Abstract
Long noncoding RNAs (lncRNAs) are defined as a class of non-protein-coding RNAs that are longer than 200 nucleotides. Previous studies have shown that lncRNAs play a vital role in the progression of multiple diseases, which highlights their potential for medical applications. The lncRNA hepatocyte nuclear factor 1 homeobox A (HNF1A) antisense RNA 1 (HNF1A-AS1) is known to be abnormally expressed in multiple cancers. HNF1A-AS1 exerts its oncogenic roles through a variety of molecular mechanisms. Moreover, aberrant HNF1A-AS1 expression is associated with diverse clinical features in cancer patients. Therefore, HNF1A-AS1 is a promising biomarker for tumor diagnosis and prognosis and thus a potential candidate for tumor therapy. This review summarizes current studies on the role and the underlying mechanisms of HNF1A-AS1 various cancer types, including gastric cancer, liver cancer, glioma, lung cancer, colorectal cancer, breast cancer, bladder cancer, osteosarcoma, esophageal adenocarcinoma, hemangioma, oral squamous cell carcinoma, laryngeal squamous cell carcinoma, cervical cancer, as well as gastroenteropancreatic neuroendocrine neoplasms. We also describe the diagnostic, prognostic, and therapeutic value of HNF1A-AS1 for multiple cancer patients.
Keywords: HNF1A-AS1, lncRNA, expression, function, mechanism, cancer biomarker
Introduction
Despite rapid improvements in diagnostic and therapeutic strategies, cancer remains with high incidence and mortality, as well as a low cure rate [1,2]. An estimated 18.1 million new cancer cases were diagnosed in 2018, associated with nearly 9.6 million deaths across 185 countries, according to the GLOBOCAN 2018 database [3,4]. Therefore, greater insight into cancer pathogenesis and the identification of reliable biomarkers for early cancer diagnosis and treatment are urgently required [5,6].
Long noncoding RNAs (lncRNAs) are a type of non-protein-coding RNA with transcripts more than 200 nucleotides in length [7-11]. Increasing evidence has revealed that lncRNAs are closely involved in modulating the progression of multiple diseases, including cancers [12-16]. LncRNAs are known to work as competitive endogenous RNAs (ceRNAs) that inhibit the expression of mRNA or interact with various protein components, regulating diverse cell biological processes [17-21].
The lncRNA hepatic nuclear factor 1α (HNF1A) antisense RNA 1 (HNF1A-AS1) [22-25] is an antisense RNA for HNF1A located on human chromosome 12q24.31. HNF1A-AS1 is 2455 nucleotides long with a start site approximately 5 kb downstream of HNF1A. Emerging studies have revealed that HNF1A-AS1 is dysregulated in many cancers. Furthermore, abnormal HNF1A-AS1 expression has been associated with various cancers. HNF1A-AS1 is also a critical regulator of cancer development by regulating multiple biological processes. Given these properties, HNF1A-AS1 is increasingly recognized as a promising biomarker for diagnosis and prognosis and an attractive therapeutic target for numerous cancers. Here, we summarize current knowledge regarding the roles of HNF1A-AS1 in cancers, which is comprised of dysregulated expression, associated clinical features, regulatory functions, related molecules, as well as clinical practice.
The expression and regulatory function of HNF1A-AS1 in cancers
Abnormal HNF1A-AS1 expression has been observed in various types of human cancer, including gastric cancer [24,26-28], liver cancer [23,29-31], glioma [32-34], lung cancer [35-39], colorectal cancer [40-44], breast cancer [45-47], bladder cancer [48-50], osteosarcoma [51-53], esophageal adenocarcinoma [25], hemangioma [54], oral squamous cell carcinoma [55], laryngeal squamous cell carcinoma [22], cervical cancer [56,57], and gastroenteropancreatic neuroendocrine neoplasms (Figure 1) [58]. Additionally, numerous studies have shown that HNF1A-AS1 expression is related to several clinical characteristics of cancer patients, such as tumor size, tumor (T) node (N) and metastasis (M) (TNM) stage, lymph node metastasis (LNM), distant metastasis, disease-free survival, and overall survival (Table 1). HNF1A-AS1 is also involved in regulating diverse cancer cell processes through multiple mechanisms, containing cell proliferation, apoptosis, invasion, migration, glycolysis, cell chemoresistance, as well as radioresistance (Table 2).
Figure 1.
The involvement of HNF1A-AS1 in different types of cancer. HNF1A-AS1 h ancer and participates in the regulation of the progression of cancer, including gastric cancer, liver cancer, glioma, lung cancer, colorectal cancer, breast as an aberrant expression in various types of c cancer, bladder cancer, osteosarcoma, esophageal adenocarcinoma, hemangioma, oral squamous cell carcinoma, laryngeal squamous cell carcinoma, cervical cancer, and gastroenteropancreatic neuroendocrine neoplasms.
Table 1.
The expression and clinical characteristics of HNF1A-AS1 in human cancers
| Disease type | Expression | Clinical characteristics | Refs |
|---|---|---|---|
| gastric cancer | upregulated | lymph node metastasis | [26-28] |
| downregulated | tumor size, levels of serum carcinoembryonic antigen (CEA), carbohydrate antigen 19-9 (CA19-9), and RRM1 expression | [24] | |
| gastric cancer | upregulated | / | [28] |
| liver cancer | upregulated | tumor size, tumor stage, multiple lesions, and poor differentiation | [29-31] |
| downregulated | / | [23] | |
| glioma | upregulated | overall survival | [32-34] |
| lung cancer | upregulated | overall survival, pathological stage, TNM stage, tumor size, and lymph node metastasis | [35-39] |
| colorectal cancer | upregulated | tumor size, TNM stage, lymph nodes metastasis, distant metastasis, overall survival, disease-free survival, and vascular invasion | [40-44] |
| colorectal cancer | upregulated | / | [88] |
| breast cancer | upregulated | overall survival | [45-47] |
| bladder cancer | upregulated | histological grade, TNM stage, lymph nodes metastasis, and overall survival | [48-50] |
| osteosarcoma | upregulated | clinical stage, distance metastasis, chemotherapy resistance, and poor overall survival | [51-53] |
| cervical cancer | upregulated | / | [56,57] |
| esophageal adenocarcinoma | upregulated | / | [25] |
| hemangioma | upregulated | [54] | |
| oral squamous cell carcinoma | upregulated | nodal invasion, tumor stage, and tissue differentiation | [55] |
| laryngeal squamous cell carcinoma | downregulated | / | [22] |
| gastroenteropancreatic neuroendocrine neoplasms | downregulated | / | [58] |
Table 2.
The roles and mechanisms of HNF1A-AS1 in human cancers
| Disease type | Role | Cell lines | Functions | Related mechanisms | Refs |
|---|---|---|---|---|---|
| Gastric cancer | tumor promoter | HGC-27, MKN-45, AGS, NCI-N87, and BGC-823 | cell proliferation, migration, invasion, and 5-FU resistance | miR-30b-5p, EIF5A2, miR-30b-3p, PIK3CD, PI3K, AKT, miR-661, CDC34, | [26-28] |
| tumor suppressor | AGS, BGC-823, and MKN-45 | / | / | [24] | |
| Gastric cancer | tumor promoter | BGC-823, and MKN-45 | cell proliferation, migration, and invasion | EGR1, and miR-661 | [28] |
| liver cancer | tumor promoter | SMMC-7721, MHCC97L, Huh7 and HepG2 | cell proliferation, apoptosis, and autophagy | miR-30b-5p, ATG5, EZH2, NKD1, and p21 | [29-31] |
| Tumor suppressor | Huh-7, MHCC-97L, MHCC-97H, MHCC-LM3, SMMC-7721, and YY-8103 | cell proliferation, migration, and invasion | SHP-1 | [23] | |
| glioma | tumor promoter | U251, SHG-44, LN229, LN18, U87, T98G, and A172 | cell proliferation, apoptosis, migration, and invasion | EGR1, miR-22-3p, ENO1, MYC, miR-32-5p, SOX4, miR-363-3p, MAP2K4, and JNK | [32-34] |
| lung cancer | tumor promoter | A549, SPC-A1, PC9, H1299, H1563, H1437, H520, H2023, H1650, H1703, SK-MES-1, and Calu-1 | cell proliferation, apoptosis, migration, and invasion | miR-92a-3p, MAP2K4, miR-149-5p, Cdk6, miR-17-5p, and DNMT1 | [35-39] |
| colorectal cancer | tumor promoter | LoVo, SW620, HT-29, DLD-1, Ls-174T, HCT 116, and SW480 | cell proliferation, migration, invasion, and glycolysis | PBX3, OTX1, ERK, MAPK, miR-124, MYO6, Wnt, β-catenin, miRNA-34a, SIRT1, and p53 | [40-44] |
| colorectal cancer | tumor promoter | HT-29, HTC116, RKO, and SW480 | / | / | [88] |
| breast cancer | tumor promoter | MDA-MB-453, MDA-MB-231, MDA-MB-468, MDA-MB-436, MCF-7, BT-20, BT549, ZR-75-30, and HCC1937 | cell proliferation, apoptosis, migration, invasion, and TAM resistance | GATA1, miR-32-5p, RNF38, miR-363, SERTAD3, TGF-β, Smad, miRNA-20a-5p, and TRIM32 | [45-47] |
| bladder cancer | tumor promoter | T24, J82, UMUC3S, W780, SV-HUC-1, and 5637 | cell proliferation, apoptosis, migration, and invasion | miR-101-3p, and Bcl-2 | [48-50] |
| osteosarcoma | tumor promoter | U2OS, SAOS-2, 143B, HOS, SOSP-9607, and MG63 | cell proliferation, apoptosis, migration, and invasion | miR-32-5p, HMGB1, Wnt, and β-catenin | [51-53] |
| cervical cancer | tumor promoter | HeLa/DDP | cell proliferation, apoptosis, and cisplatin resistance | / | [56,57] |
| esophageal adenocarcinoma | tumor promoter | jhu-esoad1, FLO-1, SKGT-4, and OE33 | cell proliferation, and migration | H19, chromatin and nucleosome assembly | [25] |
| hemangioma | tumor promoter | endothelial cells | cell proliferation, migration, and invasion | / | [54] |
| oral squamous cell carcinoma | tumor promoter | CAL-27, HN5, SCC-15, SCC-9, and Tca8113 | cell proliferation, apoptosis, migration, and invasion | STAT3, and Notch | [55] |
| laryngeal squamous cell carcinoma | Tumor suppressor | TU-177, AMC-HN-8, TU-686, and 293T | cell proliferation, migration, and invasion | / | [22] |
| gastroenteropancreatic neuroendocrine neoplasms | Tumor suppressor | QGP-1, and STC-1 | cell proliferation, migration, and invasion | TCF3, and Oncostatin M | [58] |
We present specific expression profiles, clinicopathological features, and biological roles of HNF1A-AS1 in various cancer types.
