Abstract
The rapidly evolving Coronavirus Disease 2019 (COVID-19) pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread worldwide with thousands of deaths and infected cases. For the identification of effective treatments against this disease, the main protease (Mpro) of SARS‑CoV‑2 was found to be an attractive drug target, as it played a central role in viral replication and transcription. Here, we report the results of high-throughput molecular docking with 1,045,468 ligands’ structures from 116 kinds of traditional Chinese medicine (TCM). Subsequently, 465 promising candidates were obtained, showing high binding affinities. The dynamic simulation, ADMET (absorption, distribution, metabolism, excretion and toxicity) and drug-likeness properties were further analyzed the screened docking results. Basing on these simulation results, 23 kinds of Chinese herbal extracts were employed to study their inhibitory activity for Mpro of SARS‑CoV‑2. Plants extracts from Forsythiae Fructus, Radix Puerariae, Radix astragali, Anemarrhenae Rhizoma showed acceptable inhibitory efficiencies, which were over 70%. The best candidate was Anemarrhenae Rhizoma, reaching 78.9%.
Keyword: SARS‑CoV‑2, Molecular docking, Screening, TCM, Main protease (Mpro)
Graphical abstract
1.Introduction
SARS-CoV-2, as the single-stranded positive RNA virus, has been declared as a pandemic disease (COVID-19) by the World Health Organization (WHO) [1,2]. Since then, this global public-health emergency has spread for over 2 years, crossing six continents and affected 218 countries [3]. So far, 24 June 2022, over 539.9 million cases with the infection have been confirmed and about 6.32 million patients have been died worldwide [4].
Recently, some special vaccines have been developed by many countries, such as messenger RNA vaccine (USA), inactivated vaccine (China) and etc. [5]. However, the insufficient production capacity always limits the application of these vaccines. Although SARS-CoV-2 vaccinations process has been ongoing for more than half year, the vaccination coverage is only about 15% in the worldwide. Moreover, most vaccines were controlled by a few of developed countries [6]. In fact, the vaccination coverage in many developing countries is even significantly below the average level. Moreover, the function of vaccines is to prevent this disease, but not treat it. Unfortunately, no drug has yet been approved for efficiently treating human coronaviruses [7]. The development of new therapies will take months to years. Thus, more attention should be paid on existing resources for treating or preventing infections. The molecular docking provided an efficiently high-throughput method to virtually screen or even design the potential drug against SARS-CoV-2 [8,9]. It will also provide more possibilities for further animal and clinic trials.
TCM dates back at least 2200 years. The earliest record of Chinese medicine with the written form is the Huang Di Nei Jing (Inner Canon of the Yellow Emperor) from the 3rd century C.E [10]. Traditional Chinese herbal medicines and their extracts combined thousands of years of experience and the scientific basis provided by modern experimental research [11]. TCM is widely used in the East and Southeast Asian; subsequently, it has been also practiced in the world due to its broad-spectrum activities such as the antivirals, antioxidant therapeutics, anti-inflammatory effects [12]. Coincidentally, the epidemic situation of SARS-CoV-2 in China was also intense, but recovered very quickly in several months [13]. Based on their synergistic and additive biological effects, it is accepted that the mixture of different phytochemicals may be more efficient in terms of disease prevention compared to a single phytochemical [14].
Interestingly, bioassays indicated that several phytochemicals (such as abietane, apigenin, bavachinin and etc.) derived from TCM showed inhibitory activity against Mpro of SARS, which was also caused by a kind of coronaviruses and broke out in November 2002 [15]. Indeed, SARS-CoV-2 and SARS showed a very high phylogenetic similarity [16]. Basic Local Alignment Search Tool (BLAST) was used to compare Mpro sequences of these two coronaviruses. BLAST results indicated that the percent identity reached 96%. Mpro of SARS-CoV-2 and SARS both belong to 3C-like protease and play the crucial role in the viral replication and transcription through posttranslational processing of RNA replicase [17]. Moreover, Mpro of SARS-CoV-2 may provide a conserved molecular target to design a broad spectrum anti-CoV drug [18].
Therefore, the special attention has been paid to the virtually high-throughput screening (HTS) of TCM against Mpro of SARS-CoV-2 [6]. In this work, 1045,468 ligands’ structures derived from 116 kinds of TCM were used to study their interactions with Mpro of SARS-CoV-2 through molecular docking. Then, the molecular dynamic simulation and pharmacokinetics were employed to further analyze screened compounds after docking. 465 candidates of inhibitors for Mpro of SARS-CoV-2 were screened and could be divided into 11 classes by their scaffolds, including biflavones, flavonoids, anthraquinones, polyphenols, phenylpropanoids, terpenoids, stilbenes, isoflavonoids, xanthones, flavonolignans, and alkaloids. Basing on docking results, 23 kinds of plants extract from TCMs were used to study their inhibitory activity. TCM-derived components can be used to design not only the potential drug, but also functional foods and dietary supplement, basing on the theory of “medicine and food homology” [19]. Our results suggest that some functional foods showed the potential role against SARS-CoV-2. Through adjusting our daily diet, infections with that virus may be prevented by a simple and acceptable way worldwide.
2. Materials and methods
2.1. Ligands structures
The ligands used in this work comprised a total of 1045,468 ligands’ structures, topologized by 155 different scaffolds and generated by ZINC 15 database [20]. As shown in Table 1 , we first confirmed 155 kinds of compounds with completely different scaffolds and they are widely existing in 116 kinds of TCM. For each scaffold type, all provided three-dimensional structures were extracted from the ZINC 15 database in the mol2 format. For the rapid calculation and analysis, names of these ligands were re-set in this work. Considering initial 155 compounds with different scaffolds, their names were used to represent the relevant scaffold, because the scaffold name is too long. Therefore, the name of all isomers/analogs/derivatives generated from one of initial 155 compounds were set as their initial compound names and the suffix number was used to distinguish them, such as ginkgetin 1, ginkgetin 2 and ginkgetin 3.
Table 1.
Natural compound and their source name (Latin and Chinese).
| No. | Natural compound | Traditional Chinese Medicine name | No. | Natural compound | Traditional Chinese Medicine name | |||
|---|---|---|---|---|---|---|---|---|
| Latin | Chinese | Latin | Chinese | |||||
| 1 | Ginkgetin | Ginkgo biloba | 银杏 | 67 | Nodakenin | Notopterygii Rhizoma Et Radix | 羌活 | |
| 70 | Ginkgolide A | 69 | Lycorine | Lycoris radiata | 石蒜 | |||
| 111 | Matairesinol | 71 | Carnosol | Salvia japonica Thunb. | 鼠尾草 | |||
| 135 | Bilobalide | 72 | Cephalotaxine | Cephalotaxus fortunei Hooker | 三尖杉 | |||
| 9 | Quercetin | 56 | Ellagic acid | Granati Pericarpium | 石榴 | |||
| 2 | Hypericin | Forsythia suspensa | 贯叶连翘 | 73 | Cinnamic acid | Cinnamomum cassia Presl | 肉桂 | |
| 41 | Phillyrin | 74 | Tanshiuone IIA | Salvia miltiorrhiza Bunge | 丹参 | |||
| 93 | Phillygenin | 104 | Cyptotanshinone | |||||
| 3 | Robustaflavone | Agathis dammara | 贝壳杉 | 76 | Lsochlorogenic acidB | Ganoderma lucidum | 灵芝 | |
| 5 | Agathisflavone | 89 | Ganodermanondiol | |||||
| 4 | Hinokiflavone | Selaginella tamariscina | 卷柏 | 92 | Ganoderic acid C1 | |||
| 7 | Swertianolin | Swertia bimaculata | 獐牙菜 | 101 | Ganoderic acid F | |||
| 8 | Pedunculoside | Ilex macrocarpa Oliv. var. longipedunculata S.Y.Hu | 长梗冬青 | 117 | Ganoderic acid A | |||
| 9 | Quercetin | Camellia sinensis | 茶叶 | 138 | Rhinacanthin E | |||
| 40 | Catechin | 77 | Shikonin | Lithospermum erythrorhizon Sieb. et Zucc. | 紫草 | |||
