Abstract
The biosynthetic gene clusters of the staphylococcal lantibiotics epidermin and gallidermin are distinguished by the presence of the unique genes epiH and gdmH, respectively. They encode accessory factors for the ATP-binding cassette transporters that mediate secretion of the antimicrobial peptides. Here, we show that gdmH also contributes to immunity to gallidermin but not to nisin. gdmH alone affected susceptibility to gallidermin only moderately, but it led to a multiplication of the immunity level mediated by the FEG immunity genes when cloned together with the gdmT gene, suggesting a synergistic activity of the H and FEG systems. gdmH-related genes were identified in the genomes of several bacteria, indicating an involvement in further cellular functions.
Many gram-positive bacteria produce antimicrobial peptides (bacteriocins) active against other gram-positive strains. The activity of type A lantibiotics is based on pore formation in the cytoplasmic membrane. They contain unusual thioether bridges that arise from the posttranslational modification of cysteine and serine or threonine residues (for reviews, see references 2, 3, 5, and 20). The gene clusters of lantibiotics, such as nisin (produced by Lactococcus lactis), subtilin (produced by Bacillus subtilis), epidermin, and Pep5 (both produced by Staphylococcus epidermidis) comprise very similar sets of genes; the roles of most of them have been elucidated. We have previously characterized the epidermin genes for peptide synthesis (epiA) (21), maturation (epiB, -C, and -D) (9, 10, 16), processing (epiP) (4), and regulation (epiQ) (14) in the heterologous cloning host Staphylococcus carnosus. The genes epiF, -E, and -G encode the subunits of an ATP-binding cassette ABC exporter that confers on the producer immunity to epidermin by expelling the antimicrobial peptides from the cytoplasmic membrane (12, 15).
The epidermin transporter gene epiT has been shown to be defective, since it is disrupted by a deletion causing a frameshift. The gdmT gene of the closely related lantibiotic gallidermin from Staphylococcus gallinarum, however, is intact and mediated a considerable increase of epidermin production when cloned in epidermin-producing strains. This effect was dependent on the presence of the adjacent gene, gdmH, indicating that GdmH acts as an accessory factor for the ATP-binding cassette transporter GdmT (18). gdmH encodes a membrane protein without similarity to proteins of known function. Homologous genes are lacking in all lantibiotic gene clusters except those of the epidermin and gallidermin determinants.
GdmH contributes to producer immunity.
The sensitivities to gallidermin of S. carnosus strains bearing gdmH alone or in combination with other members of the gene clusters (Fig. 1) were analyzed. Since only part of the gdmFEG operon was available, the epiFEG operon was used. The Epi and Gdm proteins have a high degree of identity, and several genes of the two gene clusters have been shown to be interchangeable (11, 18). In order to combine functional FEG and HT genes on one plasmid, the disrupted epiT was replaced with the intact gdmT by fusing a DNA fragment including epiG and the 3′ end of epiE via a conserved BalI site with a fragment including the 5′ end of gdmE and the entire gdmF, gdmH, and gdmT genes, as illustrated in Fig. 1. The resulting plasmid, pRB-FEGHT, conferred resistance to gallidermin (Fig. 2A), demonstrating that the in-frame fusion of epiE and gdmE had led to a functional hybrid immunity system. Efficient expression of the three transcription units (FEG, H, and T) from the epi and gdm gene clusters has been shown to be dependent on the presence of the transcriptional activator EpiQ (15, 18). Therefore, the various S. carnosus strains bearing combinations of epi and gdm genes on plasmid pRB473 were transformed with the compatible plasmid pTepiQ10, encoding epiQ (14).
FIG. 1.
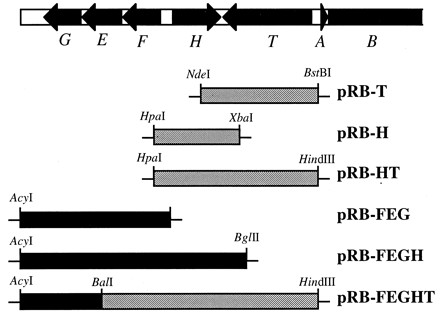
Organization of the epidermin and gallidermin gene clusters and plasmids used in this study. The conserved structure of the gallidermin and epidermin gene clusters is shown at the top. The shaded and solid bars below represent gallidermin (gdm) and epidermin (epi) sequences, respectively. pRB-FEGHT was constructed by ligating a PCR-derived DNA fragment from the gallidermin gene cluster to BalI-HindIII-digested DNA from pRB-FEG (15). The HindIII site in the PCR fragment was generated at the start codon of gdmA by modification of the primer used for PCR. The epiE and gdmE genes in the resulting plasmid were fused in frame at the conserved BalI site. pRB-T was constructed by cloning a Klenow enzyme-treated NdeI/BstBI fragment from pRB-FEGHT into SmaI-digested pRB473 (15) DNA. pRB-HT was derived from pRB-FEGHT by deleting a SmaI/HpaI fragment containing the FEG genes. Plasmid pRB-FEGH was constructed by ligating a Klenow enzyme-treated AcyI/BglII fragment from the epidermin gene cluster to SmaI-digested pRB473 DNA. The plasmids pRB-H and pRB-FEG are identical to the previously described plasmids pRBgdmH and pRBepiFEG, respectively (15, 18). Recombinant DNA techniques were used according to standard protocols (1).
