Abstract
In this study, we optimized procedures to enumerate viruses from marine sediments by epifluorescence microscopy using SYBR Green I as a stain. The highest virus yields from the bulk of the sediments were obtained by utilizing pyrophosphate and 3 min of sonication. The efficiency of extraction benthic viruses by pyrophosphate-ultrasound treatment was about 60% of the extractable virus particles. Samples treated with nucleases had increased virus counts, suggesting a masking effect of extracellular DNA. No significant differences were observed between virus counts obtained by epifluorescence microscopy and transmission electron microscopy. Both formaldehyde and glutaraldehyde gave significant reductions of virus counts after only 24 h of sediment storage, but no further loss occurred after 7 days.
Viruses are now considered to be an important component of all aquatic microbial communities. The reevaluation of the role of viruses in marine ecosystems is due to the discovery of very high virus abundance (see reference 13 for a review). Recent studies have stressed the ecological implication of viruses in the release of dissolved organic matter, nutrient recycling (18), and the pathways of organic carbon utilization, with cascade effects on marine microbial food webs and organic-matter cycling (12). The available methods for the determination of virus abundance in aquatic environments include counting by transmission electron microscopy (TEM) (1, 2, 16, 21), by flow cytometry (17), and by epifluorescence microscopy (EFM) (9, 14, 20, 26, 29). The last technique allows an accurate and easily performed enumeration, avoiding the use of expensive and bulky equipment (13). In addition, EFM is reported to be up to seven times more efficient than TEM for counting viruses (15, 28).
Available information dealing with benthic virus ecology is scant. This is due to the lack of adequate protocols for easily determining their abundance and distribution in marine sediments (6). The main objective of this work was the optimization of procedures to enumerate viruses in different marine sediments. We focused our attention on EFM counting using SYBR Green I as a stain (20) in order to address the following issues: (i) virus dislodgment from sediment particles (using surfactant and ultrasound treatments), (ii) the efficiency of virus extraction from bulk sediment (by the number of postsonication washings), and (iii) stain-counting accuracy and efficiency (by removing possible interferences due to extracellular DNA in virus counting and by comparison with TEM counts). In addition, we tested the effects of preservatives on virus abundance in long-time-course experiments carried out on fixed sediment samples.
Sediment sampling and selection of sediment.
In order to make the protocol for virus counting suitable for the widest variety of sediment samples, two different sediment types were selected in this study: shallow sands and deep-sea muds. As deep-sea samples are not generally analyzed immediately, the effects of long-term storage with preservatives were also investigated. Sandy-sediment samples (modal grain size, between 125 and 250 μm) were collected by hand coring (using Plexiglas tubes [4.7 cm inside diameter]) in June and in September 1999 near the low tidal line of a quiet tidal flat in Falconara Beach (43°6′N, 13°5′E; northern Adriatic Sea). Deep-sea sediment samples were collected in March 1998 in the Porcupine Abyssal Plain (northeastern Atlantic Ocean at 4,800-m depth; 48°50′N, 16°29′W). This area, characterized by strong seasonality and a high interannual variability in organic-matter inputs (8, 19), can be considered representative of typical deep-sea conditions (23). Undisturbed sediment samples were collected with a multicorer (Maxicorer; inside diameter, 9.0 cm; depth penetration, >20 cm). For virus analysis, immediately after sampling, subsamples of about 0.5 ml of the top 5 mm of both sediment types were taken from different cores and deployments, added to 3 ml of prefiltered (0.02-μm pore size) seawater containing 2% formalin, and stored at 4°C. All virus analyses of fixed material were performed within 3 to 4 weeks after fixation. Long time course experiments to study the effects of formaldehyde and glutaraldehyde storage were carried out on sandy sediments. The glassware utilized for virus counts was carefully cleaned by soaking it in 10% HCl overnight, rinsed with MilliQ water and subsequently autoclaved. All the solutions were prepared with MilliQ water, filtered through 0.02-μm-pore-size filters, and then autoclaved.
Treatments for virus dislodgment.
