Summary/Abstract
CRISPR-Cas9 based screening technologies enable precise, high-throughput genetic and epigenetic manipulation to study mechanisms of development and disease and identify new therapeutic targets. Here, we describe a general protocol for the generation of custom, pooled CRISPR sgRNA libraries for screening in cardiomyocyte cultures. This methodology can address variety of lab-specific research questions in cardiomyocytes and other cell types, as the genes to be modified can be curated or whole genomes can be investigated. The use of lentiviral sgRNA delivery followed by high-throughput sequencing allows for rapid comparison and identification of candidate genes and epigenetic modifiers, which can be further validated individually or in sub-pooled libraries following screening.
Keywords: CRISPR, genetic screen, cardiomyocyte, knock-out, high-throughput, proliferation, maturation, survival
1. Introduction
The application of clustered regularly interspersed palindromic repeat (CRISPR) methods to modern genome engineering has been transformative for the field, as the ability to perform precise genetic manipulations at almost any locus of interest has become both expedient and accessible [1]. One important application of CRISPR-Cas9 technology is the ability to perform efficient and high-throughput in vitro genetic screens to address a broad spectrum of scientific questions. There are several types of CRISPR screens that can be performed (Table 1), with gene knock-out (CRISPR-KO) screens being widely used [2]. However, newer techniques for epigenetic screening that involve CRISPR-based gene upregulation or repression through the use of catalytically inactive Cas9 (dCas9) fused to functional domains of chromatin-modifying proteins are also available [3]. These epigenetic activation and inhibition screens offer additional flexibility for the study of regulatory mechanisms in development and disease and for identification of novel therapeutic targets [4].
Table 1.
A summary of CRISPR screening technologies applicable to pooled CRISPR sgRNA library screening.
| Function | Type of Cas9 | Targeting Location |
Notes |
|---|---|---|---|
| Knock-Out | spCas9 | First exon | Cas9 derived from S. pyogenes is DNA endonuclease used to induce frame-shift mutations at the targeted locus, resulting in gene inactivation |
| Activation | dCas9 fused to VP64 | Promoter | VP64 is a tetrameric repeat of the herpes simplex virus protein VP16, which induces transcriptional gene activation [23, 24] |
| Activation | dCas9 fused to p300 | Promoter | p300 is a histone acetyltransferase which facilitates gene transcription [25] |
| Activation | dCas9 fused to PRDM9 | Promoter | PRDM9 is a histone methyltransferase used to stabilize gene expression via induction of the activating mark H3K4me3 |
| Inhibition or activation | dCas9 fused to HDAC3 | Promoter | HDAC3 is a histone deacetylase associated with both gene activation and repression depending on the targeted locus [26] |
| Inhibition | dCas9 fused to Dnmt3a | Promoter or CpG islands | Dnmt3a is a DNA methyltransferase used to induce targeted DNA methylation and suppress gene transcription [27, 28] |
| Inhibition | dCas9 fused to KRAB | Promoter or enhancer | Kruppel-associated box domain (KRAB) recruits a complex responsible for both histone methylation and deacetylation, resulting in heterochromatin formation and repression of gene transcription [29-31] |
| Inhibition | dCas9 fused to KRAB–MeCP2 | Promoter or CpG islands | KRAB in combination with methyl CpG binding protein 2 (MeCP2) aids in gene silencing via complex formation with histone deacetylases and via direct interaction with transcription factors [32] |
| Inhibition | dCas9 fused to LSD1 | Promoter | LSD1 is a histone demethylase used to repress enhancers by removing H3K4me2 mark from histone, resulting in reduced gene expression due to enhancer inactivation [33] |
| Mutagenesis | dCas9 fused to AIDx | Gene or regulatory region | AID is activation-induced cytidine deaminase with the ability to generate a wide array of targeted point mutations in high-throughput screens for disease-related variants [34, 35] |
In vitro CRISPR screens typically involve lentiviral delivery of a library of single guide RNA sequences (sgRNAs) targeting a subset of genes or the whole genome. As lentivirus stably integrates into the DNA, the sgRNA will be present in the cell’s genome and identifiable by high-throughput sequencing [5]. Differential prevalence of sequenced sgRNAs will point to the specific genes involved in the phenotype of interest. Library size and the desired sensitivity of detecting an effect will determine the cell number necessary for screening. Screens for cell survival or proliferation can be performed without selecting the cells for a specific phenotype, with all cells being subjected to sequencing. Screens for cell differentiation, maturation, or a particular phenotypic change will require use of fluorescent phenotypic reporters followed by cell sorting or antibiotic resistance-based selection to separate cells for sequencing. Regardless of cell selection prior to sequencing, the false positive threshold for detecting sgRNA hits should be set above the DNA replication rate since any baseline DNA synthesis will result in sgRNA amplification.