| 86 | Theophylline | 78 | Lrisflorentin | Belamcanda chinensis (Linn.) Redouté | 射干 | |||
| 10 | Camptothecin | Camptotheca acuminata | 喜树 | 79 | Geniposide | Gardenia jasminoides Ellis | 栀子 | |
| 11 | Imperialine | Fritillaria | 贝母 | 105 | Heriguard | |||
| 13 | Peimisine | 9 | Quercetin | |||||
| 12 | Emodin | Rheum | 大黄 | 83 | Hematoxylin | Caesalpinia sappan Linn. | 苏木 | |
| 14 | Batatasin I | Dioscorea oppositifolia L. | 山药 | 84 | Triptolide | Tripterygium wilfordii Hook. f. | 雷公藤 | |
| 15 | Pomiferin | Maclura pomifera (Raf.) schneid. | 橙桑 | 85 | Perillaldehyde | Perilla frutescens (Linn.) Britt. | 紫苏 | |
| 16 | Coumarin | Melilotus officinalis | 黑香豆 | 87 | Rhynchophylline | Uncaria rhynchophylla (Miq.)Miq. ex Havil. | 钩藤 | |
| 17 | Hesperetin | Citrus reticulata Blanco | 柑橘 | 88 | Decursinol | Angelica sinensis | 当归 | |
| 53 | Limonin | 100 | Columbianadin | |||||
| 68 | Folic acid | 149 | Ligustilide | |||||
| 147 | Rutin | Forsythiae Fructus | 连翘 | 91 | Sinomenine | Caulis sinomenii | 青藤 | |
| 2 | Hypericin | 63 | Sophoricoside | Sophora japonica L. | 槐树 | |||
| 18 | Ursolic acid | 66 | Esculin | Fraxinus rhynchophylla Hance | 秦皮 | |||
| 9 | Quercetin | 55 | Narciclasine | Narcissus tazetta L. var. chinensis Roem. | 水仙花 | |||
| 18 | Ursolic acid | Lonicerae Japonicae Flos | 金银花 | 37 | Colchicine | Colchicum autumnale L. | 秋水仙 | |
| 57 | Vitexin | 94 | Steviol | Stevia rebaudiana (Bertoni) Hemsl | 甜叶菊 | |||
| 19 | Rhodionin | Rhodiola rosea Linn. | 红景天 | 95 | Morroniside | Cornus officinalis Sieb. et Zucc. | 山茱萸 | |
| 114 | Salidroside | 96 | Anisodamine | Anisodus tanguticus (Maxim.) Pascher | 山莨菪 | |||
| 20 | Aloin | Aloe L. | 芦荟 | 154 | Cuscohygrine | |||
| 21 | Sarsasapogenin | Anemarrhenae Rhizoma | 知母 | 97 | Aconitine | Aconitum carmichaeli Debx. | 乌头 | |
| 82 | mangiferin | 80 | β-carotene | Hippophae Fructus | 沙棘 | |||
| 22 | Magnolol | Magnolia officinalis Rehd. et Wils. | 厚朴 | 98 | Cytisine | Parochetus communis Buch.-Ham. | 金雀花 | |
| 23 | Paclitaxel | Taxus chinensis (Pilger) Rehd. | 红豆杉 | 99 | Phloretin | Malus domestica | 苹果 | |
| 24 | Silydianin | Silybum marianum | 水飞蓟 | 102 | Cimifugin | Cimicifuga foetida Linn. | 升麻 | |
| 25 | Morusin | Morus alba Linn. | 桑根 | 105 | Heriguard | Eucommia ulmoides Oliver | 杜仲 | |
| 26 | α-Mangostin | Garcinia mangostana L. | 山竹 | 106 | Pimpinellin | Pimpinella anisum Linn. | 茴芹 | |
| 27 | Yohimibine | Corynanthe johimbe | 育亨宾 | 107 | Atractyloside | Atractylodes lancea (Thunb.) DC. | 苍术 | |
| 28 | Coclaurine | Lindera aggregate | 乌药 | 108 | Bergenin | Bergenia purpurascens (Hook. f. et Thoms.) Engl. | 岩白菜 | |
| 151 | Linderalactone | 109 | Artemether | Artemisia carvifolia | 青蒿 | |||
| 29 | Rutaecarpine | Evodia rutaecarpa | 吴茱萸 | 112 | Schisandrin B | Schisandrae Chinensis Fructus | 北五味子 | |
| 46 | Evodiamine | 113 | Schisanhenol | |||||
| 30 | Strychnine | Strychnos nux-vomica L. | 马钱子 | 132 | Calophyn | |||
| 6 | Ononin | Radix astragali | 黄芪 | 115 | Rosmanol | Rosmarinus officinalis | 迷迭香 | |
| 103 | Astilbin | 116 | Verbenalin | Verbena officinalis Linn. | 马鞭草 | |||
| 155 | Sedanolide | Ligusticum chuanxiong Hort. | 川芎 | 118 | Monocrotaline | Lilium brownii F. E. Brown ex Miellez | 野百合 | |
| 32 | Coptisine | Coptidis Rhizoma | 黄连 | 119 | Matrine | Sophora flavescens Alt. | 苦参 | |
| 36 | Berberine | Jujubae Fructus | 大枣 | 143 | Sophocarpine | |||
| 144 | Farfaratin | Tussilago farfara Linn. | 款冬花 | 120 | Paeoniflorin | Paeonia lactiflora Pall. | 芍药 | |
| 33 | Tiliroside | Tilia tuan Szyszyl. | 椴树 | 121 | Astaxanthin | Fenneropenaeus chinensis | 虾 | |
| 34 | Glabrone | Glycyrrhiza uralensis Fisch | 甘草 | 122 | Pachymic acid | Poria cocos (Schw.) Wolf. | 茯苓 | |
| 57 | Vitexin | 60 | Karanjin | Pongamia pinnata (Linn.) Pierre | 水黄皮 | |||
| 147 | Rutin | 61 | Gigantol | Dendrobium nobile Lindl. | 石斛 | |||
| 6 | Ononin | 62 | Polydatin | Polygonum cuspidatum Sieb.et Zucc. | 虎杖 | |||
| 18 | Ursolic acid | 27 | Yohimbine | Corynanthe johimbe | 育亨宾 | |||
| 31 | Medicarpin | 65 | Mollugin | Rubia cordifolia Linn. | 茜草 | |||
| 81 | Liquirtin | 142 | Poricoic acid A | Echinacea purpurea (Linn.) Moench | 松果菊 | |||
| 35 | Myristicin | Myristica fragrans | 肉豆蔻 | 124 | Parthenolide | Cyathocline purpurea (Buch.-Ham.) O. Ktunze | 杯菊 | |
| 38 | Lignans | Sesamum indicum | 芝麻 | 125 | Kirenol | Siegesbeckia orientalis L. | 豨莶草 | |
| 54 | Sesamin | 126 | Micheliolide | Michelia compressa (Maxim.) Sarg. Gard. et For. | 乌心石 | |||
| 39 | Daidzein | Glycine max (Linn.) Merr. | 黄豆 | 127 | Pogostone | Agastache rugosa (Fisch. et Mey.) O. Ktze. | 藿香 | |
| 133 | Coenzym Q10 | 128 | Swainsonine | Sphaerophysa salsula (Pall.) DC. | 苦马豆 | |||
| 42 | Gastrodin | Gastrodia elata Bl. | 天麻 | 129 | Catalpol | Rehmannia glutinosa (Gaert.) Libosch. ex Fisch. et Mey. | 地黄 | |
| 43 | Obacunone | Phellodendron amurense Rupr. | 黄柏 | 45 | Ajugol | |||
| 44 | Cordycepin | Cordyceps sinensis | 冬虫夏草 | 129 | Catalpol | |||
| 45 | Ajugol | Leonurus artemisia | 益母草 | 131 | Alnusone | Alpinia katsumadai Hayata | 草豆蔻 | |
| 113 | Stachydrine | 134 | Ellipticine | Ochrosia borbonica Gmelin | 玫瑰树 | |||
| 47 | Nuciferine | 136 | Dihydrocurcumin | Curcuma longa Linn. | 姜黄 | |||
| 48 | Bergamottin | Glehniae Radix | 北沙参 | 137 | Valepotriate | Valeriana officinalis Linn. | 缬草 | |
| 49 | Cynarin | Cirsium setosum | 蓟草 | 139 | Gentiopicroside | Gentiana scabra Bunge | 龙胆 | |
| 50 | Betulin | Betula platyphylla Suk. | 白桦 | 140 | Trilobatin | Hydrangea strigosa Rehd. | 甜茶 | |
| 51 | Gramine | Arundo donax Linn. | 芦竹 | 141 | Piperine | Piper nigrum Linn. | 胡椒 | |
| 52 | Puerarin | Radix Puerariae | 葛根 | 145 | Atractylon | Atractylodes macrocephala Koidz. | 白术 | |
| 58 | Resveratrol | Vitis vinifera Linn. | 葡萄 | 146 | Helenine | Inula helenium L. | 土木香 | |
| 90 | Cyanidin | 148 | Cedrol | Cupressus funebris Endl. | 柏木 | |||
| 59 | Protopanaxatriol | Panax ginseng | 人参 | 150 | Curcumol | Curcuma zedoaria (Christm.) Rosc. | 莪术 | |
| 75 | Panaxadiol | 152 | Fraxinellone | Cortex radicis Dictamni | 白鲜皮 | |||
| 110 | Ginsenoside Rk3 | 153 | Pilocarpine | Ruta graveolens Linn. | 芸香 | |||
2.2. Receptor structure
The crystal structure of Mpro of SARS-CoV-2 (PDB:6LU7) was reported by Jin et al. (Shanghai Technical University, China). The three-dimensional structure of Mpro of SARS-CoV-2 was shown in Figure S1 (Figure S1–14 can be found in supplementary material). In a previous study [21], DoGSiteScorer was employed to confirm the green region as the potential binding pocket of Mpro of SARS-CoV-2. Moreover, it is also the binding pocket for the Michael acceptor inhibitor, which can specifically inhibit Mpro of many coronaviruses, such as SARS-CoV and MERS-CoV [22,23]. On the other hand, the reported study indicated that Mpro of SARS-CoV-2 should have a Cys-His-catalytic dyad [24,25]. We checked all Cys-and His-residues in Mpro of SARS-CoV-2 and only residues in the green region in Figure S1 might form a Cys-His-catalytic dyad. Therefore, it was confirmed as the docking center (x: y: z, −12: 12.5: 67).
2.3. Molecular docking
Python (v.3.7) was used as the working language for all software in this work. AutoDockTools (v.1.5.6) was employed to added Gasteiger PEOE partial charges and polar hydrogen atoms for the ligands and Mpro of SARS-CoV-2. The format of the structural files was converted into PDBQT. Molecular docking was carried out by AutoDock Vina (v.1.1.2). The Broyden–Fletcher–Goldfarb–Shanno (BFGS) method was used to screen the most favorable binding sites. Semi-flexible docking method was employed, where the rigid body was Mpro of SARS-CoV-2 and rotatable bonds in all ligands were sampled. The geometric center of the docking box was set as −12: 12.5: 67 and the edge length of that was 30 Å, which can cover the whole active center (pink region in Fig. 1 ). Other parameters were used as defaults. After each docking simulation, top 20 complexes were picked according to their docking affinities. All ligands'structures and relevant docking results have been showed in the supporting Excel File2.
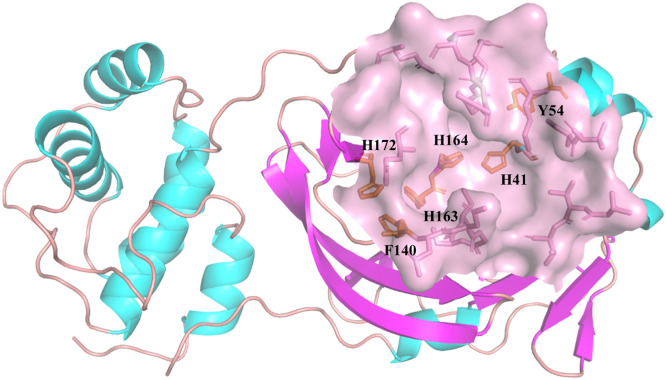
Fig. 1.
3D-structure of the substrate-binding pocket of Mpro (pink surface). The residues with aromatic ring structures in pocket were shown in orange sticks and marked in the one-letter code.