Sediment samples collected in the northeastern Atlantic (triplicate 0.5-ml samples) were added to 4.0 ml of MilliQ water and 1.0 ml of sodium pyrophosphate solution (10-mM final concentration) and incubated for 15 min. Additional sediment samples (n = 3; 0.5 ml) without pyrophosphate were added to 5 ml of MilliQ water and served as controls. After incubation, all samples were shaken manually for 1 min and then centrifuged (800 × g; 1 min) to reduce interference due to suspended particles. Aliquots of the supernatant were diluted 500 to 1,000 times and filtered through 0.02-μm-pore-size Anodisc 25 membrane filters (pressure, <100 mm of Hg). The filters were then stained with 20 μl of SYBR Green I (Lot no. 4967-30; diluted 20-fold in MilliQ water; optical density at 495 nm = 1.357) for 15 min in the dark, rinsed twice with 1 ml of MilliQ water (in order to eliminate fluorescence background noise), and analyzed by EFM using a Zeiss Axioplan microscope equipped with a 50-W lamp. Ten to 50 fields were viewed at ×1,000 magnification, and a minimum of 400 viruses were counted. Viruslike particles (VLP) were discriminated from bacteria (0.2- to 2-μm diameter) by their dimensions (0.015- to 0.2-μm diameter [20]). As sodium pyrophosphate enhanced virus extraction efficiency, any further steps were carried out with this surfactant. In order to test the combined effects of pyrophosphate and ultrasound treatments on virus extraction, muddy and sandy sediments (n = 3; 0.5 ml for both sediment types) were added to pyrophosphate and sonicated (Branson 2200 sonifier; 100 W; 47 kHz) for 0, 1, 3, 8, and 15 min in an ice bath to prevent overheating. The sonication was interrupted for 30 s every minute, during which time the samples were shaken manually. After each treatment, the sample was centrifuged, and aliquots of the supernatant were processed as described above.
Postsonication extraction efficiency.
Once the optimal sonication time was identified, the efficiency of virus detachment from sediment particles was checked by estimating the ratio of virus abundance after the first extraction with ultrasound and pyrophosphate treatment versus the cumulative virus abundance obtained by this procedure plus three further washing steps. The added steps were the following: (i) an aliquot of supernatant obtained from deep-sea sediment samples (0.5 ml of sediment plus 4.0 ml of MilliQ water and 1.0 ml of sodium pyrophosphate) after sonication (3 min) and centrifugation was withdrawn and treated for counting as described above; (ii) the remaining supernatant was carefully discharged, the pellet was resuspended with 5 ml of MilliQ water, shaken for 1 min, and centrifuged again, an aliquot of the supernatant was withdrawn, and viruses were counted as described above; and (iii) this procedure was repeated three times (since after the third washing, less than 5% of the total virus abundance was encountered).
Interference with virus enumeration due to extracellular DNA.
In order to eliminate uncertainties in virus counting due to extracellular DNA interference, we tested the effect of nuclease treatment on sediment samples. Twenty-five microliters of DNase I from bovine pancreas (1.9 U ml−1), 10 μl of nuclease P1 from Penicillium citrinum (4 U ml−1), 10 μl of nuclease S1 from Aspergillus orizae (2.3 U ml−1), and 10 μl of esonuclease 3 from Escherichia coli (1.9 U ml−1) were added to 1.0-ml aliquots of the supernatant obtained from fresh sandy sediments and incubated for 15 min at room temperature. Additional aliquots of the supernatant (1.0 ml) without enzymes were incubated under the same conditions and served as controls.
Comparison of EFM and TEM counts.
Virus enumeration performed by EFM from fixed sandy sediments was compared to TEM virus counting. For TEM analyses, we omitted the preconcentration procedure (i.e., ultracentrifugation) before mounting the sample on a grid (14), since the number of virus particles in sediment samples is expected to be very high (6, 21, 25). Subaliquots of 10 μl of the supernatant obtained as described for EFM counting were put onto 400-mesh Formvar-coated Cu grids, stained with 2% uranyl acetate, and dried under silica gel (14). The grids were examined by a Philips CM 200 TEM at a magnification of ×38,000, and view fields were counted until the total counts exceeded 200.
Effect of preservatives in long-term experiments.