This protocol describes a generalized method for designing, validating, and screening a custom CRISPR library in cultured cardiomyocytes, making a protocol for cardiomyocyte-specific high-throughput genetic screening accessible to the broad research community. While the use of a particular cardiomyocyte type (e.g. neonatal rat [6], mouse postnatal [7], human pluripotent stem cell-derived [8]) or culture model (monolayer [9] or 3D engineered tissue [10-12]) will require some adjustments, the described protocol will outline custom library design including generation of a gene list, in silico sgRNA design, and library cloning to provide flexibility when addressing lab-specific research questions. If a custom library is not desired, many pre-made pooled sgRNA screening libraries, including those targeting whole genome, are available through various resources such as Addgene.
2. Materials
2.1. Molecular Biology
LentiCRISPR V2 Plasmid (Addgene #52961, [2]).
Molecular biology grade agarose.
Tris-acetate-EDTA (TAE) buffer: 40mM Tris base, 2mM EDTA, 20mM acetic acid, pH 8.5.
Sybr Safe DNA Stain (Thermo Fisher).
DNA Gel Box and Power Supply.
Thermocycler.
Phusion High-Fidelity DNA Polymerase (NEB).
Deoxynucleoside triphosphates (dNTPs).
Restriction Enzymes.
100bp DNA Ladder.
1kb DNA Ladder.
Zymo Gel DNA Extraction Kit (Zymo Research).
Zymo Clean and Concentrator Kit (Zymo Research).
Razor blades.
UV box for viewing DNA gels.
UV face shield.
T4 DNA Ligase.
Endura Electrocompetent E. coli (Lucigen).
Gene Pulser Xcell™ Total Electroporator (Biorad).
0.1cm Gap Electroporator Cuvettes.
SOC Medium: 20g/L tryptone, 5g/L yeast extract, 0.5g/L NaCl, 20mM glucose.
10mL round bottom tubes.
Agar plates containing antibiotic.
Luria Bertani (LB) broth containing antibiotic.
Two Liter Bacterial Culture Flask.
Bacterial Shaking Incubator.
Maxi Prep Kit (Qiagen).
Centrifuge.
Molecular Biology Grade Ethanol.
NanoDrop™ 2000 (Thermo Fisher).
Custom amplification primers.
Custom pooled sgRNA oligonucleotides.
Genomic DNA Isolation Kit.
Custom Amplification Primers.
2.2. Cell Culture
HEK293T cells (ATCC).
Cardiomyocytes of choice (e.g. neonatal rat, neonatal mouse, human pluripotent stem cell-derived).
Click-iT™ EdU Alexa Fluor™ 488 Flow Cytometry Assay Kit (Invitrogen).
4′,6-diamidino-2-phenylindole (DAPI) stain.
Live/Dead Cell Viability/Cytotoxicity Kit (Invitrogen).
10cm tissue culture treated dishes.
3. Methods
3.1. Library Preparation and In Silico sgRNA Sequence Design
Generate gene list for CRISPR library using Gene Ontology resources based on specific research interests [13, 14] (see Notes 1 and 2).
Design targeting sgRNA sequences using the Broad Institute’s GPP sgRNA Designer website (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design) [15-18] or other source if desired.
Sort ranked sgRNA sequences using Excel or Matlab and export to a final list. Choose the 5 lowest overall ranked sgRNA sequences for each gene, which correspond to sequences with the best predicted on-target activity and lowest predicted off-target activity (see Notes 3 and 4).