2.4. Molecular dynamics
Molecular dynamics (MD) simulation is an efficient method to predict the stability of the inhibitor-protein complex. TIP3P was used as the model of water molecules and the size of the relevant box was set as 61 × 61 × 91 Å3. The simulation method was same with our previous work [26].
2.5. ADMET and drug-likeness screening
After virtual screening, the obtained candidates were further analyzed by SwissADME (http://www.swissadme.ch/) and admetSAR (http://lmmd.ecust.edu.cn/admetsar2) online website [27]. The drug-likeness was analyzed basing on the Lipinski rule of 5 (RO5) [28].
2.6. Mpro enzyme activity inhibition test
A fluorescence resonance energy transfer (FRET) protease assay was applied to measure the inhibitory activity of compounds against the Mpro of SARS-CoV-2. In this work, inhibitor screening kit from Beyotime (P0315S) was used to measure enzyme activity. Principle of this FRET-based protease assay: MCA (Methoxy-coumarin-acetic-acid) is a fluorescent donor, Dnp (2,4-dinitrophenyl) is a fluorescent acceptor, the absorption spectra of these two fluorescent groups have a certain overlap. When the distance between these two fluorescent groups is appropriate (generally 7–10 nm), the fluorescence energy will transfer from the donor group to the acceptor group, causing the fluorescence intensity of the donor to decay. MCA and Dnp are connected to ends of the natural substrate of Mpro, namely MCA-AVLQSGFR-Lys (Dnp)-Lys-NH2. When the Mpro protease does not cleave the substrate, the two groups are close enough that cause fluorescence resonance energy transfer, which mean Dnp can quench the fluorescence of MCA and cause no fluorescence detection.
In this work, 23 kinds of plant extracts from TCMs were used to study their inhibitory activity, which are Ginkgo biloba, Forsythia suspensa, Radix glycyrrhizae, Coptidis Rhizoma, Rhodiola rosea Linn, Lonicerae Japonicae Flos, Taxus chinensis (Pilger) Rehd., Forsythiae Fructus, Evodia rutaecarpa, Radix Puerariae, Strychni Semen, Myristica fragrans, Fritillaria, Gastrodia elata, Magnolia officinalis, Magnolia officinalis Rehd. et Wils., Rheum, Anemarrhena asphodeloides, Aloe L., Dioscorea oppositifolia L., Leonurus artemisia, Jujubae Fructu, Radix astragali, Silybum marianum. These plant extracts were gift from Shaanxi pure source bio-tech co., LTD. The concentration of plant extracts was 0.35 mg/mL.
In this way, the Mpro enzyme activity was detected very sensitively by the fluorescence detection. If the inhibitor was added to the reaction system, the generation of fluorescence would be inhibited. Since the plant extracts were mixtures without the certain molecular weight, the mass concentration was used in our work. Plant extracts solutions were prepared by DMSO with a concentration of 0.35 mg/mL. Then, 5μL of plant extracts solutions, 92μL assay buffer and 1μL of 2019-nCoV Mpro/3CLpro solution were added in the 96-well black plate. The reactions were initiated by adding 2μL of fluorogenic substrate. After that, samples were incubated in the dark at 37 °C for 10 min. Then, the fluorescence signal was measured at 325 nm (excitation)/393 nm (emission) with muti-mode plate reader in TECAN.
3. Results and discussion
3.1. Docking analysis basing on skeletons of ligands
In a previous simulation work [21], they employed the molecular docking to screen 1615 of FDA approved drugs against Mpro of SARS-CoV-2. Conivaptan was confirmed as the best candidate. However, the severe side effect limited its further study, involving blood and lymphatic system disorders, metabolism and nutrition disorders, headache, nausea, vomiting and somnolence [29]. Recently, our research group employed the molecular docking to screen 10,870 of ligands derived from functional foods compounds against Mpro of SARS-CoV-2 [26]. This study yielded 60 ligands as promising inhibitors with similar or higher docking affinities to those of conivaptan.
In this work, the screening library was enlarged for about 100 times compared to the previous work. Therefore, an automatic script for HTS by Python was compiled. Then, 1045,468 ligands’ structures derived from 116 kinds of TCM were used to study their interaction with Mpro of SARS-CoV-2 by HTS. Their docking affinities were compared with conivaptan reported as an inhibitor. Finally, 465 promising inhibitors corresponding to 669 structures (as some ligands would have more than one 3D structure) were obtained against Mpro of SARS-CoV-2, which are shown in Supporting Excel File (Promising inhibitors). All docking results were systematically analyzed. However, there were over 1 million structures. It seemed impossible to analyze them individually. In fact, these 1045,468 ligands’ structures were generated by the topology from 155 kinds of skeletons. As shown in Table 2 , only one ligand with the best docking affinity was picked to represent each type of skeleton. It is worth noting 669 promising inhibitors involved 61 kinds of different skeletons.
Table 2.
The binding affinity of 61 phytochemicals.
| Name of natural compounds | ZINC number | Binding Affinity (Kcal/mol) | Name of natural compounds | ZINC number | Binding Affinity (Kcal/mol) |
|---|---|---|---|---|---|
| Ginkgetin | 3,979,028 | −10.3 | Myristicin | 620,709,870 | −8.6 |
| Hypericin | 28,705,885 | −10.3 | Medicarpin | 13,485,420 | −8.6 |
| Robustaflavone | 40,763,477 | −9.6 | Coptisine | 6,069,258 | −8.6 |
| Hinokiflavone | 4,098,521 | −9.3 | Tiliroside | 49,888,963 | −8.6 |
| Agathisflavone | 85,566,557 | −9.2 | Glabrone | 14,658,666 | −8.6 |
| Swertianolin | 33,830,763 | −9.1 | Colchicine | 38,494,624 | −8.6 |
| Ononin | 71,789,593 | −9.1 | Lignans | 13,473,815 | −8.5 |
| Pedunculoside | 49,898,882 | −9 | Daidzein | 1,898,781 | −8.5 |
| Quercetin | 98,083,363 | −8.9 | Catechin | 66,258,345 | −8.5 |
| Camptothecin | 29,390,846 | −8.9 | Phillyrin | 1,532,044 | −8.5 |
| Imperialine | 101,084,934 | −8.9 | Gastrodin | 8,834,580 | −8.5 |
| Emodin | 12,960,987 | −8.9 | Obacunone | 100,003,095 | −8.5 |
| Peimisine | 100,037,427 | −8.9 | Cordycepin | 95,539,308 | −8.5 |
| Batatasin I | 73,224,987 | −8.9 | Ajugol | 35,455,106 | −8.4 |
| Pomiferin | 14,692,098 | −8.9 | Evodiamine | 2,031,813 | −8.4 |
| Coumarin | 150,340,486 | −8.8 | Nuciferine | 320,698 | −8.4 |
| Hesperetin | 1,660,863 | −8.8 | Bergamottin | 2,103,110 | −8.4 |
| Ursolic acid | 29,553,661 | −8.8 | Cynarin | 95,601,892 | −8.4 |
| Rhodionin | 1,531,663 | −8.8 | Betulin | 96,023,899 | −8.4 |
| Aloin | 101,529,211 | −8.8 | Gramine | 72,127,032 | −8.4 |
| Sarsasapogenin | 118,916,629 | −8.8 | Puerarin | 33,832,535 | −8.3 |
| Magnolol | 59,530,776 | −8.8 | Limonin | 1,280,471 | −8.3 |
| Paclitaxel | 257,101,405 | −8.7 | Sesamin | 38,147,404 | −8.3 |
| Silydianin | 31,457,053 | −8.7 | Narciclasine | 40,424,184 | −8.3 |
| Rutaecarpine | 85,876,671 | −8.7 | Ellagic acid | 1,227,574,377 | −8.3 |
| Morusin | 299,869,393 | −8.7 | Vitexin | 95,913,589 | −8.3 |
| α-Mangostin | 15,256,039 | −8.7 | Resveratrol | 1,661,001 | −8.3 |
| Yohimibine | 13,139,106 | −8.7 | Protopanaxatriol | 257,349,206 | −8.3 |
| Coclaurine | 11,422,321 | −8.7 | Karanjin | 14,618,800 | −8.3 |
| Strychnine | 71,782,841 | −8.7 | Gigantol | 57,770,685 | −8.3 |
| Berberine | 40,379,008 | −8.6 | Conivaptan | 12,503,187 | −8.2 |
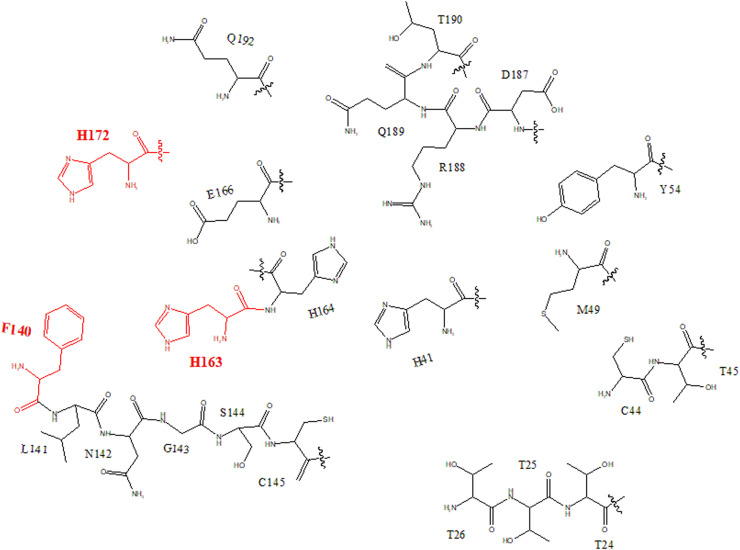
The primary analysis showed that the docking affinities against Mpro of SARS-CoV-2 exhibited a clear dependence on the ring structure of ligands. As show in Figure S2, the docking affinity increased with the number of ring structures in the ligand skeleton. There were 40 skeletons with at least four-ring structures in the promising 61 kinds of skeletons screened by HTS. For other 96 skeletons, which had the lower affinity than conivaptan, only 33 of them at least had four-ring structures. In 155 kinds of skeletons, only 13 kinds of skeletons had at least six-ring structures. Interestingly, all of them showed higher affinity against Mpro of SARS-CoV-2 than conivaptan. Moreover, the top five skeletons all had at least six-ring structures, and the best docking affinity was obtained from isomers/analogs/derivatives of hypericin, whose skeleton had the most ring number in all ligands reaching eight. In addition, the unsaturated ring might be another important indicator. There were 55 skeletons with unsaturated rings in the promising 61 kinds of skeletons. Notably, the rings structures of top five skeletons were all unsaturated state, and the best candidate of isomers/analogs/derivatives from hypericin had the most unsaturated rings, reaching eight. For example, isomers/analogs/derivatives from pomiferin I, aloin, protopanaxatriol, pachymic acid all had the four-ring structure, which had similar skeletons. Rings in the skeleton of protopanaxatriol were all saturated rings. In contrast, skeletons of pomiferin I and aloin had four and three unsaturated rings, respectively. Coincidentally, higher docking affinities were found in skeletons of pomiferin I (−8.9 Kcal/mol) and aloin (−8.8 Kcal/mol) than that of protopanaxatriol (−7.4 Kcal/mol). Another phenomenon was observed that Mpro of SARS-CoV-2 preferred to interact with multi-ring compounds in which the distance between the individual aromatic rings is narrower. For example, skeletons of quercetin and piperine both had three rings. In the skeleton of piperine, the rings were separated by the carbon chain (docking affinity −6.8 Kcal/mol), whereas in the skeleton of quercetin, ring structures were arranged closer (docking affinity −8.9 Kcal/mol). Whereby, the residues increase the mobility of delocalized electron cloud. Fig. 1 showed the binding pocket of Mpro of SARS-CoV-2, which had 6 residues with aromatic ring structures. These important 6 residues made up three of boundary lines, deciding the depth and width of binding pocket. As shown in Fig. 1, a binding triad could be confirmed as F140, H163 and H172. The projection analysis found rings of this binding triad had the compact and staggered arrangement, as shown in Fig. 2 . On the other hand, the skeleton of the inhibitor candidates with unsaturated and tight multi-rings systems and higher electronic mobility might be easily inserted into this particular pocket through the π-π stacking. On the other hand, the heterocyclic structure was another factor that influence the docking affinity.