After sampling was done, about 50 ml of fresh sandy sediment was gently mixed and carefully split into three subaliquots: one was unfixed and immediately analyzed, the second was fixed with formalin (2% final concentration), and the last was fixed with glutaraldehyde (2% final concentration). The sediment samples were processed in triplicate as described above, but the pyrophosphate final concentration was reduced to 5 mM as suggested by Epstein and Rossel (11) for benthic bacterial extraction from coarser sediments. Fixed samples stored at 4°C were analyzed after 1, 7, 30, and 90 days.
Statistical analyses.
To test differences between virus counts, the t test was employed throughout the study. Its use was justified, as (i) the density data were normally distributed (they were checked by a standard graphical test [24]) and (ii) the test for homogeneity of variances (the Fmax test [24]) showed that our samples were homoscedastic. VLP dispersion was evaluated to test the homogeneity of the virus distribution by calculating the coefficient of variation (CV) (CV = standard deviation/mean ×100).
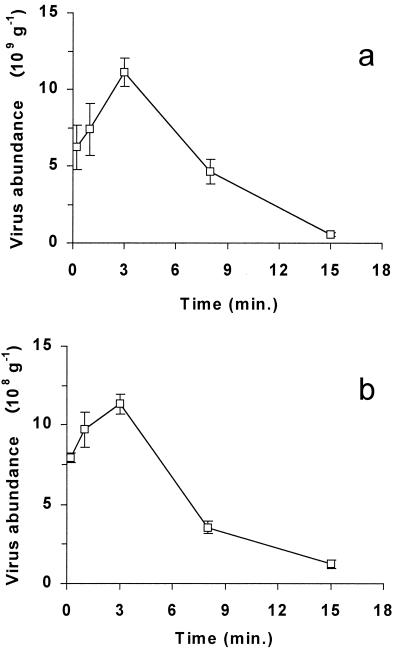
Deep-sea sediment samples incubated with sodium pyrophosphate displayed higher virus counts than untreated samples [(14.6 ± 2.79) × 109 and (9.94 ± 6.12) × 109 viruses g−1 (dry weight) of sediment, respectively], but the differences were not statistically significant (t test; P = 0.296). However, pyrophosphate-treated samples were characterized by significantly lower CVs (19.1 versus 61.6%; t test; P < 0.05). The effects of sonication on virus counts carried out on both abyssal and coastal sediments are reported in Fig. 1. The highest virus counts for both sediment types (11.1 × 109 and 1.13 × 109 viruses g−1 for muddy and sandy sediments, respectively) were obtained after 3 min of sonication and were significantly higher (two- to four fold) (t test; P < 0.01) than values obtained without sonication (i.e., with simple shaking). Further increases in the sonication time decreased virus counts, and a sonication lasting 15 min reduced virus counts by about 1 order of magnitude (t test; P < 0.01 for both sediment types).
FIG. 1.
Effect of sonication on virus abundance in deep-sea (a) and sandy (b) sediments. The standard deviations are shown.
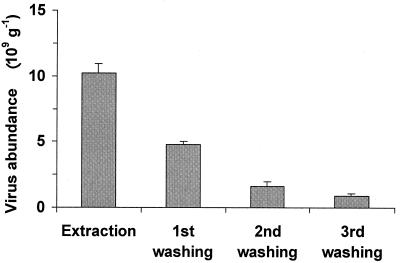
The efficiency of virus extraction by pyrophosphate-ultrasound treatment was ca. 60% (Fig. 2). The abundances of viruses extracted by this procedure were significantly lower than the total cumulative virus abundance (t test; P < 0.01). The subsequent first and second washings recovered 27.5 and 9.0% of the total virus abundance, and after the third wash step, <5% was recovered.
FIG. 2.
Postsonication extraction efficiency of virus recovery from deep-sea sediments. The standard deviations are shown.
The virus abundances obtained from nuclease-treated samples were significantly higher than those in untreated samples [(5.11 ± 0.15) × 108 and (4.62 ± 0.19) × 108 viruses g−1 (dry weight) of sediment, respectively (t test; P < 0.05)]. Counts obtained by TEM and EFM (from fixed sediment samples collected at Falconara) displayed no significant differences [(1.54 ± 0.15) × 109 and (1.46 ± 0.13) × 109 viruses g−1 (dry weight) of sediment, respectively (t test; P = 0.513)].