3.2. Sequence Design for sgRNA Oligonucleotide Cloning
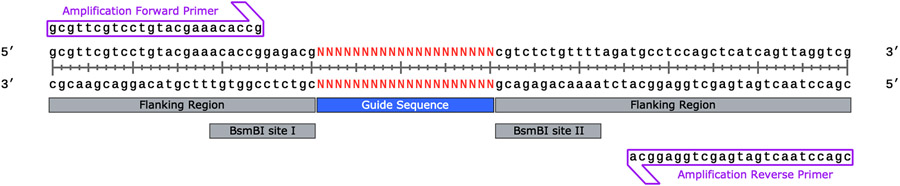
When designing the universal flanking regions for synthesized sgRNA sequences, it is recommended to include primer binding sites for oligonucleotide amplification, directional restriction sites for cloning, and sequencing primer sites for high-throughput sequencing (Figure 1) (see Notes 5 and 6).
Append universal flanking regions containing primer binding sites and restriction sites as per Figure 1 to designed sgRNA sequences using Excel or other preferred software.
Order pooled sgRNA oligonucleotides containing flanking regions.
Figure 1.
A sample sgRNA sequence denoted by “n” flanked by directional restriction sites and amplification primer binding sites.
3.3. sgRNA Library Plasmid Cloning
Upon receipt of pooled oligonucleotides containing sgRNA sequences, PCR amplify them using designed amplification primers (see Note 7).
Perform at least five 20μL PCR reactions in parallel with Phusion DNA polymerase and pool the PCR product.
Check the PCR by running 5μL of pooled PCR product on a 1% agarose gel containing Sybr Safe or other preferred DNA stain with a 100bp DNA ladder.
Confirm that a single band of the correct size is present on the gel by viewing the gel over a UV light source while wearing appropriate personal protective equipment such as a UV face shield.
Purify the remaining PCR product with a Clean and Concentrator Kit and elute DNA in 10μL sterile, nuclease-free water.
Restriction digest the 1μg of LentiCRISPR V2 plasmid and 10μL of the PCR-amplified sgRNA sequences with the restriction enzyme BsmBI overnight at 37°C as per NEB protocol.
Run a 1% agarose gel for all of the digested vectors and oligonucleotides containing sgRNA sequences. Include a 1kb DNA ladder and undigested vector as a control, making sure to skip at least one lane between the digested and undigested vectors.
Cut out bands over a UV light source with a clean razor blade, making sure to use a new blade for each band and to utilize appropriate UV protection.
Gel purify the digested oligonucleotides and digested vector with the Gel Purification kit (see Note 8).
Determine purified DNA concentrations with Nanodrop.
Ligate overnight at 4°C with T4 DNA Ligase using a molar ratio of 5 parts insert to 1 part vector.
Transform ligated DNA in triplicate into electrocompetent E. coli in a 0.1cm gap electroporation cuvette using the following settings on an electroporator: 1800V, 10μF, 600Ω.
Immediately after electroporation, quickly wash each cuvette out with 5mL SOC medium and transfer contents to a sterile 10mL round bottom culture tube.
Incubate the 10mL round bottom tubes containing the transformed library culture for 1 hour at 37°C and 250 rpm.
After one hour, take 100μL of the transformed library culture from each of the three tubes and plate 100μL of at least two dilutions (1:10 and 1:100) in LB broth on agar plates containing the appropriate antibiotic, which will allow for the calculation of transformation efficiency. Culture agar plates overnight in a stationary incubator at 37°C and count colonies the next morning (see Note 9).
Combine rest of the transformed library culture in the three 10mL round bottom tubes and transfer to a two-liter culture flask containing 250mL of LB broth with the appropriate antibiotic (see Note 10).
Incubate the two-liter flask overnight (12-16 hours) in a bacterial shaking incubator at 37°C and 250 rpm.
Following overnight incubation, spin the transformed library culture down and isolate plasmid DNA as per maxiprep protocol.
Elute DNA in sterile water inside a biological safety cabinet. This DNA will be used to make lentivirus and thus should be kept sterile.