FIG. 2.
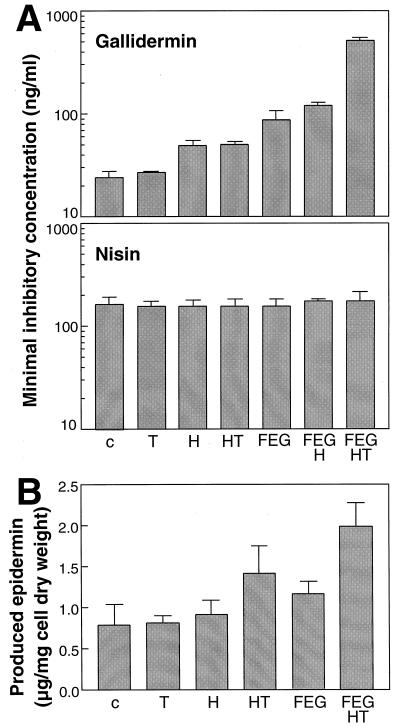
Influences of epidermin and gallidermin genes on immunity to gallidermin and nisin (A) and on epidermin production (B) in S. carnosus TM300. The epidermin or gallidermin genes expressed in the various S. carnosus strains (Fig. 1) are indicated as uppercase letters. c stands for the empty control plasmid pRB473. The various S. carnosus strains in panel B contained a second plasmid (pTepi14) bearing the genes necessary for peptide synthesis, maturation, and regulation. The means and standard deviations of at least four independent experiments are shown. The FEG and FEGH values (MIC of gallidermin) (A) and the c and FEG values (epidermin production) (B), respectively, are significantly different (P < 0.05), as calculated by Student's t test.
The MICs of gallidermin and nisin were determined by a serial dilution assay as described previously (17, 19) except that the cultures were incubated for 20 h. Plasmid pRB-H, encoding only gdmH, conferred a twofold increase in the MIC of gallidermin (Fig. 2A), indicating that gdmH also plays a role in immunity. Similar results were obtained with a plasmid bearing epiH (data not shown). In contrast to its role in secretion, the immunity conferred by gdmH was independent of the presence of gdmT, since plasmid pRB-HT conferred the same degree of immunity as pRB-H. Interestingly, plasmid pRB-FEGHT conferred a much higher level of immunity (sixfold) than pRB-FEG, indicating that GdmH acts synergistically with the FEG exporter. The immunity mediated by pRB-FEGH, however, corresponds only to an addition of the protective activities of the FEG and H determinants. Thus, though gdmT does not directly contribute to producer immunity, its presence appears to be necessary for the two systems to cooperate. The S. carnosus strain bearing pRB-FEGHT is still more than twice as susceptible to gallidermin as the natural epidermin and gallidermin producers (15; M. Hille, unpublished results). The reasons for this difference may include higher expression of the genes in the natural hosts or differences in the cell envelopes contributing to increased immunity. For example, the compositions of teichoic acids, which have a profound influence on susceptibility to lantibiotics (17), are different in S. carnosus, S. epidermidis, and S. gallinarum (7). Neither pRB-H, pRB-FEGHT, nor any of the other plasmids conferred immunity to nisin (Fig. 2A), which supports the results of previous studies showing the high specificity of the FEG systems (12) and suggests that the H proteins are similarly specific for gallidermin and epidermin. Thus, like the nisin and subtilin gene clusters, the epidermin and gallidermin determinants contain a second specific immunity system. In contrast to the H proteins, however, NisI and SpaI are lipoproteins, and there is no evidence that they are involved in the secretion of nisin and subtilin, respectively (6, 8). Some bacteriocin or lantibiotic immunity genes encode integral membrane proteins, such as PepI from the Pep5-producing strain (13). Though GdmH and EpiH are much larger and share no sequence similarity with PepI, they may have a related mode of action.
Combination of FEG and HT leads to increased epidermin production.