Fig. 2.
The projection analysis of binding triad (H163, H172, F140).
In this work, there were only two kinds of heteroatoms in all heterocyclic compounds, including oxygen and nitrogen atoms. Oxygen atoms in the heterocyclic structure were all neutral. Generally, the oxygen atom in the heterocyclic structure was a positive signal that the docking affinity would be improved. For top ten skeletons, there were eight heterocyclic compounds, in which carbon atoms were all replaced by oxygen atoms. There were two states for the nitrogen atom in the heterocyclic compound, including neutral and positive charged. The neutral nitrogen atom, like the oxygen atom, could donate electrons, so the electronic density and mobility would be increased in these heterocyclic compounds. In contrast, the nitrogen cation would withdraw electrons from the electronic cloud, decreasing the electronic density and mobility, such as sophocarpine (−6.7 Kcal/mol) and pilocarpine (−5.8 Kcal/mol).
Next, effects of properties of side chains in skeletons were systematically analyzed. First, the effect of the double bond in the side chain, which could be directly involved in the electronic conjugate effect, was analyzed. For top ten skeletons, it was observed that eight skeletons have double bonds in the side chain, which have the electronic conjugated interaction with the ring structure of the relevant skeleton. Although the double bond had the positive effect on the ligand binding, it only played a synergistic effect not the leading role. For example, the skeleton of β-carotene had two unsaturated rings and nine double bonds in the side chain. All unsaturated bonds could form the electronic conjugated effect. However, the docking affinity of the best derivate from this skeleton was only −7.9 Kcal/mol. Besides the lower number of rings in that structure, the steric resistance might be another reason. More double bonds were provided, but since the length of the side chain is enlarged, the steric resistance is increased.
Moreover, the functional group changes the docking affinity through the effect on the electronic density and mobility in the ring structure. The best example is the derivate from the skeleton of coumarin (−8.8 Kcal/mol), which has two rings. However, this ligand had many electron-donating groups, such as amine groups, which could effectively increase the electronic density and mobility. In contrast, although the derivate from the skeleton of ganoderic acid had four rings, the docking affinity was only -7.7 Kcal/mol. This could be due to the fact that four carbonyl groups, as strong electron withdrawing groups, were directly connected to aromatic rings, which decreased the density and mobility of the electronic cloud.
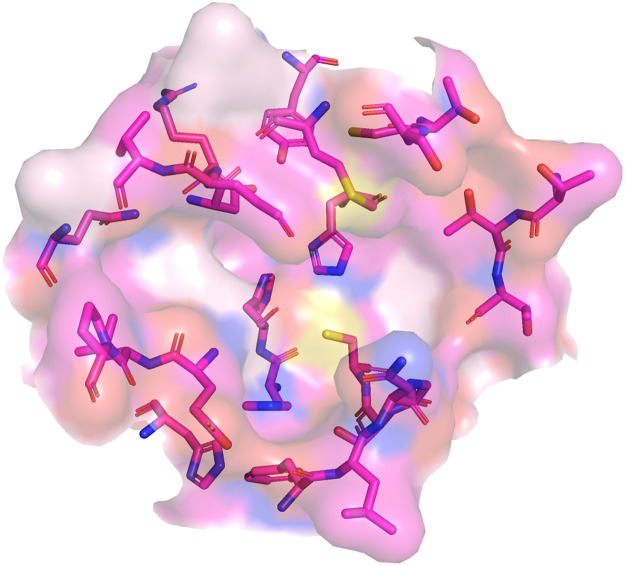
The functional group played another very crucial role by forming the hydrogen bond with Mpro of SARS-CoV-2 during the molecular docking. Hydrogen bonds significantly improve the docking affinity. The simulation results indicate that functional groups with high electronic densities would easily form hydrogen bonds with Mpro of SARS-CoV-2 and show higher binding affinities. The binding pocket of Mpro of SARS-CoV-2 was systematically studied, as shown in Fig. 3 . For Mpro of SARS-CoV-2, functional groups, which have the possibility to form hydrogen bonds, were carbonyl, amino, hydroxyl and sulfhydryl groups. For most carbonyl groups were not facing the center of the binding pocket and therefore had difficulties contacting the ligands and forming hydrogen bonds. For other functional groups, they would like to provide the hydrogen atom to form the hydrogen bond. Generally, the electron-donating group had the higher electronic density, which was easily to form stable hydrogen bonds with hydrogen donors and showed high docking affinity, such as hydroxyl group. For example, skeleton structures of agathisflavone and rutin have the same number of rings, while electron-donating groups were not hydroxyl group in the skeleton of rutin. Compared with hydroxyl group, the electronic density of ether decreased significantly and was hard to form the hydrogen bond. Thus, an obvious decline in the docking affinity was observed in rutin (−6.5 Kcal/mol) compared to that of agathisflavone (−9.2 Kcal/mol). Whereas, ligands with the strong electron-withdrawing group (particularly for ammonium group) generally showed poor docking affinities. Some typical cases could be found in derivates from skeletons of gentiopicroside (−6.9 Kcal/mol), sophocarpine (−6.7 Kcal/mol) and pilocarpine (−5.8 Kcal/mol).
Fig. 3.
3D-structure of the substrate-binding pocket, residues in pocket were shown in stick and were colored by element.
It is worth mentioning that compounds with haloalkanes, as they are strong electron-withdrawing group, were very special cases and 15 kinds of skeletons containing halogen were investigated in this work. Interestingly, nine of them (such as derivates form skeletons of hypericin, emodin and magnolol) showed higher docking affinities against Mpro of SARS-CoV-2 than conivaptan. As shown in Figure S3, although halogen didn't directly participate into the formation of the hydrogen bond, they could prompt nearby electron-donating groups to be converted into the anion by conjugated effects and benefit for the formation of hydrogen bonds. Another reason, why they didn't show higher docking affinity, might be that no electron-donating group was found around halogen. Therefore, halogen only played a role to decrease the electronic density and mobility, such as derivates form skeletons of perillaldehyde, stachydrine and swainsonine. Although electron-donating groups were close to halogen in derivates from skeletons of shikonin and cytisine, they were caught between two strong electron-withdrawing groups. It might lead to the shielding effect and lower docking affinities.
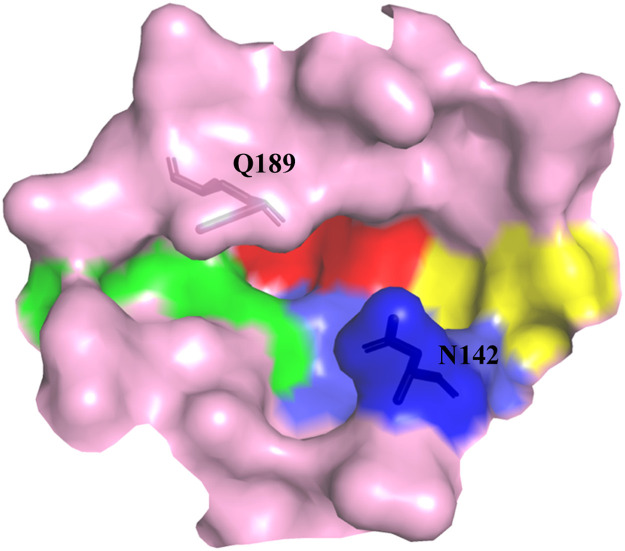
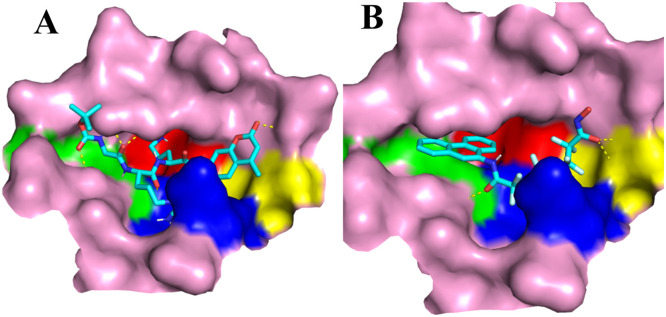
Through our analysis of the ligands' binding, we were able to identify four regions in the binding pocket that significantly influence their shape (shown in Fig. 4 ). The whole shape looked like a relevantly symmetric cross. N142 and Q189 were two extended lids to cover on this pocket. Therefore, the ligand should also fit this shape during the molecular docking. The typical example was the derivate from the skeleton of coumarin (−8.8 Kcal/mol). It only had two rings, but its structure was completely symmetric. Docking results indicated that two rings could respectively occupied the binding region I and II (Fig. 5 A). The binding region I and II had the same shape. The former was mainly surround by residues with chains, but the later was mainly surround by residues containing ring structures. Thus, some ligands consisted of a carbon chain and aromatic rings also showed higher docking affinities. For example, the derivate from the skeleton of batatasin I could be divided into a long chain and a phenanthrene ring, which would respectively occupy the binding region I and II with a high docking affinity of −8.9 Kcal/mol (Fig. 5B).