Perform high-throughput sequencing on the purified plasmid to ensure that sgRNA representation is maintained in the library following amplification and cloning (see Note 11). PAUSE POINT.
3.4. Assessment of Baseline Cardiomyocyte Proliferation
Before performing the screen, assess baseline rate of DNA synthesis of cardiomyocytes by adding 10mM EdU to culture medium for 24 hours.
Fix cells, then stain for EdU per manufacturer protocol and add DAPI stain at 1:1000 concentration. For less than 100% cardiomyocyte purity, also include a cardiomyocyte-specific antibody stain, such as cardiac troponin T or sarcomeric alpha-actinin.
Perform flow cytometry to quantify the percent EdU positive cardiomyocytes as a proportion of total cardiomyocytes (see Note 12). PAUSE POINT.
3.5. Assessment of Antibiotic Susceptibility
To determine the optimal antibiotic concentration for selection of transduced cardiomyocytes, assess puromycin susceptibility of the cardiomyocytes by treating wells with cells with a series of concentrations of puromycin in normal culture medium from 1-10μg/mL.
Apply antibiotic selection for 5 days with a media change every 48 hours and assess cardiomyocyte viability with the Live/Dead Cell Viability/Cytotoxicity Kit as per manufacturer protocol. PAUSE POINT.
3.6. sgRNA Lentivirus Library Preparation
Prepare lentivirus from the purified library plasmid generated in Step 3.2 as per Addgene protocol (https://www.addgene.org/protocols/lentivirus-production/) or any other preferred protocol in HEK293T cells [19, 20].
Determine functional (infectious) titer of the lentivirus by infecting cardiomyocytes with several concentrations of virus such that concentration can be correlated with transduction efficiency and used for determination of functional titer (see Note 13).
Deliver puromycin (or other appropriate antibiotic) 48 hours post transduction at the lowest antibiotic concentration that caused nearly 100% non-transduced cardiomyocyte death by 5 days.
Choose wells with concentrations of virus that resulted in roughly 20-40% transduction such that individual cells are easily quantifiable and calculate the percent surviving cells (percent transduction) after five days and therefore determine the functional titer of the virus in μL virus per number of cells transduced (see Notes 14 and 15).
Choose the virus concentration that generates a multiplicity of infection (MOI) of 0.2-0.4, which reduces the likelihood that cardiomyocytes will be transduced with more than one viral particle while also minimizing the number of cells necessary to ensure sufficient library coverage.
3.7. Library Delivery and Screening
To determine the cell number needed for screening, use the following formula: (Number of sgRNAs in library) * (Library coverage) * (Number of Timepoints) / (MOI).
Choose a cell number that ensures library coverage is at least 300-500X, i.e. 300-500 cells will receive each sgRNA from the library.
Plate cardiomyocytes at the desired seeding density (see Note 16). Depending on the experiment, the library lentivirus can be delivered at the time of plating or after several days to weeks.
Deliver the titered library lentivirus at a MOI of 0.2-0.4 overnight in normal culture medium.
Perform a media change 24 hours after lentiviral delivery.
Isolate genomic DNA (gDNA) from the initial timepoint at 48 hours post lentivirus delivery, which will provide the initial library representation. It is not necessary to perform FACS at this step (see Notes 17 and 18).
Determine gDNA concentration using Nanodrop or other DNA quantification method. Store the gDNA at −20°C until the other samples have been collected.
At specific time points, isolate gDNA from all cells (or selected cells with the desired phenotype) and store at −20°C. PAUSE POINT.
PCR-amplify sgRNA sequences from gDNA using custom amplification primers.
Gel-purify amplified sgRNA sequences using the Gel DNA Extraction Kit.
Prepare sequencing library and sequence on an Illumina MiSeq or NextSeq platform depending on the size of the library.
Determine sgRNA sequence representation relative to the initial time point. Select the most overrepresented sgRNA sequences for validation.
3.5. Validation of sgRNA Targets
Order individual oligonucleotides containing the sgRNA sequences of interest. Choose at least three sgRNA sequences per gene.
Clone individual sgRNA sequences into the vector and make lentivirus as outlined above (see Note 19).