Gastric cancer
Gastric cancer is one of the commonest types of gastrointestinal malignancies and contributes to the second-highest cancer mortality rate worldwide [59-61]. Despite surgical excision is the primary remedies for early-stage and locally advanced gastric cancer, patients are usually diagnosed at a terminal period that is short of effectual therapies [62-65]. Therefore, exploring highly effective, targeted therapeutic agents for advanced gastric cancers is warranted.
Several studies have reported that HNF1A-AS1 mayact as an oncogene, with upregulated expression in gastric cancer tissues and cell lines (HGC-27, MKN-45, AGS, NCI-N87, and BGC-823) [26-28]. Additionally, high HNF1A-AS1 expression is correlated with LNM and poor response to 5-fluorouracil (5-FU)-based neoadjuvant chemotherapy. HNF1A-AS1 enhances cell proliferation and invasion, as well as tumor angiogenesis and lymphangiogenesis in mouse tumor xenograft assays. However, Dang et al. [24] demonstrated HNF1A-AS1 downregulation in gastric cancer, with HNF1A-AS1 downregulation associated with tumor size, serum CEA and CA19-9 concentration, and ribonucleotide reductase catalytic subunit M1 (RRM1) expression in tissue samples. Further research is needed to clarify the reasons for differential expression of HNF1A-AS1 in gastric cancer.
Liver cancer
Hepatocellular carcinoma (HCC) is frequent liver malignancies, with high morbidity and negative prognosis [66-69]. Some studies have shown that HNF1A-AS1 levels are clearly upregulated in HCCMMC-7721, Huh7, MHCC97L, and HepG2 Cells and tissue samples. Furthermore, in these studies, HNF1A-AS1 levels were positively associated with tumor stage, multiple lesions, as well as poor differentiation [29-31]. In vitro functional assays demonstrated that HNF1A-AS1 facilitates the proliferation, apoptosis, and autophagy in HepG2, SMCC-7721, and Huh7 cells. However, other studies have shown that HNF1A-AS1 is downregulated and can inhibit the proliferative and metastatic abilities of HCC Huh-7 cells and xenograft tumors [23]. These conflicting results in liver cancer indicate that HNF1A-AS1 expression and function may be affected by tissue and cellular specificity [23]. Further studies with large sample sizes and tissues and cells from multiple sources remain necessary to define the exact roles of HNF1A-AS1 in liver cancer.
Glioma
Glioma is an aggressive, intracranial neoplasm associated with high rates of mortality, recurrence, and morbidity [70-74]. Data from the Chinese Glioma Genome Atlas showed that overall survival time remains unfavorable for malignant gliomas, despite advances in neurosurgical techniques [75-77]. HNF1A-AS1 is overexpressed in glioma tissues and LN229, LN18, U251, SHG-44, U87, T98G, and A172 cells and is correlated with lower overall patient survival. Elevated HNF1A-AS1 strengthened glioma cell proliferation, metastasis, as well as suppressing apoptosis in A172, U87, U251 and LN18 cells as well as in xenograft mouse models [32-34].
Lung cancer
Lung cancer generally causes the death of cancer patients, with a 5-year survival rate of less than 20% worldwide [78-81]. The development of an effective approach for early lung cancer detection has been a top research priority for decades [82-84]. HNF1A-AS1 expression is increased in-tumor tissues and SPC-A1, A549, PC9, H1299, H1563, H1437, H520, H2023, H1650, H1703, SK-MES-1, and Calu-1 cells [35-39]. Additionally, HNF1A-AS1 expression interferes with patients’ unfavorable overall survival, advanced pathological stage, and LNM. HNF1A-AS1 has pro-lung cancer functions by regulating cell proliferation, cell apoptosis, invasion, and migration in H1563, SKMES1, A549, PC9, and SPC-A1 cells and promoting tumor growth in mouse xenograft models.
Colorectal cancer
The incidence of colorectal cancer, a common digestive tract malignancy, has been continuously increasing worldwide [4,85-87]. HNF1A-AS1 level is increased in colorectal cancer tissue samples and LoVo, SW620, HT-29, DLD-1, Ls-174T, HCT 116, and SW480 cells [40-44,88]. Additionally, patients with higher HNF1A-AS1 levels are linked to poor disease-free survival, later TNM stage, LNM, overall survival, and vascular invasion. Additionally, HNF1A-AS1 enhances cell proliferation, migration, invasion, and glycolysis in HT29, HCT116, and SW620 cells, as well as in in vivo xenograft mouse models (Figure 2).
Figure 2.
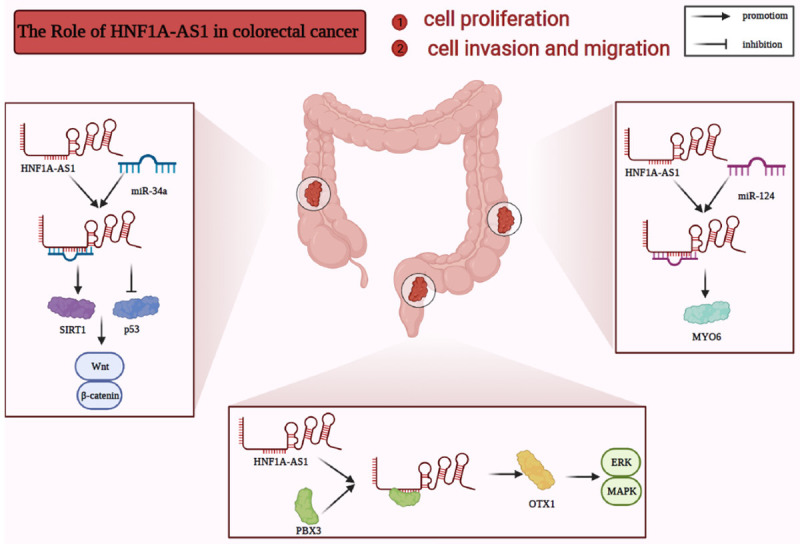
HNF1A-AS1 promotes colorectal cancer cell migration, invasion, and proliferation. In colorectal cancer, HNF1A-AS1 enhances cell proliferation, invasion, and migration by repressing miR-34a/SIRT1/p53 signaling and activating the Wnt/β-catenin signaling pathway; interacting with PBX3 and increasing OTX1 to activate the ERK/MAPK pathway; or sponging miR-124 and upregulating MYO6.
Breast cancer
Breast cancer remains the most prevalent cancer in women [89-92]. Early detection and diagnosis of breast cancer improves the patient outcomes and survival rates [93-96]. Several studies have indicated that HNF1A-AS1 is upregulated in breast cancer tissues and cell lines, including MDA-MB-453, MDA-MB-231, MDA-MB-468, MDA-MB-436, MCF-7, BT-20, BT549, ZR-75-30, and HCC1937 cells [45-47]. Additionally, high HNF1A-AS1 levels are significantly associated with lower breast cancer patient survival [46]. HNF1A-AS1 promotes cell proliferation, apoptosis, migration, invasion, and tamoxifen (TAM) sensitivity in MDA-MB231, MCF-7, MDA-MB-436, BT474, and BT-20 cells. Furthermore, HNF1A-AS1 oncogenic functions were further confirmed on the xenograft mouse model.