Fig. 4.
The binding pocket of Mpro is show in the pink surface. The different identified regions are colored differently: Region I in yellow, region II in blue, region III in green, region IV in red. The extended lids covering in this pocket are shown in sticks and marked in the one-letter code.
Fig. 5.
Representative ligand-protein complex structures of coumarin derivate (A), batatasin Ⅰ derivate (B). Region I, II, III, IV is shown in yellow, blue, green and red, respectively. The ligands are shown in sticks and the binding pocket is shown in surface.)
Moreover, this kind of cross shape of the binding pocket of Mpro of SARS-CoV-2 also limited the shape of ligands and explained why polygons showed poor docking affinities, such as derivates from skeletons of parthenolide (−7.4 Kcal/mol), schisanhenol (−7.1 Kcal/mol) and linderalactone (−5.9 Kcal/mol).
3.2. Docking analysis basing on functional classes
In order to further analyze these docking results and illustrate the relation between the binding pocket of Mpro of SARS-CoV-2 and the structure of ligand, 1045,468 ligands’ structures were firstly divided into 11 classes basing on physical and chemical properties of their skeletons, involving flavonoids, biflavones, anthraquinones, polyphenols, phenylpropanoids, terpenoids, stilbenes, isoflavonoids, xanthones, flavonolignans and alkaloids.
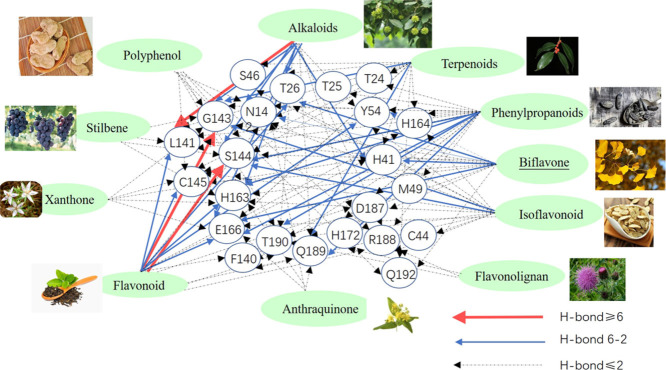
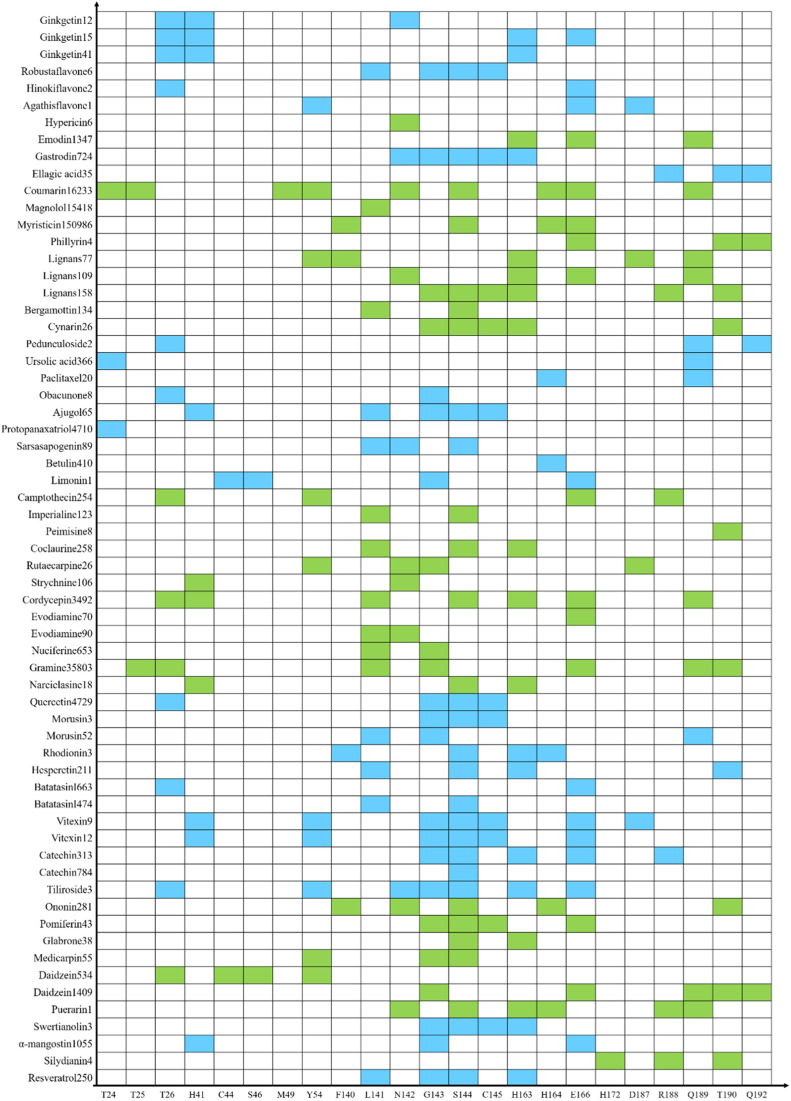
Residues that could form hydrogen bonds with ligands were shown in Fig. 6 . Docking results indicated that compounds in each class would prefer to form the hydrogen bond with some special residues basing on their unique physical and chemical properties.
Fig. 6.
The interactions between each ligand and amino acid in active pocket. 465 candidates of inhibitors for Mpro of SARS-CoV-2 were screened and could be divided into 11 classes by their scaffolds. Only the ligand with the highest affinity of each scaffold was shown. For each scaffold, the best ligand might be over 1 compound. Two kinds of colors (green and blue) were just used to only guide the eye distinguishing 11 classes of compounds.
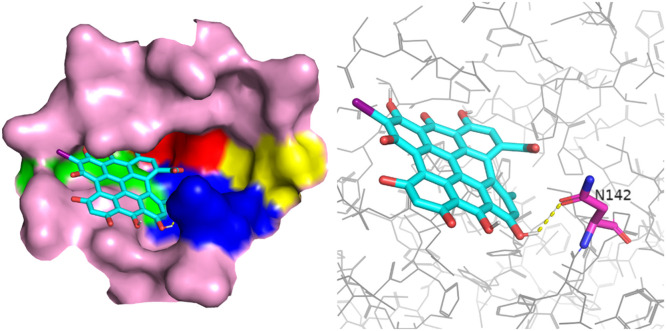
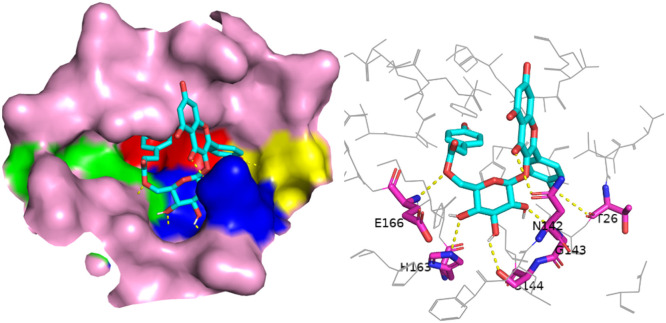
Anthraquinones were systematically analyzed, involving derivates from skeletons of hypericin and emodin. As shown in Fig. 7 , although the derivate from the skeleton of hypericin had many hydroxyl groups, only one hydrogen bond was found to connect with the carbonyl group. It was caused by the molecular steric resistance. The derivate from the skeleton of hypericin had nine tight benzene rings and showed very strong electronic conjugated effect. Therefore, it could stably occupy binding region II, III, and IV by π-π stacking. These regions mainly consisted of residues containing ring structures. Although derivates from the skeleton of emodin had less ring number, it also showed same docking results, which was the mainly characteristic of anthraquinone compounds showing in Figure S4.
Fig. 7.
Docking of a derivate from the skeleton of hypericin and the binding pocket. Left: the position of this ligand in the active pocket, the ligand is shown in sticks and the binding pocket is shown in surface. Right: the residues in pocket form hydrogen bond with this ligand, the residues are shown in pink sticks and the ligand is show in cyan sticks. Nitrogen is colored in blue, Oxygen colored in red and Iodine is colored in magenta.
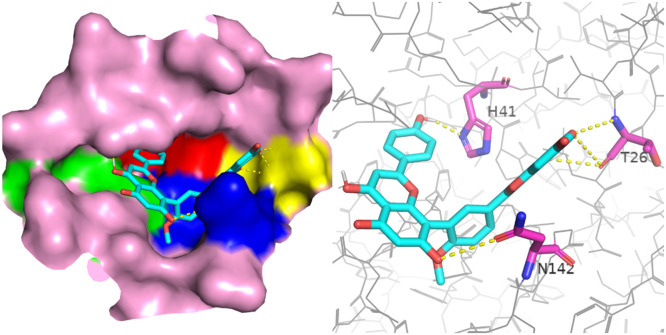
Biflavonoid should be one of best classes as promising inhibitors against Mpro of SARS-CoV-2. Bioflavonoids are generated by the condensation reaction between two flavonoid molecules [30]. Therefore, they are symmetrical molecule. Moreover, it had six unsaturated rings, thus, showed the strong conjugated effect. Except derivates from the skeleton of robustaflavone, other bioflavonoid compounds had many electron-donating (hydroxyl) groups, which could form stable hydrogen bonds with Mpro of SARS-CoV-2. For most derivates from the skeleton of ginkgetin, their carbonyl and meta‑hydroxyl groups in the terminal benzopyran could form multiple hydrogen bonds with carbonyl and amino groups in T26. Even in some cases, there was no meta‑hydroxyl group in the terminal benzopyran, but carbonyl group could still form hydrogen bond with the carbonyl group in T26. One explanation is that there was always an intramolecular hydrogen bond between carbonyl and amino groups in T26. In addition, these compounds could fully occupy all four binding regions of Mpro of SARS-CoV-2 (Fig. 8 ), implying the skeleton of ginkgetin should be the perfect inhibitor candidate. For derivates from skeletons of robustaflavone, hinokiflavone, and agathisflavone, the connection sites and directions between two flavonoid molecules made their molecular structures more rigid and longer than that of ginkgetin. Thus, these ligands couldn't perfectly fill all binding pocket regions through the reasonable bond rotation (Figure S5). For all bioflavonoid ligands, functional groups in the terminal ring structure were easy to form hydrogen bonds due to the lower steric hindrance. When the ligand structure was more rigid and straight, although the steric hindrance of the whole molecule was increased, that of the molecular middle region would be decreased, improving possibilities for the formation of hydrogen bonds of functional groups in these regions.
