Abstract
We isolated a strain of Rhodopseudomonas palustris (RCB100) by selective enrichment in light on 3-chlorobenzoate to investigate the steps that it uses to accomplish anaerobic dechlorination. Analyses of metabolite pools as well as enzyme assays suggest that R. palustris grows on 3-chlorobenzoate by (i) converting it to 3-chlorobenzoyl coenzyme A (3-chlorobenzoyl–CoA), (ii) reductively dehalogenating 3-chlorobenzoyl–CoA to benzoyl-CoA, and (iii) degrading benzoyl-CoA to acetyl-CoA and carbon dioxide. R. palustris uses 3-chlorobenzoate only as a carbon source and thus incorporates the acetyl-CoA that is produced into cell material. The reductive dechlorination route used by R. palustris for 3-chlorobenzoate degradation differs from those previously described in that a CoA thioester, rather than an unmodified aromatic acid, is the substrate for complete dehalogenation.
Dechlorination is the first critical step in the bacterial degradation of many halogenated pollutants. A number of different dechlorination reactions have been described, including hydrolytic dechlorinations and oxidative dechlorinations (13). Some anaerobic bacteria can dechlorinate aliphatic and aromatic compounds by means of reductive dehalogenases. Halorespiring microbes, many of which are related to sulfate-reducing bacteria, catalyze high rates of reductive dehalogenation using chlorinated compounds as terminal electron acceptors in anaerobic respiration (7, 12). Two strains of Rhodopseudomonas palustris, a purple nonsulfur phototrophic bacterium, have been reported to utilize 3-chlorobenzoate as a carbon source under anaerobic conditions with light as the energy source (14, 16, 28). Here we have investigated the metabolic route used by a third, newly isolated strain of this microbe to photometabolize 3-chlorobenzoate.
Isolation of RCB100, a 3-chlorobenzoate-degrading R. palustris strain.
The bacterial strain used in these studies was isolated from an anaerobic enrichment in a mineral medium (11, 15) that contained 2 mM 3-chlorobenzoate as the sole organic source and was incubated in light. The enrichment medium had been inoculated with damp soil from Cascadilla Creek in Ithaca, New York. After the enrichment culture became turbid and deep red in color, a pure isolate was obtained by repeated streaking on plates of the same medium, which were also incubated anaerobically in light. The isolated strain had the characteristic “dumbbell” morphology of R. palustris and was named RCB100. RCB100 was later also grown aerobically in mineral medium with shaking in the dark in order to test its ability to grow on various compounds in the presence of oxygen.
We amplified the 16S rRNA gene from strain RCB100 by PCR, using primers designed to amplify 16S ribosomal DNA (rDNA) from all eubacterial species (29). The DNA sequence corresponding to bases 50 to 519 of the Escherichia coli rDNA was determined at the University of Iowa DNA Facility. RCB100 was identified as a strain of R. palustris based on the 100% sequence identity of the region sequenced to the corresponding region of the type strain of R. palustris, ATCC 17001.
Growth characteristics.
The ability of RCB100 to grow on various compounds was screened by supplying a diffusion gradient of the compounds by the auxanography technique (11). Growth rates were determined in liquid medium for those compounds that supported growth on auxanographic plates. Strain RCB100 grew anaerobically in light on 3-chlorobenzoate with an average doubling time of 21 h. It also grew anaerobically in light with benzoate (12 h), 3-hydroxybenzoate (50 h), 3-bromobenzoate (40 h), and 3-chloropropionate (19 h). These compounds were supplied at a concentration of 2 mM. This strain did not grow anaerobically on 4-hydroxybenzoate, 4-chlorobenzoate, 4-bromobenzoate, 4-isopropylbenzoate, vanillate, 3,4-difluorobenzoate, 2,4-dichlorobenzoate, 2,5-dichlorobenzoate, or 3,4-dichlorobenzoate. Strain RCB100 grew aerobically with 4-hydroxybenzoate and protocatechuate but not with 3-hydroxybenzoate, 3-chlorobenzoate, or benzoate. By contrast, the R. palustris strain that we have used previously for studies of anaerobic aromatic compound degradation, CGA009, grows anaerobically on 4-hydroxybenzoate but did not grow anaerobically with 3-chlorobenzoate or 3-bromobenzoate supplied as the sole carbon source. However, when strain CG009 was supplied with both 3-chlorobenzoate and benzoate under anaerobic conditions, it grew to the yields that would be expected if both compounds were completely degraded. Thus it appears that strain CGA009 can degrade 3-chlorobenzoate if benzoate is also present, perhaps as an inducer of enzymes required for 3-chlorobenzoate degradation. A previous study has shown that benzoate is a necessary cosubstrate for 3-chlorobenzoate degradation by strain WS17 of R. palustris (14).