The amount of epidermin produced by S. carnosus strains containing plasmid pTepi14 with all of the genes for synthesis and maturation of epidermin and one of the plasmids described in Fig. 1 was determined. Tubes containing 5 ml of BM broth (1% tryptone, 0.5% yeast extract, 0.5% NaCl, 0.1% K2HPO4, 0.1% glucose) were inoculated with 1/100 volumes of precultures, which had been adjusted to the same cell density and cultivated for 20 h at 37°C. One-milliliter aliquots of the supernatants were lyophilized and dissolved in 50 to 100 μl of 10% ethanol. Filter disks loaded with the samples and subsequently dried were placed on BM agar plates containing the indicator strain S. carnosus(pT181mcs/pRB473) (18). Three samples from each culture were tested together with several filter disks containing gallidermin standards. The concentration of epidermin in the supernatants was calculated by comparing the inhibition zones from the samples and gallidermin standards.
S. carnosus(pTepi14/pRB473) produces small amounts of epidermin, though pTepi14 lacks any dedicated secretion genes. As demonstrated previously, gdmH and gdmT have only a positive influence on epidermin production when they are combined (18). When the FEG and HT genes were cloned together in S. carnosus(pTepi14/pRB-FEGHT), the specific production was considerably augmented. This increase is in agreement with earlier data showing a positive influence of epiFEG on the production of epidermin (15). It may indicate that the level of immunity limits the level of production or that the FEG exporter contributes to some extent to the secretion of epidermin. S. carnosus(pRB-FEGHT/pTepi14) produces much less epidermin than the natural producer does (Hille, unpublished). Again, different expression of the involved genes or S. epidermidis host factors lacking in S. carnosus may be responsible for this discrepancy.
gdmH-related genes occur in many bacteria.
While gdmH-related genes are lacking in most other lantibiotic gene clusters, we found hypothetical genes with significant homology in the genomes of Lactococcus lactis, Streptococcus mutans, Staphylococcus aureus, Enterococcus faecalis (bearing two corresponding genes) (Fig. 3), Neisseria meningitidis, Neisseria gonorrhoeae, Bacillus anthracis, Clostridium difficile, Clostridium acetobutylicum, Streptococcus equi, Aquifex aeolicus, and Thermotoga maritima (data not shown). Interestingly, a gdmH homologue was also identified in a putative antibiotic synthesis gene cluster of the ansamitocin-producing actinomycete Actinosynema pretiosum (Fig. 3). The adjacent genes were different in the various organisms. All GdmH-related proteins have similar hydrophobicity profiles (data not shown); the central hydrophilic domain reveals the highest degree of identity. The H proteins thus appear to be involved in several microbial processes.
FIG. 3.

Alignment of GdmH and EpiH with related proteins from various bacteria. GdmH from S. gallinarum (S.g.), EpiH from S. epidermidis (S.e.), and the related proteins from E. faecalis (E.f.1 and E.f.2), S. mutans (S.m.), L. lactis (L.1.), A. pretiosum (A.p.a.), and S. aureus (S.a.) were compared. The sequences are available under accession numbers U61158 (S.g.), U77778 (S.e.), X99710 (L.1.), U33059 (A.p.a.), and AE002359.1 (N. meningitidis) or the preliminary assignments TIGR1351|gef6162 (E.f.1), TIGR1351|gef6192 (E.f.2), UOKNOR1309 Contig871 (S.m.), or TIGR1280 2|S. aureus 2234 (S.a.). Identical or similar amino acids are highlighted by solid or shaded boxes, respectively. The deduced consensus sequence is given below.
Acknowledgments
We thank Ralph W. Jack for helpful discussions and Vera Augsburger for technical assistance.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 323).