Bladder cancer
The early diagnosis and intervention are expected to greatly improve bladder cancer patient survival rates. HNF1A-AS1 is also overexpressed in bladder cancer [97-101]. Not surprisingly, highly expressed HNF1A-AS1 is positively related to histological grade, TNM stage, LNM, and overall survival time [48-50]. Furthermore, HNF1A-AS1 promotes cell proliferation, migration, and invasion and blocks apoptosis in T24, 5637, SW780, and UM-UC-3 cells.
Osteosarcoma
Osteosarcoma is a common primary aggressive bone tumor of children and adolescents [102-106]. HNF1A-AS1 expression is increased on osteosarcoma tissue samples and143B, HOS, U2OS, SAOS-2, SOSP-9607, and MG63 cells [51-53]. Furthermore, highly expressed HNF1A-AS1 is strongly correlated with poor clinical stage, distant metastasis, chemotherapy resistance, and poor overall survival. Functional experiments revealed that HNF1A-AS1 stimulates cell proliferation, migration, and invasion, in addition to repressing apoptosis in HOS, MG63, Saos-2, and U2OS cells.
Other cancers
Many other types of cancer are affected by HNF1A-AS1. HNF1A-AS1 expression is increased in cervical cancer tissues and cisplatin (DDP)-resistant (HeLa/DDP) cells. Furthermore, HNF1A-AS1 is involved in suppressing cell apoptosis, inducing cell proliferation, and promoting cisplatin resistance [56,57]. HNF1A-AS1 is upregulated in esophageal adenocarcinoma tissues [25], as well as in JH-EsoAd1, FLO-1, SKGT-4, and OE33 cells. HNF1A-AS1 also acts as an oncogene by promoting cell proliferation and migration in the esophageal adenocarcinoma cell lines SKGT-4 and OE33. HNF1A-AS1 overexpression promotes cell proliferation, migration, and invasion in hemangioma tissues and endothelial cells [54]. Additionally, HNF1A-AS1 expression is significantly higher in oral squamous cell carcinoma tissues as well as CAL-27, HN5, SCC-15, SCC-9, and Tca8113 cells than adjacent normal tissues and cell lines [55]. HNF1A-AS1 overexpression in Tca8113 and SCC-15 cells enhances cell proliferation and migration, restrains cell apoptosis, and is associated with aggressive nodal invasion, tumor stage, and tissue differentiation. By contrast, HNF1A-AS1 is decreased in laryngeal squamous cell carcinoma tissue samples, cell lines (TU-177, AMC-HN-8, TU-686, and 293T) [22], and metastatic tumors of cervical lymph nodes. Experimental data suggest that HNF1A-AS1 represses cell proliferation and metastasis in TU-177 and TU-686 cell lines and suppresses tumor growth, epithelial-mesenchymal transition, and LNM in in vivo xenograft models. Moreover, HNF1A-AS1 is decreased in gastroenteropancreatic neuroendocrine neoplasm tissues and cells (QGP-1 and STC-1) and plays key roles in inhibiting proliferation, migration, and invasion [58].
The oncological mechanisms of HNF1A-AS1 in human cancers
HNF1A-AS1 is involved in a variety of tumor cell processes that affect cell proliferation, apoptosis, invasion, migration, glycolysis, and drug resistance. We next probe the HNF1A-AS1 oncological mechanisms for the major biological functions of diverse cancers.
Cell proliferation
The dysregulation of cell proliferation is the main hallmark of cancer. Therefore, an in-depth understanding of cell proliferation can ultimately provide insight into cancer development [107-110]. HNF1A-AS1 modulates cell proliferation, including gastric cancer [28], liver cancer [23,30,31], glioma [32-34], lung cancer [35-37,39], colorectal cancer [42-44], breast cancer [45-47], bladder cancer [49,50], osteosarcoma [51-53], cervical cancer [56], esophageal adenocarcinoma [25], hemangioma [54], oral cancer [55], laryngeal cancer [22], and gastroenteropancreatic neuroendocrine neoplasms [58].
In gastric cancer, HNF1A-AS1 is activated by early growth response 1 (EGR1) and binds to microRNA (miR)-661 (miR-661), further elevating the expression of ubiquitin-conjugating enzyme E2 R1 (CDC34) and contributing to the acceleration of cell proliferation in both MKN-45 and BGC-823 cells [28]. HNF1A-AS1 promotes the proliferation of the HCC SMCC-7721 and Huh7 cell lines by combining with histone-lysine N-methyltransferase EZH2 (EZH2) and either downregulating the protein expression of naked cuticle homolog 1 (NKD1) and cyclin-dependent kinase inhibitor 1 (p21) and sponging miR-30b-5p, which enhances autophagy protein 5 (ATG5) expression [30,31]. HNF1A-AS1 also suppresses Huh-7 cell proliferation by directly interacting with tyrosine-protein phosphatase non-receptor type 6 (SHP-1) [23]. In the glioma cell lines U87-MG, U251, LN18, and A172, HNF1A-AS1 enhances cell proliferation through the EGR1/HNF1A-AS1/miR-22-3p/enolase 1 (ENO1) axis, the myc proto-oncogene protein (MYC)/HNF1A-AS1/miR-32-5p/transcription factor SOX-4 (SOX4) axis, and the MYC/HNF1A-AS1/miR-363-3p/MAP kinase 4 (MAP2K4)/c-Jun N-terminal kinase (JNK) axis [32-34]. HNF1A-AS1 also boosts the proliferative abilities of lung cancer SKMES1, H1563, SPC-A1, A549, PC9, as well as Calu-1 through the miR-92a-3p/MAP2K4/JNK pathway, the miR-149-5p/Cdk6 axis, the DNA (cytosine-5)-methyltransferase 1 (DNMT1)/cyclin D1 axis [35-37,39], and in combination with miR-17-5p. Additionally, HNF1A-AS1 facilitates proliferation in colorectal cancer cell lines HT29, HCT-116, and SW620 by activation of the Wnt/β-catenin and by inhibiting miR-34a/sirtuin 1 (SIRT1)/cellular tumor antigen p53 (p53) pathway and subsequently activating Wnt [42-44]. HNF1A-AS1 also plays a pro-proliferative role in breast cancer MDA-MB231, and BT474, by incorporating erythroid transcription factor (GATA1), sponging miR-32-5p, upregulating E3 ubiquitin-protein ligase RNF38, integrating with miR-363 to increase SERTA domain-containing protein 3 (SERTAD3) levels, ae well as acting on miRNA-20a-5p to elevate E3 ubiquitin-protein ligase TRIM32 [45-47]. HNF1A-AS1 also sponges miR-101-3p and increases the level of apoptosis regulator Bcl-2 (Bcl-2), promoting cell proliferation in 5637, T24, SW780, and UM-UC-3 bladder cancer cell lines [50]. Additionally, HNF1A-AS1 heightens proliferation in osteosarcoma cell lines HOS, MG63, and U2OS by interacting with miR-32-5p and increasing high mobility group box 1 (HMGB1) expression and by triggering Wnt/β-catenin [52,53]. In esophageal adenocarcinoma, HNF1A-AS1 may be involved in chromatin and nucleosome assembly or in the modulation of H19 to enhance cell proliferation in SKGT-4 and OE33 cells [25]. In the oral squamous cell carcinoma cell lines SCC-15 and Tca8113 [55], STAT3 upregulates HNF1A-AS1, and HNF1A-AS1 subsequently activates the Notch signaling pathway, exerting pro-proliferative functions. HNF1A-AS1 enhances cell proliferation by mediating the microRNA-34b/TUFT1 axis in DDP-resistant cells [56], which has implications for cervical cancer. Additionally, HNF1A-AS1 induced by IL-6 weakens miR-363-3p expression, which advances the process of hemangioma endothelial cell proliferation [54]. In gastroenteropancreatic neuroendocrine neoplasms and STC-1 and QGP-1 cell lines, HNF1A-AS1 exerts anti-proliferative functions by activating transcription factor 3, subsequently inhibiting Oncostatin M and stimulating the transforming growth factor-beta (TGF-β) signaling pathway [58].
Cell migration and invasion
Aberrant cell metastasis are crucial for cancer progression [111-115]. HNF1A-AS1 affects cell migration and invasion in gastric cancer [27,28], liver cancer [23], glioma [32,33], lung cancer [35-37,39], colorectal cancer [40-44], breast cancer [47], bladder cancer [49], osteosarcoma [51-53], esophageal adenocarcinoma [25], hemangioma [54], oral cancer [55], laryngeal cancer [22], and gastroenteropancreatic neuroendocrine neoplasms [58].