Deliver lentivirus to cultured cardiomyocytes at a MOI of 1 or higher.
Assess for desired phenotype at 72 hours or later via flow cytometry or microscopy.
4. Notes
Use of Gene Ontology resources allows for the generation of a gene list that contains genes annotated to any chosen pathway or phenotype. However, it is also possible to use any other publicly available data set such as the Human Protein Atlas (http://www.proteinatlas.org) [21], which contains tissue-specific expression data.
Cost of sgRNA oligonucleotide synthesis increases with the number of oligonucleotides needed, which may factor into the choice of the number of studied genes and final library size. If the gene list is very long, it may be more cost effective to use a published whole-genome library available on Addgene rather than ordering synthesized oligonucleotides. For numbers of oligonucleotides greater than 100-200, pooled batch synthesis may be more cost effective, while for small numbers of oligonucleotides individual synthesis may be less costly.
Any sgRNA sequences that contain the restriction site used for cloning should be removed.
It is recommended to use at least five sgRNA sequences per gene with the assumption that not all sequences will necessarily be functional. It is likely that multiple sgRNAs will be functional if five are chosen. To limit numbers of needed cardiomyocytes and oligonucleotides, it is possible to use as few as three sgRNAs per gene, but less than three is not recommended.
If possible, it is recommended to use single restriction sites such as BsmBI that have recognition sites next to the cut site. This allows for two different “sticky” ends to be generated in the vector with the use of only one enzyme, allowing for directional cloning of the insert. It should be further ensured that the cloning strategy does not disrupt the guide scaffold present in the vector. To achieve high bulk cloning efficiency, primers should be carefully designed and validated with several individual sgRNAs before cloning the full library.
Amplicon length for NGS should not exceed Illumina sequencing length capacity of 50 or 75 nucleotides, depending on the exact sequencing method chosen.
This is a point at which library preparation can easily fail. Primers should be validated in advance and the optimal melting temperature empirically determined using gradient PCR. High fidelity DNA polymerase, sterile PCR grade water, and sterile filter tips should be used when working with PCR reagents to avoid contamination.
It is critical that the bands are the correct sizes and there are no additional, unexpected bands. The digested vector most likely will have two bands, and the correct band is usually the larger band. Confirm the expected band sizes and that all expected bands are present. Restriction enzymes should not be expired and should be kept at the appropriate storage temperature.
Recommended library coverage is at least 10,000X. For example, for a library of 100 genes, plasmid should be introduced into at least 106 bacteria. Number of transformed bacteria should be calculated as follows: (Total Culture Volume/Volume Plated) * (Number of Colonies Counted). If the efficiency is low, ligation and transformation should be repeated.
While the mammalian selection antibiotic for LentiCRISPR V2 is puromycin, the bacterial antibiotic is ampicillin. Ensure use of the correct antibiotic for bacterial selection.
If it is necessary to reduce cost, Sanger sequencing could be performed on many individual colonies. While this method will not provide the overall library representation, it will be more cost-effective to validate that the cloning was successful. It will be evident if library representation is not maintained following cloning if a single guide sequence or several guide sequences are predominant. If this is the case, it may be necessary to troubleshoot the library cloning steps starting with guide oligonucleotide amplification.
In addition to flow cytometry, EdU incorporation could be quantified using fluorescence microscopy. The assessment of baseline EdU incorporation rate will serve to determine how much sgRNA sequence overrepresentation could be attributed to endogenous cardiomyocyte proliferation rather than a phenotype induced by the library.
For PEG precipitated lentivirus, between 0.05μL-8μL virus per million cells has been used successfully, but the concentration should be determined empirically and can vary widely based on the lentivirus purification protocol.
LentiCRISPR V2 contains a puromycin resistance gene that is delivered and integrates in DNA along with the sgRNA sequence and Cas9. If there is no antibiotic resistance gene in the vector, fluorescence genes or other markers can be also used. If no markers are present, an ELISA-based titer determination can be performed; however, this method is not preferred as it does not always correspond to the functional titer [22].