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1990. [Google Scholar]
- 2.Bierbaum G, Götz F, Peschel A, Kupke T, van de Kamp M, Sahl H-G. The biosynthesis of the lantibiotics epidermin, gallidermin, pep5 and epilancin K7. Antonie Leeuwenhoek. 1996;69:119–127. doi: 10.1007/BF00399417. [DOI] [PubMed] [Google Scholar]
- 3.de Vos W M, Kuipers O P, van der Meer J R, Siezen R J. Maturation pathway of nisin and other lantibiotics: post-translationally modified antimicrobial peptides exported by Gram-positive bacteria. Mol Microbiol. 1995;17:427–437. doi: 10.1111/j.1365-2958.1995.mmi_17030427.x. [DOI] [PubMed] [Google Scholar]
- 4.Geißler S, Götz F, Kupke T. Serine protease EpiP from Staphylococcus epidermidis catalyzes the processing of the epidermin precursor peptide. J Bacteriol. 1996;178:284–288. doi: 10.1128/jb.178.1.284-288.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jack R W, Bierbaum G, Sahl H-G. Lantibiotics and related peptides. Berlin, Germany: Springer-Verlag; 1998. [Google Scholar]
- 6.Klein C, Entian K D. Genes involved in self-protection against the lantibiotic subtilin produced by Bacillus subtilis ATCC 6633. Appl Environ Microbiol. 1994;60:2793–2801. doi: 10.1128/aem.60.8.2793-2801.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kloos W E, Schleifer K-H, Götz F. The genus Staphylococcus. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The procaryotes. New York, N.Y: Springer-Verlag; 1992. pp. 1369–1420. [Google Scholar]
- 8.Kuipers O P, Beerthuyzen M M, Siezen R J, de Vos W M. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Eur J Biochem. 1993;216:281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
- 9.Kupke T, Götz F. Expression, purification, and characterization of EpiC, an enzyme involved in the biosynthesis of the lantibiotic epidermin, and sequence analysis of Staphylococcus epidermidis epiC mutants. J Bacteriol. 1996;178:1335–1340. doi: 10.1128/jb.178.5.1335-1340.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kupke T, Kempter C, Jung G, Götz F. Oxidative decarboxylation of peptides catalyzed by flavoprotein EpiD: determination of substrate specificity using peptide libraries and neutral loss mass spectrometry. J Biol Chem. 1995;270:11282–11289. doi: 10.1074/jbc.270.19.11282. [DOI] [PubMed] [Google Scholar]
- 11.Ottenwälder B, Kupke T, Brecht S, Gnau V, Metzger J, Jung G, Götz F. Isolation and characterization of genetically engineered gallidermin and epidermin analogs. Appl Environ Microbiol. 1995;61:3894–3903. doi: 10.1128/aem.61.11.3894-3903.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otto M, Peschel A, Götz F. Producer self-protection against the lantibiotic epidermin by the ABC transporter EpiFEG of Staphylococcus epidermidis Tü3298. FEMS Microbiol Lett. 1998;166:203–211. doi: 10.1111/j.1574-6968.1998.tb13891.x. [DOI] [PubMed] [Google Scholar]
- 13.Pag U, Heidrich C, Bierbaum G, Sahl H-G. Molecular analysis of expression of the lantibiotic Pep5 immunity phenotype. Appl Environ Microbiol. 1999;65:591–598. doi: 10.1128/aem.65.2.591-598.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peschel A, Augustin J, Kupke T, Stevanovic S, Götz F. Regulation of epidermin biosynthetic genes by EpiQ. Mol Microbiol. 1993;9:31–39. doi: 10.1111/j.1365-2958.1993.tb01666.x. [DOI] [PubMed] [Google Scholar]
- 15.Peschel A, Götz F. Analysis of the Staphylococcus epidermidis genes epiF, -E, and -G involved in epidermin immunity. J Bacteriol. 1996;178:531–536. doi: 10.1128/jb.178.2.531-536.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peschel A, Ottenwälder B, Götz F. Inducible production and cellular location of the epidermin biosynthetic enzyme EpiB using an improved staphylococcal expression system. FEMS Microbiol Lett. 1996;137:279–284. doi: 10.1111/j.1574-6968.1996.tb08119.x. [DOI] [PubMed] [Google Scholar]
- 17.Peschel A, Otto M, Jack R W, Kalbacher H, Jung G, Götz F. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins and other antimicrobial peptides. J Biol Chem. 1999;274:8405–8410. doi: 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 18.Peschel A, Schnell N, Hille M, Entian K-D, Götz F. Secretion of the lantibiotics epidermin and gallidermin: sequence analysis of the genes gdmT and gdmH, their influence on epidermin production and their regulation by EpiQ. Mol Gen Genet. 1997;254:312–318. doi: 10.1007/s004380050421. [DOI] [PubMed] [Google Scholar]
- 19.Peschel A, Vuong C, Otto M, Götz F. The d-alanine residues of Staphylococcus aureus teichoic acids alter the susceptibility to vancomycin and the activity of autolysins. Antimicrob Agents Chemother. 2000;44:2845–2847. doi: 10.1128/aac.44.10.2845-2847.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sahl H-G, Bierbaum G. Lantibiotics: biosynthesis and biological activities of uniquely modified peptides from Gram-positive bacteria. Annu Rev Microbiol. 1998;52:41–79. doi: 10.1146/annurev.micro.52.1.41. [DOI] [PubMed] [Google Scholar]
- 21.Schnell N, Entian K D, Schneider U, Götz F, Zähner H, Kellner R, Jung G. Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with four sulphide-rings. Nature. 1988;333:276–278. doi: 10.1038/333276a0. [DOI] [PubMed] [Google Scholar]