Fig. 8.
Docking between derivate from the skeleton of ginkgetin and the binding pocket. Left: the position of this ligand in the active pocket, the ligand is shown in sticks and the binding pocket is shown in surface. Right: the residues in pocket form hydrogen bond with this ligand, the residues are shown in pink sticks and the ligand is show in cyan sticks. Nitrogen is colored in blue, Oxygen colored in red.
Next, flavonoids were systematically analyzed, as shown in Figure S6. The most typical characteristic was that flavonoids consisted of benzene and benzopyran rings. Generally, steric hindrance of flavonoids was relevantly low, so that they could be easily inserted into the binding pocket of Mpro of SARS-CoV-2. Meanwhile, the small size also limited them to occupy only three binding regions (I, II, III or II, III, IV). Only derivate from the skeleton of tiliroside couldn't completely enter into this binding pocket, because it had the very large chain with 2 additional rings, as shown in Fig. 9 . A phenomenon was found that most flavonoids would like to form hydrogen bonds with Gly143 and Ser144, which located on the boundary between region I and II. On one hand, these hydrogen bonds were found in some specific structures, involving electron-donating (hydroxyl) groups in terminal benzene/benzopyran ring and branched chains without ring structures. On the other hand, the relevant small size of flavonoids (compared to the bioflavonoids) made them easily enter into the binding pocket, but it also implied the steric limitation of the binding pocket for these ligands were not very strong and stable. It might explain that although most flavonoids could completely enter into the binding pocket, their docking affinities were lower than those of biflavonoids.
Fig. 9.
Docking between derivate from the skeleton of tiliroside and the binding pocket. Left: the position of this ligand in the active pocket, the ligand is shown in sticks and the binding pocket is shown in surface. Right: the residues in pocket form hydrogen bond with this ligand, the residues are shown in pink sticks and the ligand is show in cyan sticks. Nitrogen is colored in blue, Oxygen colored in red.
Isoflavonoids are a class of special flavonoid phenolic compounds. Compared to flavonoids, the location of benzene ring was moved from meta- into ortho-position of carbonyl group in the benzopyran ring. The length of the skeleton was increased for ∼33% from 6 (flavonoid) to 9 (isoflavonoid) Å. Considering directions and sizes of four binding regions, this pocket made the ring structure of isoflavonoids like to occupy region II and III, as shown in Figure S7. Only the branched chain without ring structure could enter into the region I. Meanwhile, most isoflavonoids also liked to form hydrogen bonds with Gly143 and Ser141. The reason behind this phenomenon was same with flavonoids.
Terpenoids, also known as isoprenoids, were a class of compounds mostly forming special ring-structures. Although most rings in these ligands were saturated, the highest docking affinity was even higher than that of the flavonoids. It might be because their compact ring structures and all ring terpenoids shared their ring edges. Therefore, the density and mobility of the electronic cloud is higher in the ring-terpenoid partition, which made ring terpenoids easily interact with binding region II or III or IV showing in Figure S8. On one hand, terpenoids didn't have much branched carbon chain, most of them also liked to form hydrogen bonds with region I via electron donating groups in terminal rings. On the other hand, the number and compactness of ring structures improved the electronic intensity, but it also increased the steric hindrance. The docking results indicated that too rigidified multi-ring structures were not beneficial for binding with the pocket.
Docking results of phenylpropanoids against Mpro of SARS-CoV-2 were shown in Figure S9. A relevantly typical feature was that there were always strong electron withdrawing groups around electron donating groups, which formed hydrogen bonds with Ser144. These electron withdrawing groups would prompt the hydrogen ion in electron donating groups to delocalize and form the hydrogen bond with residues. Although the number of rings in phenylpropanoids were relevantly lower, most rings were unsaturated and showed strong conjugated effect. Thus, phenylpropanoids could also bind interact with binding region II or III or IV. A special case was the derivate from the skeleton of coumarin. It had a very large branched chain. Importantly, there were many amino, carboxyl, and ether groups, increasing the intensity and mobility of the electronic cloud in this chain, which made the branched chain form many hydrogen bonds with residues and be inserted into binding region II, III, and IV.
Compounds of the group of the alkaloids comprised most skeletons from this study. Docking results of alkaloids against Mpro of SARS-CoV-2 were shown in Figure S10. Their binding characteristics were as same as that of terpenoids, as the alkaloids also had most compact ring structures and all rings shared their ring edges. Thus, the strong conjugated effect should be the advantage for alkaloids against Mpro of SARS-CoV-2, but it also leaded into the rigidification of the whole ligand molecule, which increased their steric hindrance. The docking analysis indicated that alkaloids would like to form the hydrogen bonds with Leu141.
Stilbenes consist of a skeleton of stilbene and branched carbon chain. The former showed the strong conjugated effect and liked to interact with binding region II and III, as shown in Figure S11. Hydroxyl groups in the benzene ring formed hydrogen bonds with Leu141, Gly143, Ser144 and His163. The longest branched carbon chain would fit into binding region I. The less number of rings and higher steric hindrance limited the docking affinity of stilbenes.
The skeleton of xanthones (dibenzo-γ-pirones) has the same amount of ring as flavonoids, but their orientation is different. Although there were some xanthones showing higher docking affinities than the reference conivaptan, their performances were obviously worse than flavonoids. Docking results of xanthones against Mpro of SARS-CoV-2 were shown in Figure S12. In fact, the structure of xanthone ring was more compact and should have stronger conjugated effect. However, it also leaded to a larger and rigidified of the ring skeleton.
The skeleton of silydianin belong to flavonolignan class. The interaction between silydianin and Mpro of SARS-CoV-2 were shown in Figure S13. Silydianin formed hydrogen bonds with region III. Silydianin with polycyclic structure, could fit well with this active pocket.
Polyphenols were systematically analyzed, involving derivates from skeletons of gastrodin and ellagic acid. As shown in Figure S14, derivates from gastrodin and ellagic acid showing the strong conjugated effect, formed hydrogen bonds in region II and III, respectively. Polyphenols contain multiple hydroxyl groups. The oxygen of hydroxyl has many pairs of single electrons, which may be used to form more and stable delocalized electrons.
3.3. Dynamic simulation
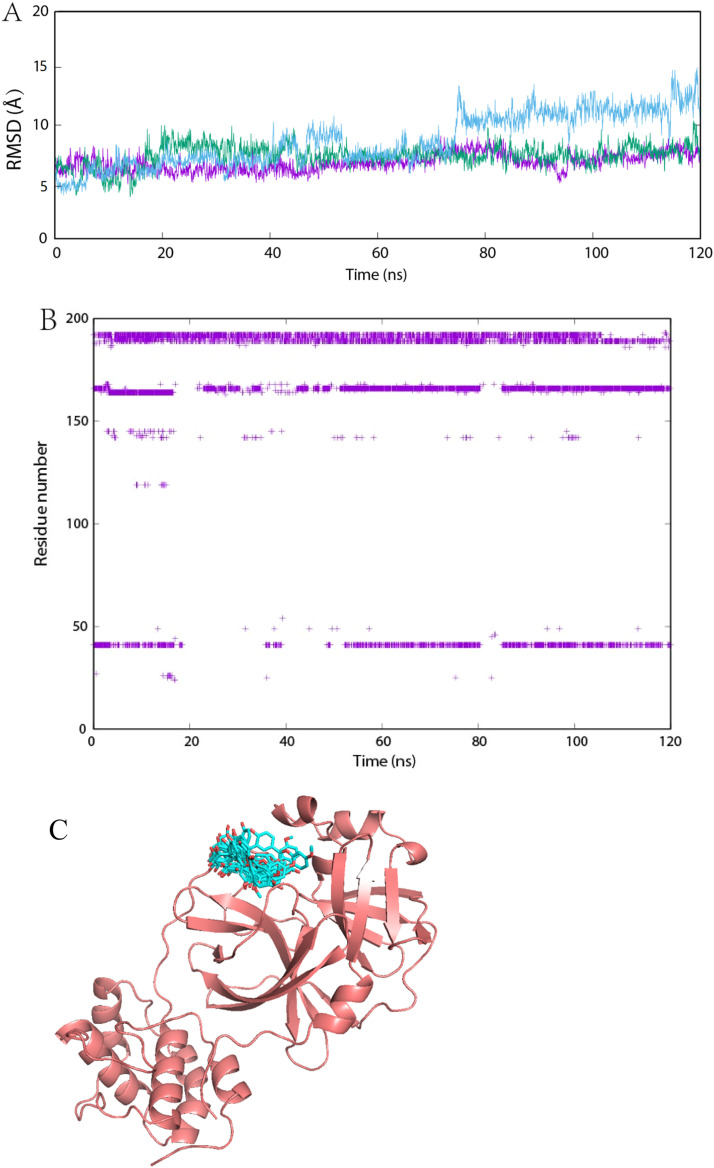
MD analysis was employed to further analyze the stability of the ligand-protein complex. Considering the current emergency situation and our limited computational power, only best candidate (derivate from ginkgetin, the first ligand in Figure S2) was used as an example for the systematically investigation. The ligand-protein complex obtained from the docking was employed as the first frame for further MD analysis. Fig. 10 indicated that screened ligand could form stable complex with Mpro of SARS-CoV-2 in each MD simulation. All of them were able to keep slight fluctuation in RMSD. Meanwhile, the molecular trajectory always stayed in the active pocket during whole MD simulation. Hydrogen bond interactions were further analyzed. The density of points in Fig. 10 represented the affinity strength. Results indicated that the derivate from ginkgetin could form stable hydrogen bonds with many residues in the active center of Mpro of SARS-CoV-2 (H41, H164, E166, Q189, T190, A191, and Q192). They all leaded to the very high stability of these ligand-protease complexes. This result was also better than our previous work [26].