Dechlorination of 3-chlorobenzoate.
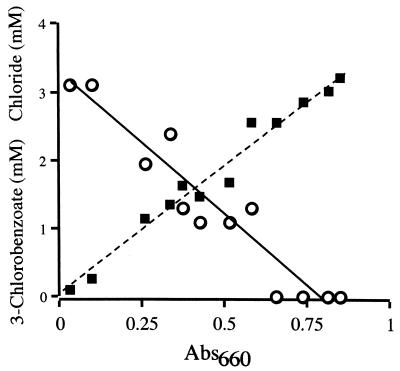
When RCB100 was grown on 3-chlorobenzoate, chloride ions, measured colorimetrically (2), accumulated in the culture medium in proportion to the amount of 3-chlorobenzoate consumed (Fig. 1). The concentration of 3-chlorobenzoate was determined by high-pressure liquid chromatography (HPLC) (21). We looked for, but did not detect at any stage during growth, free benzoate in culture supernatants. This indicates that free 3-chlorobenzoate was not reductively dechlorinated.
FIG. 1.
Consumption of 3-chlorobenzoate (open circles connected by a solid line) and release of chloride (boxes connected by a dashed line) during anaerobic growth of R. palustris strain RCB100 on 3-chlorobenzoate as the sole carbon source. Abs660, A660.
Conversion of 3-chlorobenzoate to benzoyl coenzyme A (benzoyl-CoA).
Thin-layer chromatography was used to analyze intracellular metabolites formed from 3-chlorobenzoate by whole cells of RCB100 (19). In short, illuminated, washed suspensions of 3-chlorobenzoate-grown cells of R. palustris RCB100 from logarithmic-phase cultures were incubated anaerobically for short periods of time with 500 μM [7-14C]3-chlorobenzoate (0.55 mCi/mmol; American Radiolabeled Chemicals, St. Louis, Mo.). Samples (0.1 ml) were taken at intervals ranging from 2 to 5 min after addition of labeled 3-chlorobenzoate and were added to 1 ml of boiling water to release soluble intracellular contents. This material was concentrated and chromatographed on cellulose thin-layer plates (19), which were then exposed to X-ray film.
3-Chlorobenzoyl–CoA and benzoyl-CoA, used as standards, were synthesized from the acid chlorides as previously described (17). 3-Hydroxybenzoyl–CoA was synthesized using the method described by Merkel et al. (19) with the omission of the final alkali treatment. The crude products were purified by HPLC (21), and their expected molecular weights were confirmed by electrospray mass spectrometry at the University of Iowa High Resolution Mass Spectrometry Facility.
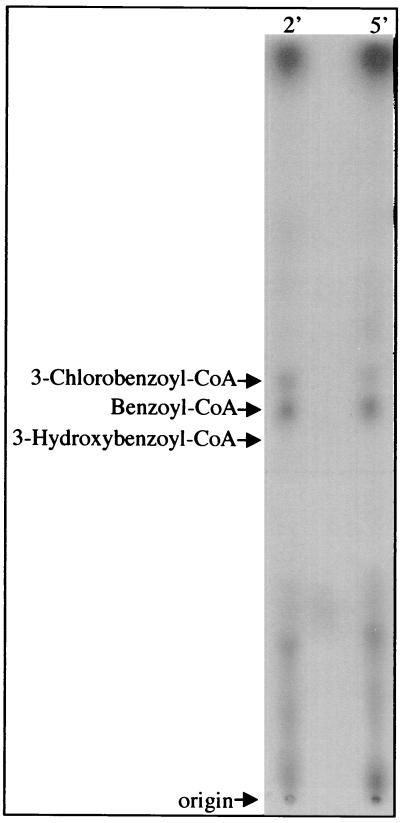
Radioactive derivatives of [7-14C]3-chlorobenzoate that comigrated with authentic 3-chlorobenzoyl–CoA (Rf, 0.56) and benzoyl-CoA (Rf, 0.52) were present in cells 2 min following their exposure to 3-chlorobenzoate, with 3-chlorobenzoyl–CoA disappearing by 5 min after exposure to radiolabeled 3-chlorobenzoate (Fig. 2). We did not observe the formation of 3-hydroxybenzoyl–CoA as an intermediate of 3-chlorobenzoate degradation. We thought that this compound might be formed because dehalogenating enzymes have been described that catalyze a hydrolytic attack on CoA-modified substrates, resulting in the substitution of the chloride with a hydroxyl group (3). To rigorously exclude the possibility that 3-hydroxybenzoyl–CoA was formed during 3-chlorobenzoate degradation, we added cold 3-hydroxybenzoate (1 mM) to cell suspensions containing radiolabeled 3-chlorobenzoate. This treatment would be expected to “trap” any radiolabeled 3-hydroxybenzoyl–CoA that might be formed transiently during 3-chlorobenzoate metabolism. Even in this case, we failed to detect radiolabeled products that comigrated with 3-hydroxybenzoyl–CoA (Rf, 0.47).