In the gastric cancer, HNF1A-AS1 induces cell migration and invasion by sponging miR-30b-3p and upregulating the phosphoinositol 3-kinase (PI3K)/AKT pathway or by interacting with miR-661 and increasing the expression of CDC34 after activation by EGR1 [27,28]. By contrast, HNF1A-AS1 blocks metastasis in the HCC cell line Huh-7 by functioning as phosphatase accelerators of SHP-1 [23]. HNF1A-AS1 can be activated by MYC, interacting with miR-32-5p to raise SOX4 levels, and activated by EGR1, influencing the miR-22-3p/ENO1 axis to promote cell invasion in glioma cell lines A172, U87, U251, and LN18 [32,33]. In several-lung cancer cell lines, HNF1A-AS1 facilitates the processes of cell migration and invasion through various mechanisms. In A549 and Calu-1 cells, HNF1A-AS1 facilitates the processes of cell migration and invasion by blocking miR-92a-3p and positively modulating the expression of MAP2K4. Furthermore, HNF1A-AS1 inhibits the expression of miR-17-5p in PC9 and A549 cells. Additionally, HNF1A-AS1 sponges miR-149-5p and upregulates Cdk6 in H1563 and SKMES1 cells [35-37]. In colon cancer, HNF1A-AS1 displays pro-metastatic functionality by binding to pre-B-cell leukemia transcription factor 3 (PBX3) and increasing homeobox protein OTX1 (OTX1) to activate the extracellular signal-regulated kinase 1/2 (ERK)/mitogen-activated protein kinase (MAPK) pathway by competitively combining with miR-124 and upregulating unconventional myosin-VI (MYO6), or by crippling the miR-34a/SIRT1/p53 axis and enhancing Wnt in HCT116 and SW620 cells [40-42,44]. HNF1A-AS1 accelerates migration and invasion in breast cancer cell lines MDA-MB231 and MCF-7 by sponging miR-20a-5p and upregulating TRIM32 expression [47]. Through different mechanisms, HNF1A-AS1 boosts cell migration and invasion in osteosarcoma cell lines. In MG63 and U2OS cells, HNF1A-AS1 binds to miR-32-5p, elevating HMGB1 expression. Furthermore, HNF1A-AS1 also promotes the Wnt/β-catenin signaling pathway in HOS and MG-63 cells [52,53]. In esophageal adenocarcinoma, HNF1A-AS1 is hypothesized to affect cell migration in SKGT-4 and OE33 cells by modulating chromatin and nucleosome assembly as well as H19 induction [25]. In the oral squamous cell carcinoma cell lines SCC-15 and Tca8113, HNF1A-AS1 enhances migration and invasion by the activation of the Notch signaling pathway [55], mediated by STAT3-linked upregulation of HNF1A-AS1. In hemangioma cell lines, IL-6 increases HNF1A-AS1 expression, which accelerates cell migration and invasion by sponging miR-363-3p [54].
Clinical significance of HNF1A-AS1 in cancer management
The diagnostic and prognostic value of HNF1A-AS1
Given the shortage of reliable diagnostic and prognostic biomarkers, the prognosis of cancer patients remains poor. Several findings demonstrate that HNF1A-AS1 levels of cancer cells and tissue samples are highly significant for the clinical diagnosis and prognosis of cancers evaluated by receiver operating characteristic (ROC) curve analysis, Kaplan-Meier analysis, or univariate and multivariate analyses.
For example, HNF1A-AS1 is regarded as a biomarker in gastric cancer for predicting LNM and is able to distinguish patients with LNM from those without with 0.7650 AUC value [27]. Further supporting the powerful diagnostic abilities of HNF1A-AS1, the AUC value of HNF1A-AS1 has been reported as high as 0.8714 in colorectal cancer [42]. Additionally, Kaplan-Meier [40], univariate, and multivariate analyses [44] suggest colon cancer patients with higher HNF1A-AS1 levels have unsatisfactory prognostic outcomes and higher disease relapse rates, further validating the prognostic potential of HNF1A-AS1. The diagnostic value of HNF1A-AS1 was also validated in cervical cancer by ROC curve analysis, with an AUC of 0.774 [57]. Univariate and multivariate analyses [38,48,53] have shown HNF1A-AS1 can represent an independent marker of poor prognosis in non-small cell lung cancer, bladder cancer, and osteosarcoma (P<0.05). More importantly, HNF1A-AS1 expression in serum correlates with patient status, revealing that HNF1A-AS1 has more effective diagnostic value than alkaline phosphatase (ALP) for differentiating osteosarcoma patients from healthy cases, with a high AUC value of 0.849 [52]. Blood specimens have several advantages, including easy availability, reduced trauma, lower risk, and high cost-effectiveness associated with sample collection compared with obtaining a cell or tissue biopsy, making blood samples convenient for long-term and continuous monitoring of cancer [116-119]. Simple, quantitative blood assays would provide greater opportunities for cancer identification and earlier intervention and would ultimately increase therapeutic effectiveness [120-124]. Research has focused mostly on HNF1A-AS1 expression in cancer tissues and cells, which is limited for several inherent factors, such as trauma, complicated operation, and high cost, making it unsuitable for early disease diagnosis and the evaluation of disease prognosis. Further study is required to investigate HNF1A-AS1 expression as a diagnostic or prognostic biomarker in less invasive biological specimens, such as blood.
The therapeutic value of HNF1A-AS1
HNF1A-AS1 is dysregulated in different cancers and exerts pro-oncogenic or tumor-suppressing effects. Simultaneously, HNF1A-AS1 modulates a wide range of the vital biological processes of cancer through diverse molecular mechanisms, especially resistance to chemotherapy and radiotherapy (Figure 3). Based on the above characteristics, upregulating or downregulating HNF1A-AS1 expression as well as targeting HNF1A-AS1-related molecules and pathways may ultimately pave the way for novel cancer treatments.
Figure 3.

HNF1A-AS1-mediated chemoresistance and radioresistance in cancers. HNF1A-AS1 increases 5-FU resistance in the gastric cancer HGC-27 and MKN-45 by sponging miR-30b-5p and upregulating EIF5A2. HNF1A-AS1 also enhances TAM resistance in breast cancer cell lines MCF-7 and BTF474 via reducing miR-363 and promoting SERTAD3 expression to activate the TGF-β/Smad pathway. In-lung cancer cells, HNF1A-AS1 strengthened radiotherapy resistance of A549 and Calu-1 cells by modulating the miR-92a-3p/MAP2K4/JNK pathway.
For example, HNF1A-AS1 reduces tumor cell sensitivity to 5-FU by obstructing miR-30b-5p expression and upregulating eukaryotic translation initiation factor 5A-2 (EIF5A2) in the gastric cancer, suggesting that HNF1A-AS1 knockout may result in the remission of cell chemoresistance [26]. Similarly, in breast cancer cells [46], HNF1A-AS1 increases TAM resistance by sponging miR-363 and promoting SERTAD3 expression to activate the TGF-β/Smad pathway. HNF1A-AS1 also impairs the radiotherapy sensitivity of A549 and Calu-1 lung cancer cell lines by regulating the miR-92a-3p/MAP2K4/JNK pathway, suggesting that inhibiting HNF1A-AS1 could strengthen the efficacy of radiotherapy in non-small cell lung cancer [35]. Furthermore, HNF1A-AS1 enhances ritonavir (RTV)-induced hepatotoxicity mainly by inflencing cytochrome P450 3A4 (CYP3A4) expression in hepatoma cells, which provides direction for subsequent research and therapeutics based on the mechanisms of RTV-induced liver damage [29]. Although clinical research of HNF1A-AS1 remains in its infancy, it is a promising field of study for further exploring the clinical value of HNF1A-AS1 in non-invasive cancer detection, prognosis, diagnosis, and treatment effectiveness.
Conclusion
HNF1A-AS1 expression is dysregulated in diverse cancer types, including gastric cancer, liver cancer, glioma, lung cancer, colorectal cancer, breast cancer, bladder cancer, osteosarcoma, esophageal adenocarcinoma, hemangioma, oral cancer, laryngeal cancer, cervical cancer, and gastroenteropancreatic neuroendocrine neoplasms. Furthermore, HNF1A-AS1 is correlated with a series of clinical characteristics, including TNM stage, tumor size, lymphatic and distant metastasis, and disease-free and overall survival. HNF1A-AS1 participates in multiple critical biological processes in cancer cells by regulating cell proliferation, invasion, and migration, all of which can affect cancer development. Therefore, HNF1A-AS1 could be utilized in promising medicinal applications for cancer, such as diagnosis, prognosis, and treatment.
Altogether, we currently have a rudimentary understanding of the functions and applicational value of HNF1A-AS1 in cancers. Further research into the mechanisms and clinical applications of HNF1A-AS1 is warranted to assess the expression, stability, and presence of HNF1A-AS1 in non-invasive bodily fluids, as well as the effectiveness and safety of targeted-HNF1A-AS1 therapies.