Fig. 10.
MD study on ginkgetin-protein complexes. Time evolution of RMSD for ginkgetin molecules (A). Interactions plot between the active pocket of the main protease residues and ginkgetin (B). Visualization showing snapshots of ginkgetin during trajectories (C). Snapshots of ginkgetin in the structure of Mpro (PDB:6LU7), taken from the first MD simulation at a time interval of 120 ns, are shown. Ginkgetin is colored in cyan, Mpro in red.
3.4. ADMET and drug-likeness properties analyses
Finally, ADMET and drug-likeness analyses were shown in Supporting excel file (ADMET and drug-likeness). ADMET analysis was employed to predict the water solubility, acute oral toxicity, binding efficiency for plasma protein, lipophilicity, TPSA, bioavailability score, fraction Csp3, P-glycoprotein substrate, Caco-2 and human oral bioavailability of screened 465 compounds. Nine derivates from skeletons of imperialine, emodin, batatasin I, peimisine, quercetin, magnolol, evodiamine, glabrone and strychnine showed positive scores in P-glycoprotein substrate, Caco-2 and human oral bioavailability. According to the RO5, 465 candidates of inhibitors for Mpro of SARS-CoV-2 were further analyzed via molecular weight, hydrogen bond donor, hydrogen bond acceptor, rotatable bonds and calculated Log P. Results indicated that there were 403 kinds of candidates meeting the Lipinski's rule. Although hypericin had a high docking affinity (−10.3 kcal/mol), the whole molecule was very rigid and only had one rotatable bond. It is worth noting that ADMET and drug-likeness analyses only provided the reference due to their relevant low accuracy. This detail results were not shown, but could be found in Supporting excel file (ADMET and drug-likeness).
3.5. Enzyme inhibitory activity of plant extracts against Mpro of SARS-CoV-2
Next, relevant experiments were carried out to prove our docking results. Theoretically, the inhibition activity should be measured for every screened compound and a high purity will be provided to maximize the reaction efficiency. However, most of these compounds were very expensive derivatives and their availabilities were not easy. Some of them even needed to wait over half year. Given the urgency of the SARS-CoV-2 outbreak, we should focus on the use of existing resources for treating or preventing infections.
Plants extracts provided a relevantly rapid and effective way to prove partial docking results, which allows us to screen and predict the potential drug against SARS-CoV-2. Although these plants extracts were mixtures, they also showed strong relevance with these screened compounds. The Later derivates from the structural scaffolder of the former. Thus, they had same physicochemical properties and close docking affinities. The most advantage is that these TCMs and their plants extracts have been widely used as medicine or functional foods in many countries (such as China, Japan, South Korea). However, there are over thousands kinds of TCMs. It's impossible to test all of them in a work. Fortunately, this work provided much useful information for functions of ingredients of these TCMs to inhibit Mpro of SARS-CoV-2.
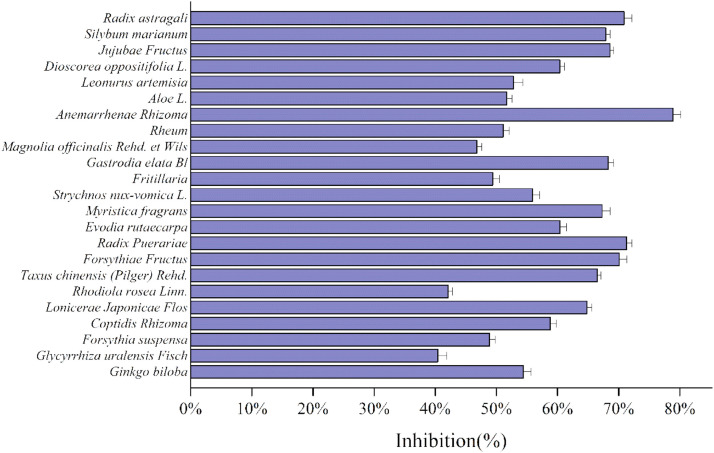
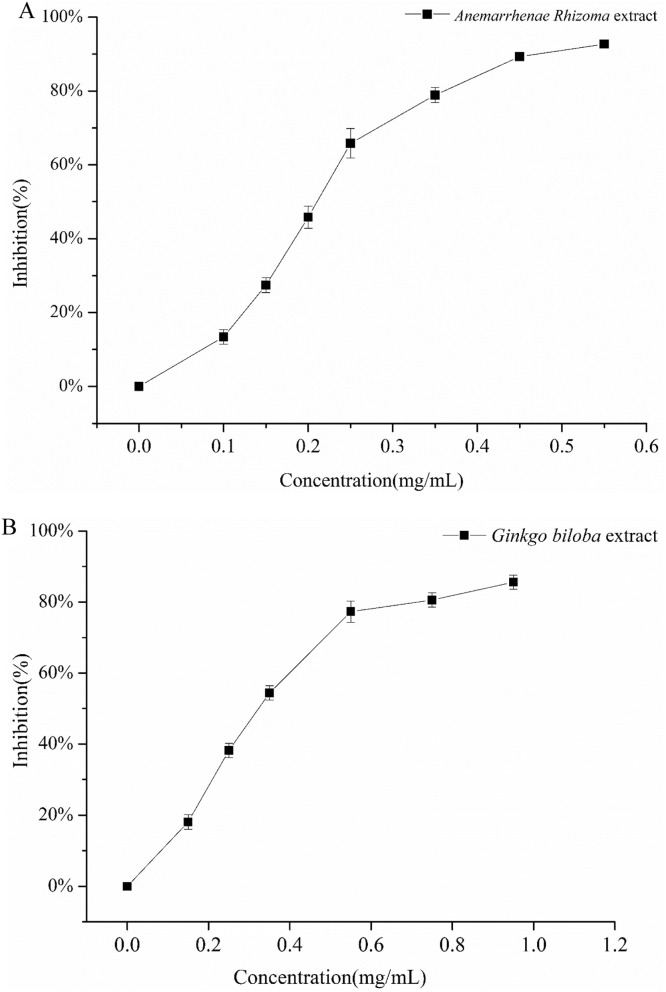
Basing on docking results, 23 kinds of plants extract from TCMs, which included most/all screened scaffolds, were used to study their inhibitory activity, as shown in Fig. 11 . Plants extracts from Anemarrhenae Rhizoma has the highest inhibitory efficiency in all cases. The median inhibitory concentration (IC50) of Anemarrhenae Rhizoma and Ginkgo biloba plants extracts were shown in Fig. 12 , reaching 0.23 mg/mL and 0.34 mg/mL, respectively.
Fig. 11.
Inhibitory activities of 23 kinds of Chinese herbal extracts against Mpro of SARS-CoV-2 by inhibitor screening kits with FRET protease assay.
Fig. 12.
The median inhibitory concentration (IC50) of plants extracts. Anemarrhenae Rhizoma(A). Ginkgo biloba (B).
Although Anemarrhenae Rhizoma might not contain the best candidate compound (ginkgetin), it still had many functional compounds, including kampferol (which shared the same scaffold with quercetin), sarsasapogenin, mangiferin. The docking affinities of these three phytochemistries and their derivates were −8.9 Kcal/mol, −8.8 Kcal/mol, −7.9 Kcal/mol, respectively. Quercetin widely exists in the nature and has many functional isomers/analogs/derivatives. Quercetin had good water solubility and satisfied Lipinski rule of 5 (Log S (ESOL)=−3.16, TPSA=131.36Ų). The most typical characteristic was quercetin consisted of benzene and benzopyran rings, belonging to flavonoids. Quercetin had three ring structures, which were arranged closely. Therefore, steric hindrance of quercetin was relevantly low, which they could be easily inserted into the binding pocket of Mpro of SARS-CoV-2 and formed the strong π–π stacking interaction with binding region II or III or IV showing in Figure S8. Sarsasapogenin as a kind of saponins, was the main component in Anemarrhenae Rhizoma and reached 6.0%. In addition, Sarsasapogenin has the good drug-like properties, high polar, and good bioavailability according to ADMET analysis (AlogP=5.79, Caco-2 = 0.5138(positive), TPSA=38.69 Ų). Sarsasapogeninan and its isomers/analogs/derivatives had six saturation rings. Their compact ring structures shared their ring edges. Therefore, the density and mobility of the electronic cloud is high, which made ring terpenoids easily interact with binding region II or III or IV showing in Figure S8. The previous analysis showed that the docking affinities against Mpro of SARS-CoV-2 exhibited a clear dependence on the number of ring structures of ligands. Meanwhile, xanthones were another main chemical component in Anemarrhenae Rhizoma, and their content were higher than 0.7%. Mangiferin, as a kind of bifenpyrone compounds, had good water solubility and good lipophiles (Log S (ESOL)=−2.65, Log Po/w (XLOGP3) = −0.07). The binding score of mangiferin (−7.9 Kcal/mol) was also close to the compare drug (conivaptan, −8.2 Kcal/mol). Although ginkgetin had the best docking score in molecular docking, the experimental result of Ginkgo biloba was not. It might be caused by its poor water solubility. On the other hand, contents of target compounds in these plant extracts were other limitations. The content of ginkgetin only reached 1.0%. For the moment, the plant extracts of Anemarrhenae Rhizoma without deep purification showed the acceptable inhibitory activity. The plant extracts are cheaper and more available, which were easier to be widely used in the functional food, adjuvant and medicine fields.