FIG. 2.
Autoradiogram showing intracellular products formed by R. palustris incubated with [7-14C]3-chlorobenzoate for 2 or 5 min (2′ or 5′). The arrows mark the positions of radiolabeled metabolites that comigrated with unlabeled benzoyl-CoA and 3-chlorobenzoyl–CoA standards. The position of a nonradioactive 3-hydroxybenzoyl–CoA standard is also indicated.
3-Chlorobenzoate–CoA ligase from RCB100.
Extracts of 3-chlorobenzoate-grown cells had a 3-chlorobenzoate–CoA ligase (specific activity of 8.7 nmol/min/mg of protein) as determined with an isotopic assay (8), using 200 μM substrate. This activity was induced eightfold over the levels seen in succinate-grown cells (1.1 nmol/min/mg of protein). 3-Chlorobenzoate–CoA ligase activity was also induced by growth on benzoate (6.6 nmol/min/mg of protein). RCB100 cells grown on benzoate had a benzoate-CoA ligase activity of 26.4 nmol/min/mg of protein when the isotopic assay was carried out with 10 μM benzoate (8).
R. palustris strain CGA009 can, as mentioned above, cometabolize 3-chlorobenzoate in the presence of benzoate. The genome of this strain has recently been sequenced by the U.S. Department of Energy Joint Genome Institute (http://spider.jgi-psf.org/JGI_microbial/html/). A search of the genome sequence indicates that R. palustris has at least 35 acyl-CoA ligase genes, at least one of which must encode an enzyme that is active with 3-chlorobenzoate. We have purified three CoA ligases from strain CGA009 that are active with various aromatic acids (8, 9, 17). One of these, benzoate-CoA ligase, encoded by the gene badA, has a low level of activity with 3-chlorobenzoate as a substrate (8). To see if this enzyme could be responsible for 3-chlorobenzoyl–CoA formation by strain RCB100, we mutated the RCB100 version of badA using a gene knockout construction that we had used successfully in the past to mutate the badA gene in strain CGA009 (6). Southern hybridization was used to verify that the badA gene had been disrupted in this strain, designated RCB101. Furthermore, cell extracts prepared from wild-type strain RCB100 reacted with antiserum that was specific to the purified BadA protein from strain CGA009, but extracts from the mutant RCB101 did not react with anti-BadA antiserum (data not shown). RCB101 was impaired in its ability to synthesize benzoyl-CoA. When assayed at 10 μM benzoate, this strain had a benzoate-CoA ligase activity of 0.97 nmol of benzoyl-CoA formed/min/mg of protein versus 26.4 nmol of benzoyl-CoA formed/min/mg of protein for its wild-type parent. However, RCB101 grew at wild-type rates on 3-chlorobenzoate and had wild-type levels of 3-chlorobenzoate–CoA ligase activity. This demonstrates that 3-chlorobenzoyl–CoA formation by R. palustris strain RCB101 is catalyzed by an enzyme that is distinct from benzoate-CoA ligase.
Reductive dehalogenation.
We failed to detect the conversion of 3-chlorobenzoyl–CoA to benzoyl-CoA or to any other CoA-modified compound in cell extracts of RCB100 that were prepared and incubated anaerobically with NADPH, NADH, reduced methyl viologen, or reduced glutathione as possible electron donors. HPLC was used to detect product appearance and substrate disappearance. Cell extracts had a high thioesterase activity that converted 3-chlorobenzoyl–CoA to 3-chlorobenzoate. This may have masked our ability to detect 3-chlorobenzoyl–CoA dehalogenation.