Acknowledgements
This work was funded by Henan Medical Science and Technology Joint Building Program (LHGJ20210308 and LHGJ20210328).
Disclosure of conflict of interest
None.
References
- 1.Chen W, Zheng R, Zhang S, Zeng H, Xia C, Zuo T, Yang Z, Zou X, He J. Cancer incidence and mortality in China, 2013. Cancer Lett. 2017;401:63–71. doi: 10.1016/j.canlet.2017.04.024. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, Znaor A, Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 4.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 5.Yang Z, Wu L, Wang A, Tang W, Zhao Y, Zhao H, Teschendorff AE. dbDEMC 2.0: updated database of differentially expressed miRNAs in human cancers. Nucleic Acids Res. 2017;45:D812–D818. doi: 10.1093/nar/gkw1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C, Shu L, Zou W. Role of long non-coding RNA TP73-AS1 in cancer. Biosci Rep. 2019;39:BSR20192274. doi: 10.1042/BSR20192274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng J, Li Y, Wang Y, Xie G, Feng Q, Yang Y, Feng J. lncRNA 00312 attenuates cell proliferation and invasion and promotes apoptosis in renal cell carcinoma via miR-34a-5p/ASS1 axis. Oxid Med Cell Longev. 2020;2020:5737289. doi: 10.1155/2020/5737289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patrushev LI, Kovalenko TF. Functions of noncoding sequences in mammalian genomes. Biochemistry (Mosc) 2014;79:1442–1469. doi: 10.1134/S0006297914130021. [DOI] [PubMed] [Google Scholar]
- 9.Mosca L, Vitiello F, Borzacchiello L, Coppola A, Tranchese RV, Pagano M, Caraglia M, Cacciapuoti G, Porcelli M. Mutual correlation between non-coding RNA and S-adenosylmethionine in human cancer: roles and therapeutic opportunities. Cancers (Basel) 2021;13:3264. doi: 10.3390/cancers13133264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pant T, Dhanasekaran A, Zhao M, Thorp EB, Forbess JM, Bosnjak ZJ, Benjamin IJ, Ge ZD. Identification and analysis of circulating long non-coding RNAs with high significance in diabetic cardiomyopathy. Sci Rep. 2021;11:2571. doi: 10.1038/s41598-021-82345-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganesh S, Horvat F, Drutovic D, Efenberkova M, Pinkas D, Jindrova A, Pasulka J, Iyyappan R, Malik R, Susor A, Vlahovicek K, Solc P, Svoboda P. The most abundant maternal lncRNA Sirena1 acts post-transcriptionally and impacts mitochondrial distribution. Nucleic Acids Res. 2020;48:3211–3227. doi: 10.1093/nar/gkz1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng Q, Lin Z, Xu J, Lu Y, Meng Q, Wang C, Yang Y, Xin X, Li X, Pu H, Gui X, Li T, Xiong W, Lu D. Long noncoding RNA MEG3 suppresses liver cancer cells growth through inhibiting β-catenin by activating PKM2 and inactivating PTEN. Cell Death Dis. 2018;9:253. doi: 10.1038/s41419-018-0305-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pu FF, Shi DY, Chen T, Liu YX, Zhong BL, Zhang ZC, Liu WJ, Wu Q, Wang BC, Shao ZW, He TC, Liu JX. SP1-induced long non-coding RNA SNHG6 facilitates the carcinogenesis of chondrosarcoma through inhibiting KLF6 by recruiting EZH2. Cell Death Dis. 2021;12:59. doi: 10.1038/s41419-020-03352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi S, Noro R, Seike M, Zeng C, Matsumoto M, Yoshikawa A, Nakamichi S, Sugano T, Hirao M, Matsuda K, Hamada M, Gemma A. Long non-coding RNA CRNDE is involved in resistance to EGFR tyrosine kinase inhibitor in EGFR-mutant lung cancer via eIF4A3/MUC1/EGFR signaling. Int J Mol Sci. 2021;22:4005. doi: 10.3390/ijms22084005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng ZN, Huang GZ, Wu QQ, Ye HY, Zeng WS, Lv XZ. NF-κB-mediated lncRNA AC007271.3 promotes carcinogenesis of oral squamous cell carcinoma by regulating miR-125b-2-3p/Slug. Cell Death Dis. 2020;11:1055. doi: 10.1038/s41419-020-03257-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, Yang J, Wang HN, Fu RY, Liu XD, Piao YS, Wei LQ, Wang JW, Zhang L. LncRNA BCYRN1-induced autophagy enhances asparaginase resistance in extranodal NK/T-cell lymphoma. Theranostics. 2021;11:925–940. doi: 10.7150/thno.46655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han L, Li Z, Jiang Y, Jiang Z, Tang L. SNHG29 regulates miR-223-3p/CTNND1 axis to promote glioblastoma progression via Wnt/β-catenin signaling pathway. Cancer Cell Int. 2019;19:345. doi: 10.1186/s12935-019-1057-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Chen Y, Liu W, Liu T, Sun D. Hsa_circ_0128846 promotes tumorigenesis of colorectal cancer by sponging hsa-miR-1184 and releasing AJUBA and inactivating Hippo/YAP signalling. J Cell Mol Med. 2020;24:9908–9924. doi: 10.1111/jcmm.15590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong F, Ruan S, Wang J, Xia Y, Le K, Xiao X, Hu T, Wang Q. M2 macrophage-induced lncRNA PCAT6 facilitates tumorigenesis and angiogenesis of triple-negative breast cancer through modulation of VEGFR2. Cell Death Dis. 2020;11:728. doi: 10.1038/s41419-020-02926-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He J, Xue Y, Wang Q, Zhou X, Liu L, Zhang T, Shang C, Ma J, Ma T. Long non-coding RNA MIAT regulates blood tumor barrier permeability by functioning as a competing endogenous RNA. Cell Death Dis. 2020;11:936. doi: 10.1038/s41419-020-03134-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Li Z, Cui Y, Cui X, Chen C, Wang Z. Exosomes isolated from bone marrow mesenchymal stem cells exert a protective effect on osteoarthritis via lncRNA LYRM4-AS1-GRPR-miR-6515-5p. Front Cell Dev Biol. 2021;9:644380. doi: 10.3389/fcell.2021.644380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi Y, Zhang Q, Xie M, Feng Y, Ma S, Yi C, Wang Z, Li Y, Liu X, Liu H, Yang H, Yan Y, Zhang Y, Ren X, Luo H. Aberrant methylation-mediated decrease of lncRNA HNF1A-AS1 contributes to malignant progression of laryngeal squamous cell carcinoma via EMT. Oncol Rep. 2020;44:2503–2516. doi: 10.3892/or.2020.7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding CH, Yin C, Chen SJ, Wen LZ, Ding K, Lei SJ, Liu JP, Wang J, Chen KX, Jiang HL, Zhang X, Luo C, Xie WF. The HNF1α-regulated lncRNA HNF1A-AS1 reverses the malignancy of hepatocellular carcinoma by enhancing the phosphatase activity of SHP-1. Mol Cancer. 2018;17:63. doi: 10.1186/s12943-018-0813-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dang Y, Lan F, Ouyang X, Wang K, Lin Y, Yu Y, Wang L, Wang Y, Huang Q. Expression and clinical significance of long non-coding RNA HNF1A-AS1 in human gastric cancer. World J Surg Oncol. 2015;13:302. doi: 10.1186/s12957-015-0706-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang X, Song JH, Cheng Y, Wu W, Bhagat T, Yu Y, Abraham JM, Ibrahim S, Ravich W, Roland BC, Khashab M, Singh VK, Shin EJ, Yang X, Verma AK, Meltzer SJ, Mori Y. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut. 2014;63:881–890. doi: 10.1136/gutjnl-2013-305266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang L, Zhang Y, Su P, Ma Z, Ye X, Kang W, Liu Y, Yu J. Long non-coding RNA HNF1A-AS1 induces 5-FU resistance of gastric cancer through miR-30b-5p/EIF5A2 pathway. Transl Oncol. 2022;18:101351. doi: 10.1016/j.tranon.2022.101351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu HT, Ma RR, Lv BB, Zhang H, Shi DB, Guo XY, Zhang GH, Gao P. LncRNA-HNF1A-AS1 functions as a competing endogenous RNA to activate PI3K/AKT signalling pathway by sponging miR-30b-3p in gastric cancer. Br J Cancer. 2020;122:1825–1836. doi: 10.1038/s41416-020-0836-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu HT, Liu S, Liu L, Ma RR, Gao P. EGR1-mediated transcription of lncRNA-HNF1A-AS1 promotes cell-cycle progression in gastric cancer. Cancer Res. 2018;78:5877–5890. doi: 10.1158/0008-5472.CAN-18-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Yu Y, Wang P, Yang K, Wang Y, Yan L, Zhong XB, Zhang L. Long noncoding RNAs hepatocyte nuclear factor 4A antisense RNA 1 and hepatocyte nuclear factor 1A antisense RNA 1 are involved in ritonavir-induced cytotoxicity in hepatoma cells. Drug Metab Dispos. 2022;50:704–715. doi: 10.1124/dmd.121.000693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang C, Mou L, Chai HX, Wang F, Yin YZ, Zhang XY. Long non-coding RNA HNF1A-AS1 promotes hepatocellular carcinoma cell proliferation by repressing NKD1 and P21 expression. Biomed Pharmacother. 2017;89:926–932. doi: 10.1016/j.biopha.2017.01.031. [DOI] [PubMed] [Google Scholar]