Low titer lentivirus should not be used for screening. If lentiviral titer is low, it may be necessary to optimize lentivirus production protocols and/or remake the virus.
Seeding densities of 130,000/cm2 for neonatal rat ventricular myocytes (NRVMs) and 210,000/cm2 for human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) are recommended for proliferation-related screens. However, a lower seeding density can also be used if necessary to observe a phenotype of interest.
It is essential to isolate gDNA at an initial timepoint following transduction, but before sgRNA sequence representation changes due to the occurrence of the phenotype of interest. Forty-eight hours is a reasonable timepoint for a lentivirally delivered library to allow for viral transduction and genomic integration of the sgRNA sequences to occur without changes in library representation. This timing may be experiment-dependent and should be adjusted for alternative methods of library delivery, such as with adeno-associated viruses which take longer to express than lentivirus.
The manufacturer-recommended cell number per column and the column capacity should not be exceeded, as the resulting DNA loss will lead to a loss of library coverage.
If many targets are being validated, it is possible to make a sub-pooled library and re-screen. For a small number of targets, each target should be validated individually.
Acknowledgements
This work was supported by NIH grants U01HL134764, U01EB028901, HL132389, and a grant from Foundation Leducq to NB and the T32 Developmental and Stem Cell Biology Training Grant 5T32HD040372-18 to SD.
5 References
- 1.Ishino Y, Krupovic M, Forterre P (2018) History of CRISPR-Cas from Encounter with a Mysterious Repeated Sequence to Genome Editing Technology. J Bacteriol 200:. 10.1128/JB.00580-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanjana NE, Shalem O, Zhang F (2014) Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11:783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuster A, Erasimus H, Fritah S, et al. (2019) RNAi/CRISPR Screens: from a Pool to a Valid Hit. Trends Biotechnol 37:38–55. 10.1016/j.tibtech.2018.08.002 [DOI] [PubMed] [Google Scholar]
- 4.Doench JG (2018) Am I ready for CRISPR? A user’s guide to genetic screens. Nat Rev Genet 19:67–80. 10.1038/nrg.2017.97 [DOI] [PubMed] [Google Scholar]
- 5.Aregger M, Chandrashekhar M, Tong AHY, et al. (2019) Pooled Lentiviral CRISPR-Cas9 Screens for Functional Genomics in Mammalian Cells. Methods Mol Biol Clifton NJ: 1869:169–188. 10.1007/978-1-4939-8805-1_15 [DOI] [PubMed] [Google Scholar]
- 6.Bian W, Badie N, Himel HD, Bursac N (2014) Robust T-tubulation and maturation of cardiomyocytes using tissue-engineered epicardial mimetics. Biomaterials 35:3819–3828. 10.1016/j.biomaterials.2014.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bassat E, Mutlak YE, Genzelinakh A, et al. (2017) The extracellular matrix protein agrin promotes heart regeneration in mice. Nature 547:179–184. 10.1038/nature22978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackman CP, Carlson AL, Bursac N (2016) Dynamic culture yields engineered myocardium with near-adult functional output. Biomaterials 111:66–79. 10.1016/j.biomaterials.2016.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang D, Shadrin IY, Lam J, et al. (2013) Tissue-engineered cardiac patch for advanced functional maturation of human ESC-derived cardiomyocytes. Biomaterials 34:5813–5820. 10.1016/j.biomaterials.2013.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackman C, Li H, Bursac N (2018) Long-term contractile activity and thyroid hormone supplementation produce engineered rat myocardium with adult-like structure and function. Acta Biomater 78:98–110. 10.1016/j.actbio.2018.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shadrin IY, Allen BW, Qian Y, et al. (2017) Cardiopatch platform enables maturation and scale-up of human pluripotent stem cell-derived engineered heart tissues. Nat Commun 8:1–15. 10.1038/s41467-017-01946-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackman CP, Ganapathi AM, Asfour H, et al. (2018) Engineered cardiac tissue patch maintains structural and electrical properties after epicardial implantation. Biomaterials 159:48–58. 10.1016/j.biomaterials.2018.