Although we were unable to obtain information about the type of reductive dehalogenase that is involved in 3-chlorobenzoyl–CoA dechlorination, the activity that we have observed in whole cells differs from reductive dehalogenases described to date. Two reductive dehalogenases that mediate halorespiration and that are active with aromatic compounds have been described. A membrane-bound 3-chlorobenzoate reductive dehalogenase has been purified from Desulfomonile tiedjei (20). A 3-chloro-4-hydroxyphenylacetate reductive dehalogenase has been purified from Desulfitobacterium sp., and its corresponding gene has been sequenced (4, 26). Both enzymes are active with free aromatic acids rather than with CoA thioesters. Two reductive dehalogenases that are active with aromatic compounds or quinones under aerobic conditions have been described. An activity that catalyzes an NADPH-dependent reductive dechlorination of 2,4-dichlorobenzoyl–CoA to 4-chlorobenzoyl–CoA has been described in two strains of coryneform bacteria (22), but the aromatic ring is not completely dechlorinated. This is accomplished by the hydrolytic dehalogenation of 4-chlorobenzoyl–CoA (22). Finally, a tetrachlorohydroquinone dehalogenase has been described from Sphingomonas chlorophenolica that has weak sequence similarity to members of the glutathione S-transferase superfamily (1). Again, the substrate for this enzyme is not modified with CoA.
The R. palustris strain CGA009 genome does not include any genes that resemble known reductive dehalogenase genes. Nor are there any genes with significant similarity to the hydrolytic 4-chlorobenzoyl–CoA dehalogenase gene that has been described in aerobic systems (23). The R. palustris genome does have a gene with 55% deduced amino acid sequence identity to a hydrolytic haloacid dehalogenase (27). We are currently investigating whether this gene may play a role in chloropropionate dehalogenation. It does not appear to be involved in 3-chlorobenzoate degradation.
Conclusions.
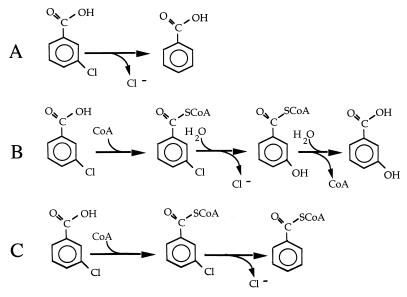
Our results indicate that strain RCB100 metabolizes 3-chlorobenzoate by converting it to 3-chlorobenzoyl–CoA and then reductively dehalogenating 3-chlorobenzoyl–CoA to benzoyl-CoA (Fig. 3). R. palustris degrades benzoyl-CoA to acetyl-CoA and carbon dioxide by means of a reductive pathway that has been well studied in this organism (10). RCB100 differs from several other bacteria that have been reported to dechlorinate aromatic compounds anaerobically in that it uses these compounds as a carbon source rather than as a terminal electron acceptor in an anaerobic respiration. A second difference is that a CoA-modified compound, rather than the free acid, is the substrate for complete dechlorination.
FIG. 3.
Three routes for 3-chlorobenzoate dehalogenation. (A) Reductive dechlorination of 3-chlorobenzoate as it occurs in D. tiedjei (18). 3-Chlorobenzoate is the terminal electron acceptor in halorespiration, and thus benzoate is the final product of the reaction sequence. (B) A proposed reaction series for 3-chlorobenzoate degradation that has not been observed but is analogous to the 4-chlorobenzoate utilization pathway of several species of aerobic bacteria (5). The free aromatic acid that is formed following dechlorination is degraded to tricarboxylic acid cycle intermediates by means of oxygen-requiring pathways for aromatic compound utilization. (C) Formation of 3-chlorobenzoyl–CoA followed by reductive dechlorination to form benzoyl-CoA as proposed here for R. palustris. Benzoyl-CoA is further metabolized to acetyl-CoA and carbon dioxide.
Nitrate-reducing bacteria have been described that grow on 3-chlorobenzoate under anaerobic but not aerobic conditions (24, 25). These isolates also utilize 3-chlorobenzoate as a carbon source rather than as a terminal electron acceptor. One isolate that was tested had 3-chlorobenzoate–CoA ligase activity (D. M. Eby, O. T. Harriott, M. C. Stanton, and M. M. Häggblom, Abstr. 97th Gen. Meet. Am. Soc. Microbiol., abstr. Q-120, p. 475, 1997). Thus the route for 3-chlorobenzoate degradation described here may be present in other bacteria in addition to R. palustris.