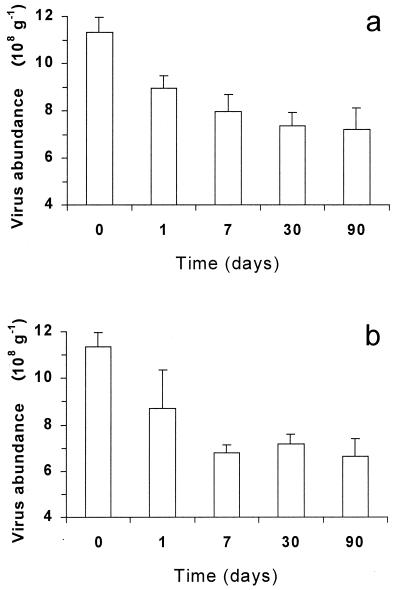
Sediments fixed with both formaldehyde and glutaraldehyde showed a significant decrease in virus numbers after only 24 h of preservation (21 and 23%, respectively [t test; P < 0.01]) (Fig. 3). After 7 days of formaldehyde and glutaraldehyde storage, virus abundance decreased from 30 to 40%, respectively, compared to that in fresh sediment samples and then remained constant for up to 90 days in storage.
FIG. 3.
Effects of preservatives on virus abundance in sandy sediments. Reported are formaldehyde (a) and glutaraldehyde (b) long-term storage. The standard deviations are shown.
Virus identification by EFM has been attempted since the early 1990s (14, 22, 26), and the uncertainty in recognizing viruses has induced scientists to refer to the products of these counts as VLP (2, 3). In this study, we employed SYBR Green I, which has the highest staining efficiency under EFM and flow cytometry (17) compared to other fluorochromes (such as DAPI [4′,6′-diamidino-2-phenylindole] and Yo-Pro I [20]). Since all these fluorochromes bind to nucleic acids, the possibility that some small bacteria (<0.3-μm diameter) may be counted as viruses cannot be excluded. Noble and Fuhrman (20) reported that even if all small bacteria were counted as viruses, the overestimation of the total virus counts would be negligible. In this study, utilizing SYBR Green I at a concentration higher than that previously reported for virus counts in water samples (17, 20), we noticed that virus-size fluorescent particles displayed a higher brightness than bacterial fluorescence. Also, the confidence in virus counts reported in this study is based on two factors: (i) TEM and EFM provided almost identical estimates of virus density and (ii) extracellular-DNA removal increased virus counts. In this regard, Drake et al. (9) reported no significant differences between the VLP densities of DNase-treated pore water samples and control samples. In this study, surprisingly, we found significantly higher virus densities in nuclease-treated samples. As sediments might contain large amounts of extracellular DNA (5, 7, 8), such increased yields due to nuclease treatment could be related to reduction of the fluorescence noise (i.e., reducing the masking effect) due to SYBR staining of extracellular DNA, facilitating virus counting.
Previous studies proved that the use of 10 mM pyrophosphate allowed the highest virus yields from formaldehyde-preserved freshwater sediments (16). In accordance with this, we found that the use of higher pyrophosphate concentrations made the optical field opalescent under EFM, thus making virus counting difficult. Pyrophosphate treatment did not significantly increase virus extraction, but the use of this surfactant at a final concentration not exceeding 10 mM significantly lowered CVs (about threefold lower than in untreated samples). Similar results have been reported for benthic bacteria (11), indicating that the use of pyrophosphate increases counting accuracy under EFM so that a smaller number of replicate analyses is required to obtain acceptable CV values.
Maranger and Bird (16) utilized 45s of sonication to dislodge viruses from sediment samples. The results of the present study confirm that sonication is crucial for maximizing virus recovery from marine sediments. However, the optimal time of sonication to achieve the highest virus yields was never tested before. In this study, the highest extraction efficiency from both sediment types was obtained with 3 min of sonication. Similar results have been reported for bacterial extraction from different sediment types (4, 11). As observed for benthic bacteria, longer sonication times (up to 15 min) reduced virus abundance to 1/10.