- 31.Liu Z, Wei X, Zhang A, Li C, Bai J, Dong J. Long non-coding RNA HNF1A-AS1 functioned as an oncogene and autophagy promoter in hepatocellular carcinoma through sponging hsa-miR-30b-5p. Biochem Biophys Res Commun. 2016;473:1268–1275. doi: 10.1016/j.bbrc.2016.04.054. [DOI] [PubMed] [Google Scholar]
- 32.Ma C, Wang H, Zong G, He J, Wang Y, Yang F, Yang Z, Bian E, Zhao B. EGR1 modulated LncRNA HNF1A-AS1 drives glioblastoma progression via miR-22-3p/ENO1 axis. Cell Death Discov. 2021;7:350. doi: 10.1038/s41420-021-00734-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu J, Li R, Li L, Gu Y, Zhan H, Zhou C, Zhong C. MYC-activated lncRNA HNF1A-AS1 overexpression facilitates glioma progression via cooperating with miR-32-5p/SOX4 axis. Cancer Med. 2020;9:6387–6398. doi: 10.1002/cam4.3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bi Y, Mao Y, Su Z, Du J, Ye L, Xu F. Long noncoding RNA HNF1A-AS1 regulates proliferation and apoptosis of glioma through activation of the JNK signaling pathway via miR-363-3p/MAP2K4. J Cell Physiol. 2021;236:1068–1082. doi: 10.1002/jcp.29916. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z, Liu L, Du Y, Mi Y, Wang L. The HNF1A-AS1/miR-92a-3p axis affects the radiosensitivity of non-small cell lung cancer by competitively regulating the JNK pathway. Cell Biol Toxicol. 2021;37:715–729. doi: 10.1007/s10565-021-09595-z. [DOI] [PubMed] [Google Scholar]
- 36.Liu L, Chen Y, Li Q, Duan P. lncRNA HNF1A-AS1 modulates non-small cell lung cancer progression by targeting miR-149-5p/Cdk6. J Cell Biochem. 2019;120:18736–18750. doi: 10.1002/jcb.29186. [DOI] [PubMed] [Google Scholar]
- 37.Zhang G, An X, Zhao H, Zhang Q, Zhao H. Long non-coding RNA HNF1A-AS1 promotes cell proliferation and invasion via regulating miR-17-5p in non-small cell lung cancer. Biomed Pharmacother. 2018;98:594–599. doi: 10.1016/j.biopha.2017.12.080. [DOI] [PubMed] [Google Scholar]
- 38.Ma YF, Liang T, Li CR, Li YJ, Jin S, Liu Y. Long non-coding RNA HNF1A-AS1 up-regulation in non-small cell lung cancer correlates to poor survival. Eur Rev Med Pharmacol Sci. 2016;20:4858–4863. [PubMed] [Google Scholar]
- 39.Wu Y, Liu H, Shi X, Yao Y, Yang W, Song Y. The long non-coding RNA HNF1A-AS1 regulates proliferation and metastasis in lung adenocarcinoma. Oncotarget. 2015;6:9160–9172. doi: 10.18632/oncotarget.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu J, Meng X, Jia Y, Chai J, Wang J, Xue X, Dang T. Long non-coding RNA HNF1A-AS1 upregulates OTX1 to enhance angiogenesis in colon cancer via the binding of transcription factor PBX3. Exp Cell Res. 2020;393:112025. doi: 10.1016/j.yexcr.2020.112025. [DOI] [PubMed] [Google Scholar]
- 41.Guo X, Zhang Y, Liu L, Yang W, Zhang Q. HNF1A-AS1 Regulates Cell Migration, Invasion and Glycolysis via Modulating miR-124/MYO6 in Colorectal Cancer Cells. Onco Targets Ther. 2020;13:1507–1518. doi: 10.2147/OTT.S231249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X, Xiong Y, Tang F, Bian Y, Chen Y, Zhang F. Long noncoding RNA HNF1A-AS1 indicates a poor prognosis of colorectal cancer and promotes carcinogenesis via activation of the Wnt/β-catenin signaling pathway. Biomed Pharmacother. 2017;96:877–883. doi: 10.1016/j.biopha.2017.10.033. [DOI] [PubMed] [Google Scholar]
- 43.Zhu W, Zhuang P, Song W, Duan S, Xu Q, Peng M, Zhou J. Knockdown of lncRNA HNF1A-AS1 inhibits oncogenic phenotypes in colorectal carcinoma. Mol Med Rep. 2017;16:4694–4700. doi: 10.3892/mmr.2017.7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fang C, Qiu S, Sun F, Li W, Wang Z, Yue B, Wu X, Yan D. Long non-coding RNA HNF1A-AS1 mediated repression of miR-34a/SIRT1/p53 feedback loop promotes the metastatic progression of colon cancer by functioning as a competing endogenous RNA. Cancer Lett. 2017;410:50–62. doi: 10.1016/j.canlet.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 45.Yang J, Niu H, Chen X. GATA1-Activated HNF1A-AS1 facilitates the progression of triple-negative breast cancer via sponging miR-32-5p to upregulate RNF38. Cancer Manag Res. 2021;13:1357–1369. doi: 10.2147/CMAR.S274204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Liu L, Lv Y, Zhang Y, Zhang L, Yu H, Tian W, Zhang Z, Cui S. Silencing long non-coding RNA HNF1A-AS1 inhibits growth and resistance to TAM of breast cancer cells via the microRNA-363/SERTAD3 axis. J Drug Target. 2021;29:742–753. doi: 10.1080/1061186X.2021.1878362. [DOI] [PubMed] [Google Scholar]
- 47.Meng Q, Wang L, Lv Y, Wu J, Shi W. Deletion of HNF1A-AS1 suppresses the malignant phenotypes of breast cancer cells in vitro and in vivo through targeting miRNA-20a-5p/TRIM32 axis. Cancer Biother Radiopharm. 2021;36:23–35. doi: 10.1089/cbr.2019.3168. [DOI] [PubMed] [Google Scholar]
- 48.Wang YH, Liu YH, Ji YJ, Wei Q, Gao TB. Upregulation of long non-coding RNA HNF1A-AS1 is associated with poor prognosis in urothelial carcinoma of the bladder. Eur Rev Med Pharmacol Sci. 2018;22:2261–2265. doi: 10.26355/eurrev_201804_14813. [DOI] [PubMed] [Google Scholar]
- 49.Feng Z, Wang B. Long non-coding RNA HNF1A-AS1 promotes cell viability and migration in human bladder cancer. Oncol Lett. 2018;15:4535–4540. doi: 10.3892/ol.2018.7878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhan Y, Li Y, Guan B, Wang Z, Peng D, Chen Z, He A, He S, Gong Y, Li X, Zhou L. Long non-coding RNA HNF1A-AS1 promotes proliferation and suppresses apoptosis of bladder cancer cells through upregulating Bcl-2. Oncotarget. 2017;8:76656–76665. doi: 10.18632/oncotarget.20795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lou P, Ding T, Zhan X. Long noncoding RNA HNF1A-AS1 regulates osteosarcoma advancement through modulating the miR-32-5p/HMGB1 axis. Cancer Biother Radiopharm. 2021;36:371–381. doi: 10.1089/cbr.2019.3486. [DOI] [PubMed] [Google Scholar]
- 52.Cai L, Lv J, Zhang Y, Li J, Wang Y, Yang H. The lncRNA HNF1A-AS1 is a negative prognostic factor and promotes tumorigenesis in osteosarcoma. J Cell Mol Med. 2017;21:2654–2662. doi: 10.1111/jcmm.12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao H, Hou W, Tao J, Zhao Y, Wan G, Ma C, Xu H. Upregulation of lncRNA HNF1A-AS1 promotes cell proliferation and metastasis in osteosarcoma through activation of the Wnt/β-catenin signaling pathway. Am J Transl Res. 2016;8:3503–3512. [PMC free article] [PubMed] [Google Scholar]
- 54.Xu W, Tao S, Zhu X. HNF1A-AS1 inhibits proliferation, migration and invasion of IL-6-induced hemangioma endothelial cells by targeting miR-363-3p. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2021;38:1091–1096. doi: 10.3760/cma.j.cn511374-20200830-00633. [DOI] [PubMed] [Google Scholar]