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashburner M, Ball CA, Blake JA, et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25:25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The Gene Ontology Consortium (2019) The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 47:D330–D338. 10.1093/nar/gky1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doench JG, Fusi N, Sullender M, et al. (2016) Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34:184–191. 10.1038/nbt.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanson KR, Hanna RE, Hegde M, et al. (2018) Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat Commun 9:1–15. 10.1038/s41467-018-07901-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanson KR, DeWeirdt PC, Sangree AK, et al. (2019) Optimization of AsCas12a for combinatorial genetic screens in human cells. bioRxiv 747170. 10.1101/747170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim HK, Min S, Song M, et al. (2018) Deep learning improves prediction of CRISPR–Cpf1 guide RNA activity. Nat Biotechnol 36:239–241. 10.1038/nbt.4061 [DOI] [PubMed] [Google Scholar]
- 19.Tiscornia G, Singer O, Verma IM (2006) Production and purification of lentiviral vectors. Nat Protoc 1:241–245. 10.1038/nprot.2006.37 [DOI] [PubMed] [Google Scholar]
- 20.Wang X, McManus M (2009) Lentivirus Production. J Vis Exp JoVE. 10.3791/1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uhlén M, Fagerberg L, Hallström BM, et al. (2015) Proteomics. Tissue-based map of the human proteome. Science 347:1260419. 10.1126/.1260419 [DOI] [PubMed] [Google Scholar]
- 22.Geraerts M, Willems S, Baekelandt V, et al. (2006) Comparison of lentiviral vector titration methods. BMC Biotechnol 6:34. 10.1186/1472-6750-6-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perez-Pinera P, Kocak DD, Vockley CM, et al. (2013) RNA-guided gene activation by CRISPR-Cas9–based transcription factors. Nat Methods 10:973–976. 10.1038/nmeth.2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seipel K, Georgiev O, Schaffner W (1992) Different activation domains stimulate transcription from remote ('enhancer’) and proximal ('promoter’) positions. EMBO J 11:4961–4968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hilton IB, D’Ippolito AM, Vockley CM, et al. (2015) Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 33:510–517. 10.1038/nbt.3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon DY, Zhao Y-T, Lamonica JM, Zhou Z (2017) Locus-specific histone deacetylation using a synthetic CRISPR-Cas9-based HDAC. Nat Commun 8:15315. 10.1038/ncomms15315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu XS, Wu H, Ji X, et al. (2016) Editing DNA methylation in the mammalian genome. Cell 167:233–247.e17. 10.1016/j.cell.2016.08.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vojta A, Dobrinić P, Tadić V, et al. (2016) Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res 44:5615–5628. 10.1093/nar/gkw159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thakore PI, D’Ippolito AM, Song L, et al. (2015) Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods 12:1143–1149. 10.1038/nmeth.3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polstein LR, Perez-Pinera P, Kocak DD, et al. (2015) Genome-wide specificity of DNA binding, gene regulation, and chromatin remodeling by TALE- and CRISPR/Cas9-based transcriptional activators. Genome Res 25:1158–1169. 10.1101/gr.179044.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parsi KM, Hennessy E, Kearns N, Maehr R (2017) Using an Inducible CRISPR-dCas9-KRAB Effector System to Dissect Transcriptional Regulation in Human Embryonic Stem Cells. Methods Mol Biol Clifton NJ: 1507:221–233. 10.1007/978-1-4939-6518-2_16 [DOI] [PubMed] [Google Scholar]
- 32.Yeo NC, Chavez A, Lance-Byrne A, et al. (2018) An enhanced CRISPR repressor for targeted mammalian gene regulation. Nat Methods 15:611–616. 10.1038/s41592-018-0048-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kearns NA, Pham H, Tabak B, et al. (2015) Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods 12:401–403. 10.1038/nmeth.3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma Y, Zhang J, Yin W, et al. (2016) Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat Methods 13:1029–1035. 10.1038/nmeth.4027 [DOI] [PubMed] [Google Scholar]
- 35.Hess GT, Frésard L, Han K, et al. (2016) Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat Methods 13:1036–1042. 10.1038/nmeth.4038 [DOI] [PMC free article] [PubMed] [Google Scholar]