Lianhua Qingwen (LHQW) granule/capsule may be another good example, which was an innovative Chinese medicine for the treatment of COVID-19. Inhibitory activities of five TCMs ingredients from LHQW were also tested in our works (Fig. 11), involving Forsythiae Fructus, Rhodiola rosea Linn., Rheum, Radix glycyrrhizae, Lonicerae Japonicae Flos. As show in the Table 1, these TCMs had many functional compounds, which showed higher docking affinities than comparison. For example, the inhibitory activity of Forsythiae Fructus reached 70.1% for Mpro of SARS‑CoV‑2 (under our experimental conditions), which had many functional compounds with good binding scores including rutin, hypericin, ursolic acid, quercetin and its isomers/analogs/derivatives. The clinical researches showed that patient who used LHQW capsule had significantly higher recovery rate, and shorter median time to symptom recovery. Besides, on April 14, 2020, the National Medical Products Administration of China approved adding a new indication in LHQW (Yiling Pharmaceutical Inc.) for COVID-19 treatment in mild and normal cases [31].
4. Discussion
In this work, 1045,468 ligands’ structures derived from 116 kinds of TCM were employed to study their interaction with Mpro of SARS-CoV-2 by the molecular docking. 465 candidates of inhibitors for Mpro of SARS-CoV-2 could be divided into 11 classes by their scaffolds, involving biflavones, flavonoids, anthraquinones, polyphenols, phenylpropanoids, terpenoids, stilbenes, isoflavonoids, xanthones, flavonolignans and alkaloids. Basing on these simulation results, 23 kinds of TCMs plants extracts were employed to study their inhibitory activity. The inhibitory activity of 18 kinds of Chinese herbal extracts were higher than 50% at the concentration of 0.35 mg/mL, which involved Ginkgo biloba, Coptidis Rhizoma, Lonicerae Japonicae Flos, Taxus chinensis (Pilger) Rehd., Forsythiae Fructus, Radix Puerariae, Evodia rutaecarpa, Myristica fragrans, Strychnos nux-vomica L., Gastrodia elata Bl., Rheum, Anemarrhenae Rhizoma, Aloe L., Leonurus artemisia, Dioscorea oppositifolia L., Jujubae Fructus, Silybum marianum, Radix astragali. Plants extracts from Anemarrhenae Rhizoma has the highest inhibitory efficiency reaching 78.9% for Mpro of SARS‑CoV‑2. Many functional compounds were closely related to our daily life. If some (functional) foods show the potential role against this virus, SARS-CoV-2, these functional foods can be used for adjusting our daily diet to prevent infections as an easier and accepted operation in the world.
Significance statement
This study reports that 465 promising candidates against the main protease of SARS-CoV-2 were obtained from over one million compounds by high-throughput virtual screening. The dynamic simulation, ADMET and drug-likeness properties analyses were employed to further analyze the screened docking results. The structural functional of the main protease of SARS-CoV-2 were systematically investigated basing on these docking results. 23 kinds of TCMs plants extracts were chose to study their inhibitory activity for Mpro of SARS‑CoV‑2. This work would provide much possibilities for the drug design against SARS-CoV-2.
Funding
This work was supported by National Natural Science Foundation of China for Young Scholars (No. 22108227). Key Research and Invention Program in Shaanxi Province of China (Program No. 2020NY-127 and 2018NY-131). Shaanxi Association for Science and Technology for Young Scholars (No. 20220201)
CRediT authorship contribution statement
Xinyu Qi: Data curation, Formal analysis, Investigation, Software, Validation, Visualization, Writing – original draft. Binglin Li: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Writing – original draft, Writing – review & editing. Alejandra B. Omarini: Data curation, Formal analysis, Investigation, Validation, Writing – original draft, Writing – review & editing. Martin Gand: Investigation, Validation, Writing – original draft, Writing – review & editing. Xiaoli Zhang: Conceptualization, Funding acquisition, Project administration, Resources, Supervision. Jiao Wang: Conceptualization, Methodology, Project administration, Resources, Software, Supervision.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.molstruc.2022.133709.
Appendix. Supplementary materials
Data Availability
I have shared my data at supporting file
References
- 1.Shang J., et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kashyap K., Kakkar R. Pharmacophore-enabled virtual screening, molecular docking and molecular dynamics studies for identification of potent and selective histone deacetylase 8 inhibitors. Comput. Biol. Med. 2020;123(October 2019) doi: 10.1016/j.compbiomed.2020.103850. [DOI] [PubMed] [Google Scholar]
- 3.C S., S D.K.D.K., Ragunathan V., Tiwari P., A S., Brindha Devi B.D.B.D. Molecular docking, validation, dynamics simulations, and pharmacokinetic prediction of natural compounds against the SARS-CoV-2 main-protease. J. Biomol. Struct. Dyn. 2020;0(0):1–27. doi: 10.1080/07391102.2020.1815584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.“https://www.who.int/emergencies/diseases/novel-coronavirus-2019”.
- 5.Li D.D.D.D., Li Q.H.Q.H. SARS-CoV-2: vaccines in the pandemic era. Mil. Med. Res. 2021;8(1):1–15. doi: 10.1186/s40779-020-00296-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanjanasirirat P., et al. High-content screening of Thai medicinal plants reveals Boesenbergia rotunda extract and its component Panduratin A as anti-SARS-CoV-2 agents. Sci. Rep. 2020;10(1):1–12. doi: 10.1038/s41598-020-77003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang F., et al. A review of therapeutic agents and Chinese herbal medicines against SARS-COV-2 (COVID-19. Pharmacol. Res. 2020;158(May) doi: 10.1016/j.phrs.2020.104929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hosseini F.S.F.S., Amanlou M. Anti-HCV and anti-malaria agent, potential candidates to repurpose for coronavirus infection: virtual screening, molecular docking, and molecular dynamics simulation study. Life Sci. 2020;258(March) doi: 10.1016/j.lfs.2020.118205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keretsu S., Bhujbal S.P.S.P., Cho S.J.S.J. Rational approach toward COVID-19 main protease inhibitors via molecular docking, molecular dynamics simulation and free energy calculation. Sci. Rep. 2020;10(1):1–14. doi: 10.1038/s41598-020-74468-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ren W., et al. Research progress of traditional Chinese medicine against COVID-19. Biomed. Pharmacother. 2021;137 doi: 10.1016/j.biopha.2021.111310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pei Z., Lou Z., Zhang B., Wang H., Li Y. Development of a compound oral liquid containing herbal extracts and its effect on immunity and gastric mucosa. J. Food Sci. 2021;86(6):2684–2699. doi: 10.1111/1750-3841.15761. [DOI] [PubMed] [Google Scholar]
- 12.Li C., Wang L., Ren L. Antiviral mechanisms of candidate chemical medicines and traditional Chinese medicines for SARS-CoV-2 infection. Virus Res. 2020;286(May) doi: 10.1016/j.virusres.2020.198073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.hai Zhang D., lun Wu K., Zhang X., qiong Deng S., Peng B. In silico screening of Chinese herbal medicines with the potential to directly inhibit 2019 novel coronavirus. J. Integr. Med. 2020;18(2):152–158. doi: 10.1016/j.joim.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donno D., Beccaro G.L.G.L., Mellano M.G.M.G., Bonvegna L., Bounous G. Castanea spp. buds as a phytochemical source for herbal preparations: botanical fingerprint for nutraceutical identification and functional food standardisation. J. Sci. Food Agric. 2014;94(14):2863–2873. doi: 10.1002/jsfa.6627. [DOI] [PubMed] [Google Scholar]
- 15.Swain S.S.S.S., Panda S.K.S.K., Luyten W. Phytochemicals against SARS-CoV as potential drug leads. Biomed. J. 2021;44(1):74–85. doi: 10.1016/j.bj.2020.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.H. Su et al., “Discovery of baicalin and baicalein as novel, natural product inhibitors of SARS-CoV-2 3CL protease in vitro,” pp. 1–29, 2020, doi: 10.1101/2020.04.13.038687.
- 17.Jin Z., et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 18.Marinho E.M.E.M., et al. Virtual screening based on molecular docking of possible inhibitors of Covid-19 main protease. Microb. Pathog. 2020;148(June):1–6. doi: 10.1016/j.micpath.2020.104365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang C., et al. Screening and identifying of α-amylase inhibitors from medicine food homology plants: insights from computational analysis and experimental studies. J. Food Biochem. 2020;44(12):1–12. doi: 10.1111/jfbc.13536. [DOI] [PubMed] [Google Scholar]
- 20.“(https://zinc15.docking.org/).”
- 21.Odhar H.A.H.A. Molecular docking and dynamics simulation of FDA approved drugs with the main protease from 2019 novel coronavirus. Bioinformation. 2020;16(3):236–244. doi: 10.6026/97320630016236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang H., et al. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3(10) doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang F Y.H.Y.H., Chen C., Tan W., Yang K. Structure of main protease from human coronavirus NL63: insights for wide spectrum anti-coronavirus drug design. Sci. Rep. 2016 doi: 10.1038/srep22677. [Online]. Available: doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L., et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (80-.). 2020;3405(March):1–9. doi: 10.1126/science.abb3405. [Online]. Available2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Günther S., Reinke P., Fernández-García Y., Lieske J, et al. X-ray screening idfentifies active site and allosteric inhibitors of SARS-CoV-2 main protease. Science (80-.). 2021;7945(April) doi: 10.1126/science.abf7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang B., Zhang J., Omarini X., Li A. Virtual screening for functional foods against the main protease of 2019 novel coronavirus. J. Food Biochem. 2020 doi: 10.1111/jfbc.13481. [Online]. Available. [DOI] [PubMed] [Google Scholar]
- 27.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7(March):1–13. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lipinski C.A.C.A. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov. Today Technol. 2004;1(4):337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 29.“https://www.drugs.com/.” .
- 30.Mercader A.G.A.G., Pomilio A.B.A.B. 2D- and 3D-QSAR Studies of Flavonoids, Biflavones and Chalcones: antiviral, Antibacterial, Antifungal, and Antimycobacterial Activities. Anti-Infective Agents. 2013;10(1):41–54. doi: 10.2174/2211362611201010041. [DOI] [Google Scholar]
- 31.Xia Q.D.Q.D., et al. Network pharmacology and molecular docking analyses on Lianhua Qingwen capsule indicate Akt1 is a potential target to treat and prevent COVID-19. Cell Prolif. 2020;53(12):1–13. doi: 10.1111/cpr.12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
I have shared my data at supporting file