The extraction efficiency of benthic viruses after pyrophosphate and sonication treatment was approximately 60% of the total cumulative virus number. Similarly low extraction efficiencies were reported for benthic bacteria by Ellery and Schleyer (10), which suggested the need for a correction factor (of 1.44) for inadequacy in bacterial extraction.
Xenopoulos and Bird (29) observed a dramatic decrease in virus abundance (up to 75%) in water samples during the first 4 weeks of formaldehyde storage. A significant decrease in virus counts after glutaraldehyde preservation has also been observed in samples of Phaeocystis infected by viruses (17). Similarly, Turley and Hughes (27) reported a bacterial decrease of 39% during the first 40 days of seawater sample storage with glutaraldehyde. In accordance with these findings, we found that both formaldehyde and glutaraldehyde storage caused significant reductions in virus counts after only 24 h of preservation. After 7 to 90 days of preservation, no further virus loss was observed.
Acknowledgments
We acknowledge the support from the European Commission's Marine Science and Technology Program (MAST III) under Mass Transfer and Ecosystem Response (MATER; MAS3-CT-960051) and High-Resolution Spatial and Temporal Study of the Benthic Biology and Geochemistry in a NorthEastern Atlantic Abyssal Locality (BENGAL; MAS3-CT-950018).
We thank Monica Armeni (University of Ancona) for her technical assistance and Tanya Hall (University of Athens) for improving the English form.
REFERENCES
- 1.Berg O/, Borsheim K Y, Bratbak G, Heldal M. High abundance of viruses found in aquatic environments. Nature. 1989;340:467–468. doi: 10.1038/340467a0. [DOI] [PubMed] [Google Scholar]
- 2.Borsheim K Y, Bratbak G, Heldal M. Enumeration and biomass estimation of planktonic bacteria and viruses by transmission electron microscopy. Appl Environ Microbiol. 1990;56:352–356. doi: 10.1128/aem.56.2.352-356.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brussaard C P D, Kempers R S, Kop A J, Riegman R, Heldal M. Virus-like particles in a summer bloom of Emiliana huleyi in the North Sea. Aquat Microb Ecol. 1996;10:105–113. [Google Scholar]
- 4.Danovaro R, Feminò A, Fabiano M. Comparison between different methods for bacterial counting in marine sandy sediments. Boll Mus Ist Biol Univ Genova. 1994;58:142–152. [Google Scholar]
- 5.Danovaro R, Dell'Anno A, Pusceddu A, Fabiano M. Nucleic acid concentrations (DNA, RNA) in the continental and deep-sea sediments of the Eastern Mediterranean: relationships with seasonally varying organic inputs and bacterial dynamics. Deep-Sea Res. 1999;46:1077–1094. [Google Scholar]
- 6.Danovaro R, Serresi M. Viral density and virus-to-bacterium ratio in deep-sea sediments of the Eastern Mediterranean. Appl Environ Microbiol. 2000;66:1857–1861. doi: 10.1128/aem.66.5.1857-1861.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dell'Anno A, Fabiano M, Duineveld G C A, Kok A, Danovaro R. Nucleic acid (DNA, RNA) quantification and RNA/DNA ratio determination in marine sediments: comparison of spectrophotometric, fluorometric, and high-performance liquid chromatography methods and estimation of detrital DNA. Appl Environ Microbiol. 1998;64:3238–3245. doi: 10.1128/aem.64.9.3238-3245.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dell'Anno A, Fabiano M, Mei M L, Danovaro R. Pelagic-benthic coupling of nucleic acids in an abyssal location of the northeastern Atlantic Ocean. Appl Environ Microbiol. 1999;65:4451–4457. doi: 10.1128/aem.65.10.4451-4457.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drake L A, Choi K H, Haskell A G E, Dobbs F C. Vertical profiles of virus-like particles and bacteria in the water column and sediments of Chesapeake Bay, USA. Aquat Microb Ecol. 1998;16:17–25. [Google Scholar]