- 55.Liu Z, Li H, Fan S, Lin H, Lian W. STAT3-induced upregulation of long noncoding RNA HNF1A-AS1 promotes the progression of oral squamous cell carcinoma via activating Notch signaling pathway. Cancer Biol Ther. 2019;20:444–453. doi: 10.1080/15384047.2018.1529119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luo X, Wei J, Yang FL, Pang XX, Shi F, Wei YX, Liao BY, Wang JL. Exosomal lncRNA HNF1A-AS1 affects cisplatin resistance in cervical cancer cells through regulating microRNA-34b/TUFT1 axis. Cancer Cell Int. 2019;19:323. doi: 10.1186/s12935-019-1042-4. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Xu J, Zou J, Wu L, Lu W. Transcriptome analysis uncovers the diagnostic value of miR-192-5p/HNF1A-AS1/VIL1 panel in cervical adenocarcinoma. Sci Rep. 2020;10:16584. doi: 10.1038/s41598-020-73523-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xue J, Bai J, Long Q, Wei Y, Pan J, Li X, Tang Q. TCF-3-mediated transcription of lncRNA HNF1A-AS1 targeting oncostatin M expression inhibits epithelial-mesenchymal transition via TGFβ signaling in gastroenteropancreatic neuroendocrine neoplasms. Aging (Albany NY) 2021;13:14065–14077. doi: 10.18632/aging.203024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23:700–713. doi: 10.1158/1055-9965.EPI-13-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yuan L, Xu ZY, Ruan SM, Mo S, Qin JJ, Cheng XD. Long non-coding RNAs towards precision medicine in gastric cancer: early diagnosis, treatment, and drug resistance. Mol Cancer. 2020;19:96. doi: 10.1186/s12943-020-01219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Giraud J, Bouriez D, Seeneevassen L, Rousseau B, Sifré E, Giese A, Mégraud F, Lehours P, Dubus P, Gronnier C, Varon C. Orthotopic patient-derived xenografts of gastric cancer to decipher drugs effects on cancer stem cells and metastatic dissemination. Cancers (Basel) 2019;11:560. doi: 10.3390/cancers11040560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Z, Pi J, Zou D, Wang X, Xu J, Yu S, Zhang T, Li F, Zhang X, Zhao H, Wang F, Wang D, Ma Y, Yu J. microRNA arm-imbalance in part from complementary targets mediated decay promotes gastric cancer progression. Nat Commun. 2019;10:4397. doi: 10.1038/s41467-019-12292-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen X, Zhang S, Du K, Zheng N, Liu Y, Chen H, Xie G, Ma Y, Zhou Y, Zheng Y, Zeng L, Yang J, Shen L. Gastric cancer-secreted exosomal X26nt increases angiogenesis and vascular permeability by targeting VE-cadherin. Cancer Sci. 2021;112:1839–1852. doi: 10.1111/cas.14740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Petrelli F, Zaniboni A, Ghidini A, Ghidini M, Turati L, Pizzo C, Ratti M, Libertini M, Tomasello G. Timing of adjuvant chemotherapy and survival in colorectal, gastric, and pancreatic cancer. A systematic review and meta-analysis. Cancers (Basel) 2019;11:550. doi: 10.3390/cancers11040550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen M, Fan L, Zhang SM, Li Y, Chen P, Peng X, Liu DB, Ma C, Zhang WJ, Zou ZW, Li PD. LINC01939 inhibits the metastasis of gastric cancer by acting as a molecular sponge of miR-17-5p to regulate EGR2 expression. Cell Death Dis. 2019;10:70. doi: 10.1038/s41419-019-1344-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li M, Gao J, Li D, Yin Y. CEP55 promotes cell motility via JAK2-STAT3-MMPs cascade in hepatocellular carcinoma. Cells. 2018;7:99. doi: 10.3390/cells7080099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Q, Zhang P, Li Z, Feng X, Lv C, Zhang H, Xiao H, Ding J, Chen X. Evaluation of polymer nanoformulations in hepatoma therapy by established rodent models. Theranostics. 2019;9:1426–1452. doi: 10.7150/thno.31683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang Y, Song S, Meng Q, Wang L, Li X, Xie S, Chen Y, Jiang X, Wang C, Lu Y, Xin X, Pu H, Gui X, Li T, Xu J, Li J, Jia S, Lu D. miR24-2 accelerates progression of liver cancer cells by activating Pim1 through tri-methylation of Histone H3 on the ninth lysine. J Cell Mol Med. 2020;24:2772–2790. doi: 10.1111/jcmm.15030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Davis GL, Dempster J, Meler JD, Orr DW, Walberg MW, Brown B, Berger BD, O’Connor JK, Goldstein RM. Hepatocellular carcinoma: management of an increasingly common problem. Proc (Bayl Univ Med Cent) 2008;21:266–280. doi: 10.1080/08998280.2008.11928410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun J, Sun Z, Gareev I, Yan T, Chen X, Ahmad A, Zhang D, Zhao B, Beylerli O, Yang G, Zhao S. Exosomal miR-2276-5p in plasma is a potential diagnostic and prognostic biomarker in glioma. Front Cell Dev Biol. 2021;9:671202. doi: 10.3389/fcell.2021.671202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shao W, Gu J, Huang C, Liu D, Huang H, Huang Z, Lin Z, Yang W, Liu K, Lin D, Ji T. Malignancy-associated metabolic profiling of human glioma cell lines using 1H NMR spectroscopy. Mol Cancer. 2014;13:197. doi: 10.1186/1476-4598-13-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang X, Sun CL, Hageman L, Smith K, Singh P, Desai S, Hawkins DS, Hudson MM, Mascarenhas L, Neglia JP, Oeffinger KC, Ritchey AK, Robison LL, Villaluna D, Landier W, Bhatia S. Clinical and genetic risk prediction of subsequent cns tumors in survivors of childhood cancer: a report from the COG ALTE03N1 study. J. Clin. Oncol. 2017;35:3688–3696. doi: 10.1200/JCO.2017.74.7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhan W. Effects of focused-ultrasound-and-microbubble-induced blood-brain barrier disruption on drug transport under liposome-mediated delivery in brain tumour: a pilot numerical simulation study. Pharmaceutics. 2020;12:69. doi: 10.3390/pharmaceutics12010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mangiola A, Anile C, Pompucci A, Capone G, Rigante L, De Bonis P. Glioblastoma therapy: going beyond Hercules Columns. Expert Rev Neurother. 2010;10:507–514. doi: 10.1586/ern.09.158. [DOI] [PubMed] [Google Scholar]
- 75.Cheng C, Feng S, Jiao J, Huang W, Huang J, Wang L, Jiang W, Jiang C, Dai M, Li Z, Zhang R, Sun J, Shao J. DLC2 inhibits development of glioma through regulating the expression ratio of TAp73α/TAp73β. Am J Cancer Res. 2018;8:1200–1213. [PMC free article] [PubMed] [Google Scholar]
- 76.Yang P, Wang Y, Peng X, You G, Zhang W, Yan W, Bao Z, Wang Y, Qiu X, Jiang T. Management and survival rates in patients with glioma in China (2004-2010): a retrospective study from a single-institution. J Neurooncol. 2013;113:259–266. doi: 10.1007/s11060-013-1103-9. [DOI] [PubMed] [Google Scholar]
- 77.Patel CK, Vemaraju R, Glasbey J, Shires J, Northmore T, Zaben M, Hayhurst C. Trends in peri-operative performance status following resection of high grade glioma and brain metastases: the impact on survival. Clin Neurol Neurosurg. 2018;164:67–71. doi: 10.1016/j.clineuro.2017.11.016. [DOI] [PubMed] [Google Scholar]
- 78.Wu JE, Wu YY, Tung CH, Tsai YT, Chen HY, Chen YL, Hong TM. DNA methylation maintains the CLDN1-EPHB6-SLUG axis to enhance chemotherapeutic efficacy and inhibit lung cancer progression. Theranostics. 2020;10:8903–8923. doi: 10.7150/thno.45785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hu C, Zhang Q, Tang Q, Zhou H, Liu W, Huang J, Liu Y, Wang Q, Zhang J, Zhou M, Sheng F, Lai W, Tian J, Li G, Zhang R. CBX4 promotes the proliferation and metastasis via regulating BMI-1 in lung cancer. J Cell Mol Med. 2020;24:618–631. doi: 10.1111/jcmm.14771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Koller M, Hjermstad MJ, Tomaszewski KA, Tomaszewska IM, Hornslien K, Harle A, Arraras JI, Morag O, Pompili C, Ioannidis G, Georgiou M, Navarra C, Chie WC, Johnson CD, Himpel A, Schulz C, Bohrer T, Janssens A, Kuliś D, Bottomley A. An international study to revise the EORTC questionnaire for assessing quality of life in lung cancer patients. Ann Oncol. 2017;28:2874–2881. doi: 10.1093/annonc/mdx453. [DOI] [PubMed] [Google Scholar]
- 81.Rosenzweig KE, Gomez JE. Concurrent chemotherapy and radiation therapy for inoperable locally advanced non-small-cell lung cancer. J. Clin. Oncol. 2017;35:6–10. doi: 10.1200/JCO.2016.69.9678. [DOI] [PubMed] [Google Scholar]