Acknowledgments
This work was supported by the Division of Energy Biosciences, U.S. Department of Energy (grant DE-FG02-95ER20184), and by the U.S. Army Research Office (grant DAAG55-98-0083). Preliminary sequence data were obtained from the DOE Joint Genome Institute (JGI) at the website http://spider.jgi-psf.org/JGI_microbial/html/.
REFERENCES
- 1.Anandarajah K, Kiefer P M, Jr, Donohoe B S, Copley S D. Recruitment of a double bond isomerase to serve as a reductive dehalogenase during biodegradation of pentachlorophenol. Biochemistry. 2000;39:5303–5311. doi: 10.1021/bi9923813. [DOI] [PubMed] [Google Scholar]
- 2.Bergmann J G, Sanik J., Jr Determination of trace amounts of chlorine in naphtha. Anal Chem. 1957;29:241–243. [Google Scholar]
- 3.Chang K H, Liang P H, Beck W, Scholten J D, Dunaway-Mariano D. Isolation and characterization of the three polypeptide components of 4-chlorobenzoate dehalogenase from Pseudomonas sp. strain CBS-3. Biochemistry. 1992;31:5605–5610. doi: 10.1021/bi00139a025. [DOI] [PubMed] [Google Scholar]
- 4.Christiansen N, Ahring B K, Wohlfarth G, Diekert G. Purification and characterization of the 3-chloro-4-hydroxy-phenylacetate reductive dehalogenase of Desulfitobacterium hafniense. FEBS Lett. 1998;436:159–162. doi: 10.1016/s0014-5793(98)01114-4. [DOI] [PubMed] [Google Scholar]
- 5.Dunaway-Mariano D, Babbitt P C. On the origins and functions of the enzymes of the 4-chlorobenzoate to 4-hydroxybenzoate converting pathway. Biodegradation. 1994;5:259–276. doi: 10.1007/BF00696464. [DOI] [PubMed] [Google Scholar]
- 6.Egland P G, Gibson J, Harwood C S. Benzoate-coenzyme A ligase, encoded by badA, is one of three ligases able to catalyze benzoyl-coenzyme A formation during anaerobic growth of Rhodopseudomonas palustris on benzoate. J Bacteriol. 1995;177:6545–6551. doi: 10.1128/jb.177.22.6545-6551.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.EI Fantroussi S, Naveau H, Agathos S N. Anaerobic dechlorinating bacteria. Biotechnol Prog. 1998;14:167–188. doi: 10.1021/bp980011k. [DOI] [PubMed] [Google Scholar]
- 8.Geissler J F, Harwood C S, Gibson J. Purification and properties of benzoate-coenzyme A ligase, a Rhodopseudomonas palustris enzyme involved in the anaerobic degradation of benzoate. J Bacteriol. 1988;170:1709–1714. doi: 10.1128/jb.170.4.1709-1714.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibson J, Dispensa M, Fogg G C, Evans D T, Harwood C S. 4-Hydroxybenzoate-coenzyme A ligase from Rhodopseudomonas palustris: purification, gene sequence, and role in anaerobic degradation. J Bacteriol. 1994;176:634–641. doi: 10.1128/jb.176.3.634-641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harwood C S, Burchhardt G, Herrmann H, Fuchs G. Anaerobic metabolism of aromatic compounds via the benzoyl-CoA pathway. FEMS Microbiol Rev. 1999;22:439–458. [Google Scholar]
- 11.Harwood C S, Gibson J. Anaerobic and aerobic metabolism of diverse aromatic compounds by the photosynthetic bacterium Rhodopseudomonas palustris. Appl Environ Microbiol. 1988;54:712–717. doi: 10.1128/aem.54.3.712-717.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holliger C, Wohlfarth G, Diekert G. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol Rev. 1999;22:383–398. [Google Scholar]
- 13.Janssen D B, Pries F, van der Ploeg J R. Genetics and biochemistry of dehalogenating enzymes. Annu Rev Microbiol. 1994;48:163–191. doi: 10.1146/annurev.mi.48.100194.001115. [DOI] [PubMed] [Google Scholar]
- 14.Kamal V S, Wvndham R C. Anaerobic phototrophic metabolism of 3-chlorobenzoate by Rhodopseudomonas palustris WS17. Appl Environ Microbiol. 1990;56:3871–3873. doi: 10.1128/aem.56.12.3871-3873.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim M-K, Harwood C S. Regulation of benzoate-CoA ligase in Rhodopseudomonas palustris. FEMS Microbiol Lett. 1991;83:199–204. [Google Scholar]