- 10.Ellery W N, Scheyer M H. Comparison of homogenization and ultrasonication as techniques in extracting attached sedimentary bacteria. Mar Ecol Prog Ser. 1984;15:247–250. [Google Scholar]
- 11.Epstein S E, Rossel J. Enumeration of sandy sediment bacteria: search for optimal protocol. Mar Ecol Prog Ser. 1995;117:289–298. [Google Scholar]
- 12.Fuhrman J A, Noble R T. Viruses and protists cause similar bacterial mortality in costal seawater. Limnol Oceanogr. 1995;40:1236–1242. [Google Scholar]
- 13.Fuhrman J A. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399:541–548. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- 14.Hara S, Terauchi K, Koike I. Abundance of viruses in marine waters: assessment by epifluorescence and transmission electron microscopy. Appl Environ Microbiol. 1991;57:2731–2734. doi: 10.1128/aem.57.9.2731-2734.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hennes J E, Suttle C A. Direct counts of viruses in natural waters and laboratory cultures by epifluorescence microscopy. Limnol Oceanogr. 1995;40:1050–1055. [Google Scholar]
- 16.Maranger R, Bird D E. High concentrations of viruses in the sediments of Lac Gilbert, Quebec. Microb Ecol. 1996;31:141–151. doi: 10.1007/BF00167860. [DOI] [PubMed] [Google Scholar]
- 17.Marie D, Brussaard C P D, Thyrhaug R, Bratbak G, Vaulot D. Enumeration of marine viruses in culture and natural samples by flow cytometry. Appl Environ Microbiol. 1999;65:45–52. doi: 10.1128/aem.65.1.45-52.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Middelboe M, Jørgensen N O G, Kroer N. Effects of viruses on nutrient turnover and growth efficiency of noninfected marine bacterioplankton. Appl Environ Microbiol. 1996;62:1991–1997. doi: 10.1128/aem.62.6.1991-1997.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newton P R, Lampitt R S, Jickell T D, King P, Boutle C. Temporal and spatial variability of biogenic particle fluxes during the JGOFS NE-Atlantic process studies at 47°N 20°W. Deep-Sea Res. 1994;41:1617–1642. [Google Scholar]
- 20.Noble R T, Fuhrman J A. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat Microb Ecol. 1998;14:113–118. [Google Scholar]
- 21.Paul J H, Rose J B, Jiang S C, Kellogg C A, Dickson L. Distribution of viral abundance in the reef environment of Key Largo, Florida. Appl Environ Microbiol. 1993;59:718–724. doi: 10.1128/aem.59.3.718-724.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Proctor L M, Fuhrman J A. Mortality of marine bacteria in response to enrichments of the virus size fraction from seawater. Mar Ecol Prog Ser. 1992;87:283–293. [Google Scholar]
- 23.Rice A L, Thurston M H, Bett B J. The IOSDL DEEPSEAS programme: introduction and photographic evidence for the presence and absence of a seasonal input of phytodetritus at contrasting abyssal sites in the northern-eastern Atlantic. Deep-Sea Res. 1994;41:1305–1320. [Google Scholar]
- 24.Sokal R R, Rohlf F J. Introduction to biostatistics. 2nd ed. San Francisco, Calif: W. H. Freeman; 1987. [Google Scholar]
- 25.Steward G F, Smith D C, Azam F. Abundance and production of bacteria and viruses in the Bering and Chukchi Sea. Mar Ecol Prog Ser. 1996;131:287–300. [Google Scholar]
- 26.Suttle C A, Chan A M, Cottrell M T. Infection of phytoplankton by viruses and reduction of primary productivity. Nature. 1990;387:467–469. [Google Scholar]
- 27.Turley C M, Hughes D J. Effects of storage on direct estimates of bacterial numbers of preserved seawater samples. Deep-Sea Res. 1992;39:375–394. [Google Scholar]
- 28.Weinbauer M G, Suttle C A. Comparison of epifluorescence and transmission electron microscopy for counting viruses in natural marine waters. Aquat Microb Ecol. 1997;13:225–232. [Google Scholar]
- 29.Xenopoulos M A, Bird D F. Virus à la sauce Yo-Pro: microwave enhanced staining for counting viruses by epifluorescence microscopy. Limnol Oceanogr. 1997;42:1648–1650. [Google Scholar]