute to the synthesis of an iron-binding metabolite. Work aimed at further understanding sodA expression under anaerobic arginine-fermenting conditions is currently under way.
Acknowledgments
We thank Irwin Fridovich for insightful discussions.
J.R.F. was supported by an undergraduate research fellowship from the National Science Foundation Center for Biofilm Engineering (EEC-8907039), Montana State University. This work was also supported by National Institutes of Health grant AI-40541 to D.J.H.
REFERENCES
- 1.Brown R W M, Anwar H, Lambert P A. Evidence that mucoid Pseudomonas aeruginosa in the cystic fibrosis lung grows under iron-restricted conditions. FEMS Microbiol Lett. 1984;21:113–117. [Google Scholar]
- 2.Brown S M, Howell M L, Vasil M L, Anderson A, Hassett D J. Cloning and characterization of the katB gene of Pseudomonas aeruginosa encoding a hydrogen peroxide-inducible catalase: purification of KatB, cellular localization, and demonstration that it is essential for optimal resistance to hydrogen peroxide. J Bacteriol. 1995;177:6536–6544. doi: 10.1128/jb.177.22.6536-6544.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costerton J W, Lewandowski Z, Caldwell D E, Korbe D R, Lappin-Scott H M. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 4.Cunin R, Glansdorff N, Pierard A, Stalon V. Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev. 1986;50:314–352. doi: 10.1128/mr.50.3.314-352.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elkins J G, Hassett D J, Stewart P S, Schweizer H P, McDermott T R. Protective role of catalase in Pseudomonas aeruginosa biofilm resistance to hydrogen peroxide. Appl Environ Microbiol. 1999;65:4594–4600. doi: 10.1128/aem.65.10.4594-4600.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emery T. Iron metabolism in humans and plants. Am Sci. 1982;70:626–631. [PubMed] [Google Scholar]
- 7.Haas B, Kraut J, Marks J, Zanker S C, Castigenetti D. Siderophore presence in sputa of cystic fibrosis patients. Infect Immun. 1991;59:3997–4000. doi: 10.1128/iai.59.11.3997-4000.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hassett D J. Anaerobic production of alginate by Pseudomonas aeruginosa: alginate restricts the diffusion of oxygen. J Bacteriol. 1996;178:7322–7325. doi: 10.1128/jb.178.24.7322-7325.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hassett D J, Charniga L, Bean K A, Ohman D E, Cohen M S. Antioxidant defense mechanisms in Pseudomonas aeruginosa: resistance to the redox-active antibiotic pyocyanin and demonstration of a manganese-cofactored superoxide dismutase. Infect Immun. 1992;60:328–336. doi: 10.1128/iai.60.2.328-336.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hassett D J, Woodruff W A, Wozniak D J, Vasil M L, Cohen M S, Ohman D E. Cloning and characterization of the Pseudomonas aeruginosa sodA and sodB genes encoding manganese- and iron-cofactored superoxide dismutase: demonstration of increased manganese superoxide dismustase activity in alginate-producing bacteria. J Bacteriol. 1993;175:7658–7665. doi: 10.1128/jb.175.23.7658-7665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hassett D J, Schweizer H P, Ohman D E. Pseudomonas aeruginosa sodA and sodB mutants defective in manganese- and iron-cofactored superoxide dismutase activity demonstrate the importance of the iron-cofactored form in aerobic metabolism. J Bacteriol. 1995;177:6330–6337. doi: 10.1128/jb.177.22.6330-6337.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hassett D J, Sokol P, Howell M L, Ma J-F, Schweizer H P, Ochsner U, Vasil M L. Ferric uptake regulator (Fur) mutants of Pseudomonas aeruginosa demonstrate defective siderophore-mediated iron uptake and altered aerobic metabolism. J Bacteriol. 1996;178:3996–4003. doi: 10.1128/jb.178.14.3996-4003.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hassett D J, Howell M L, Sokol P A, Vasil M L, Dean G E. Fumarase C activity is elevated in response to iron deprivation and in mucoid, alginate-producing Pseudomonas aeruginosa: cloning and characterization of fumC and purification of the native FumC. J Bacteriol. 1997;179:1442–1450. doi: 10.1128/jb.179.5.1442-1451.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hassett D J, Howell M L, Ochsner U, Johnson Z, Vasil M, Dean G E. An operon containing fumC and sodA encoding fumarase C and manganese superoxide dismutase is controlled by the ferric uptake regulator (Fur) in Pseudomonas aeruginosa: fur mutants produce elevated alginate levels. J Bacteriol. 1997;179:1452–1459. doi: 10.1128/jb.179.5.1452-1459.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassett D J, Elkins J G, Ma J F, McDermott T R. Pseudomonas aeruginosa biofilm sensitivity to biocides: use of hydrogen peroxide as model antimicrobial agent for examining resistance mechanisms. Methods Enzymol. 1999;310:599–608. doi: 10.1016/s0076-6879(99)10046-6. [DOI] [PubMed] [Google Scholar]
- 16.Hassett D J, Ma J F, Elkins J G, McDermott T R, Ochsner U A, West S E H, Huang C-T, Fredericks J, Burnett S, Stewart P S, McFeters G, Passador L, Iglewski B H. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol Microbiol. 1999;34:1082–1093. doi: 10.1046/j.1365-2958.1999.01672.x. [DOI] [PubMed] [Google Scholar]
- 17.Hassett, D. J., U. A. Ochsner, T. de Kievit, B. H. Iglewski, L. Passador, T. S. Livinghouse, J. A. Whitsett, and T. R. McDermott. Genetics of quorum sensing circuitry in Pseudomonas aeruginosa: implications for control of pathogenesis, biofilm formation and antibiotic/biocide resistance. In U. Streips and R. Yasbin (ed.), Modern molecular genetics. Wiley-Liss, New York, N.Y., in press.
- 18.Holloway B W. Genetics of Pseudomonas. Bacteriol Rev. 1969;33:419–443. doi: 10.1128/br.33.3.419-443.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matzanke B F. Structures, coordination chemistry and functions of microbial iron chelates. In: Winkelmann G, editor. CRC handbook of microbial iron chelates. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 15–64. [Google Scholar]
- 20.Mercenier A, Simon J-P, Vander Wauven C, Haas D, Stalon V. Regulation of enzyme synthesis in the arginine deiminase pathway of Pseudomonas aeruginosa. J Bacteriol. 1980;144:159–163. doi: 10.1128/jb.144.1.159-163.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu K D, Stewart P S, Xia F, Huang C-T, McFeters G A. Physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl Environ Microbiol. 1998;64:4035–4039. doi: 10.1128/aem.64.10.4035-4039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]