- 16.Krooneman J, van den Akker S, Pedro Gomes T M, Forney L J, Gottschal J C. Degradation of 3-chlorobenzoate under low-oxygen conditions in pure and mixed cultures of the anoxygenic photoheterotroph Rhodopseudomonas palustris DCP3 and an aerobic Alcaligenes species. Appl Environ Microbiol. 1999;65:131–137. doi: 10.1128/aem.65.1.131-137.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Küver J, Xue J Y, Gibson J. Metabolism of cyclohexane carboxylic acid by the photosynthetic bacterium Rhodopseudomonas palustris. Arch Microbiol. 1995;164:337–345. doi: 10.1007/BF02529980. [DOI] [PubMed] [Google Scholar]
- 18.Louie T M, Mohn W W. Evidence for a chemiosmotic model of dehalorespiration in Desulfomonile tiedjei DCB-1. J Bacteriol. 1999;181:40–46. doi: 10.1128/jb.181.1.40-46.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merkel S M, Eberhard A E, Gibson J, Harwood C S. Involvement of coenzyme A thioesters in anaerobic metabolism of 4-hydroxybenzoate by Rhodopseudomonas palustris. J Bacteriol. 1989;171:1–7. doi: 10.1128/jb.171.1.1-7.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ni S, Fredrickson J K, Xun L. Purification and characterization of a novel 3-chlorobenzoate-reductive dehalogenase from the cytoplasmic membrane of Desulfomonile tiedjei DCB-1. J Bacteriol. 1995;177:5135–5139. doi: 10.1128/jb.177.17.5135-5139.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pelletier D A, Harwood C S. 2-Hydroxycyclohexanecarboxyl coenzyme A dehydrogenase, an enzyme characteristic of the anaerobic benzoate degradation pathway used by Rhodopseudomonas palustris. J Bacteriol. 2000;182:2753–2760. doi: 10.1128/jb.182.10.2753-2760.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romanov V, Hausinger R P. NADPH-dependent reductive ortho dehalogenation of 2,4-dichlorobenzoic acid in Corynebacterium sepedonicum KZ-4 and coryneform bacterium strain NTB-1 via 2,4-dichlorobenzoyl coenzyme A. J Bacteriol. 1996;178:2656–2661. doi: 10.1128/jb.178.9.2656-2661.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scholten J D, Chang K H, Babbitt P C, Charest H, Sylvestre M, Dunaway-Mariano D. Novel enzymatic hydrolytic dehalogenation of a chlorinated aromatic. Science. 1991;253:182–185. doi: 10.1126/science.1853203. [DOI] [PubMed] [Google Scholar]
- 24.Song B, Palleroni N J, Häggblom M M. Description of strain 3CB-1, a genomovar of Thauera aromatica, capable of degrading 3-chlorobenzoate coupled to nitrate reduction. Int J Syst Evol Microbiol. 2000;50:551–558. doi: 10.1099/00207713-50-2-551. [DOI] [PubMed] [Google Scholar]
- 25.Song B, Palleroni N J, Häggblom M M. Isolation and characterization of diverse halobenzoate-degrading denitrifying bacteria from soils and sediments. Appl Environ Microbiol. 2000;66:3446–3453. doi: 10.1128/aem.66.8.3446-3453.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van de Pas B A, Smidt H, Hagen W R, van der Oost J, Schraa G, Stams A J, de Vos W M. Purification and molecular characterization of ortho-chlorophenol reductive dehalogenase, a key enzyme of halorespiration in Desulfitobacterium dehalogenans. J Biol Chem. 1999;274:20287–20292. doi: 10.1074/jbc.274.29.20287. [DOI] [PubMed] [Google Scholar]
- 27.van der Ploeg J, van Hall G, Janssen D B. Characterization of the haloacid dehalogenase from Xanthobacter autotrophicus GJ10 and sequencing of the dhlB gene. J Bacteriol. 1991;173:7925–7933. doi: 10.1128/jb.173.24.7925-7933.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Woude B J, de Boer M, van der Put N M, van der Geld F M, Prins R A, Gottschal J C. Anaerobic degradation of halogenated benzoic acids by photoheterotrophic bacteria. FEMS Microbiol Lett. 1994;119:199–208. doi: 10.1111/j.1574-6968.1994.tb06889.x. [DOI] [PubMed] [Google Scholar]
- 29.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]