- 82.Aberle DR, Abtin F, Brown K. Computed tomography screening for lung cancer: has it finally arrived? Implications of the national lung screening trial. J. Clin. Oncol. 2013;31:1002–1008. doi: 10.1200/JCO.2012.43.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fontana RS, Sanderson DR, Woolner LB, Taylor WF, Miller WE, Muhm JR, Bernatz PE, Payne WS, Pairolero PC, Bergstralh EJ. Screening for lung cancer. A critique of the Mayo Lung Project. Cancer. 1991;67:1155–1164. doi: 10.1002/1097-0142(19910215)67:4+<1155::aid-cncr2820671509>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 84.Poulet C, Njock MS, Moermans C, Louis E, Louis R, Malaise M, Guiot J. Exosomal long non-coding RNAs in lung diseases. Int J Mol Sci. 2020;21:3580. doi: 10.3390/ijms21103580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Singh MP, Rai S, Singh NK, Srivastava S. Transcriptomic landscape of early age onset of colorectal cancer identifies novel genes and pathways in Indian CRC patients. Sci Rep. 2021;11:11765. doi: 10.1038/s41598-021-91154-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Deng YN, Xia Z, Zhang P, Ejaz S, Liang S. Transcription factor RREB1: from target genes towards biological functions. Int J Biol Sci. 2020;16:1463–1473. doi: 10.7150/ijbs.40834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ferlay J, Colombet M, Soerjomataram I, Dyba T, Randi G, Bettio M, Gavin A, Visser O, Bray F. Cancer incidence and mortality patterns in Europe: estimates for 40 countries and 25 major cancers in 2018. Eur J Cancer. 2018;103:356–387. doi: 10.1016/j.ejca.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 88.Sepehri Z, Banerjee A, Vizeacoumar FS, Freywald A, Vizeacoumar FJ, Dolinsky VW, Davie JR. Differential expression of HNF1A and HNF1A-AS1 in colon cancer cells. IUBMB Life. 2022;74:496–507. doi: 10.1002/iub.2609. [DOI] [PubMed] [Google Scholar]
- 89.Bohan S, Ramli Hamid MT, Chan WY, Vijayananthan A, Ramli N, Kaur S, Rahmat K. Diagnostic accuracy of tomosynthesis-guided vacuum assisted breast biopsy of ultrasound occult lesions. Sci Rep. 2021;11:129. doi: 10.1038/s41598-020-80124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang L, Liu Q, Lang GT, Cao AY, Shao ZM. Concordance of hormone receptor status and BRCA1/2 mutation among women with synchronous bilateral breast cancer. Front Oncol. 2020;10:27. doi: 10.3389/fonc.2020.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Forouzanfar M, Lachinani L, Dormiani K, Nasr-Esfahani MH, Ghaedi K. Increased expression of MUSASHI1 in epithelial breast cancer cells is due to down regulation of miR-125b. BMC Mol Cell Biol. 2021;22:10. doi: 10.1186/s12860-021-00348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Butti R, Das S, Gunasekaran VP, Yadav AS, Kumar D, Kundu GC. Receptor tyrosine kinases (RTKs) in breast cancer: signaling, therapeutic implications and challenges. Mol Cancer. 2018;17:34. doi: 10.1186/s12943-018-0797-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Teh YC, Tan GH, Taib NA, Rahmat K, Westerhout CJ, Fadzli F, See MH, Jamaris S, Yip CH. Opportunistic mammography screening provides effective detection rates in a limited resource healthcare system. BMC Cancer. 2015;15:405. doi: 10.1186/s12885-015-1419-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wilkinson L, Thomas V, Sharma N. Microcalcification on mammography: approaches to interpretation and biopsy. Br J Radiol. 2017;90:20160594. doi: 10.1259/bjr.20160594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cummings MC, Simpson PT, Reid LE, Jayanthan J, Skerman J, Song S, McCart Reed AE, Kutasovic JR, Morey AL, Marquart L, O’Rourke P, Lakhani SR. Metastatic progression of breast cancer: insights from 50 years of autopsies. J Pathol. 2014;232:23–31. doi: 10.1002/path.4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Qian X, Tucker A, Gidcumb E, Shan J, Yang G, Calderon-Colon X, Sultana S, Lu J, Zhou O, Spronk D, Sprenger F, Zhang Y, Kennedy D, Farbizio T, Jing Z. High resolution stationary digital breast tomosynthesis using distributed carbon nanotube x-ray source array. Med Phys. 2012;39:2090–2099. doi: 10.1118/1.3694667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guo C, Li X, Xie J, Liu D, Geng J, Ye L, Yan Y, Yao X, Luo M. Long noncoding RNA SNHG1 activates autophagy and promotes cell invasion in bladder cancer. Front Oncol. 2021;11:660551. doi: 10.3389/fonc.2021.660551. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 98.Al-Saraireh YM, Alshammari F, Youssef AMM, Al-Sarayreh S, Almuhaisen GH, Alnawaiseh N, Al Shuneigat JM, Alrawashdeh HM. Profiling of CYP4Z1 and CYP1B1 expression in bladder cancers. Sci Rep. 2021;11:5581. doi: 10.1038/s41598-021-85188-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lin JT, Tsai KW. Circulating miRNAs act as diagnostic biomarkers for bladder cancer in urine. Int J Mol Sci. 2021;22:4278. doi: 10.3390/ijms22084278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ma W, Zhang W, Shen L, Liu J, Yang F, Maskey N, Wang H, Zhang J, Yan Y, Yao X. Can smoking cause differences in urine microbiome in male patients with bladder cancer? A retrospective study. Front Oncol. 2021;11:677605. doi: 10.3389/fonc.2021.677605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Antoni S, Ferlay J, Soerjomataram I, Znaor A, Jemal A, Bray F. Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol. 2017;71:96–108. doi: 10.1016/j.eururo.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 102.Assi T, Watson S, Samra B, Rassy E, Le Cesne A, Italiano A, Mir O. Targeting the VEGF Pathway in Osteosarcoma. Cells. 2021;10:1240. doi: 10.3390/cells10051240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shen S, Yao T, Xu Y, Zhang D, Fan S, Ma J. CircECE1 activates energy metabolism in osteosarcoma by stabilizing c-Myc. Mol Cancer. 2020;19:151. doi: 10.1186/s12943-020-01269-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Whelan JS, Jinks RC, McTiernan A, Sydes MR, Hook JM, Trani L, Uscinska B, Bramwell V, Lewis IJ, Nooij MA, van Glabbeke M, Grimer RJ, Hogendoorn PC, Taminiau AH, Gelderblom H. Survival from high-grade localised extremity osteosarcoma: combined results and prognostic factors from three European Osteosarcoma Intergroup randomised controlled trials. Ann Oncol. 2012;23:1607–1616. doi: 10.1093/annonc/mdr491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li S, Pei Y, Wang W, Liu F, Zheng K, Zhang X. Extracellular nanovesicles-transmitted circular RNA has_circ_0000190 suppresses osteosarcoma progression. J Cell Mol Med. 2020;24:2202–2214. doi: 10.1111/jcmm.14877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Berlanga P, Muñoz L, Piqueras M, Sirerol JA, Sánchez-Izquierdo MD, Hervás D, Hernández M, Llavador M, Machado I, Llombart-Bosch A, Cañete A, Castel V, Font de Mora J. miR-200c and phospho-AKT as prognostic factors and mediators of osteosarcoma progression and lung metastasis. Mol Oncol. 2016;10:1043–1053. doi: 10.1016/j.molonc.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mazurek A, Luo W, Krasnitz A, Hicks J, Powers RS, Stillman B. DDX5 regulates DNA replication and is required for cell proliferation in a subset of breast cancer cells. Cancer Discov. 2012;2:812–825. doi: 10.1158/2159-8290.CD-12-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Martinez-Carreres L, Nasrallah A, Fajas L. Cancer: linking powerhouses to suicidal bags. Front Oncol. 2017;7:204. doi: 10.3389/fonc.2017.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Laengsri V, Nantasenamat C, Schaduangrat N, Nuchnoi P, Prachayasittikul V, Shoombuatong W. TargetAntiAngio: a sequence-based tool for the prediction and analysis of anti-angiogenic peptides. Int J Mol Sci. 2019;20:2950. doi: 10.3390/ijms20122950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hanna A, Shevde LA. Hedgehog signaling: modulation of cancer properies and tumor mircroenvironment. Mol Cancer. 2016;15:24. doi: 10.1186/s12943-016-0509-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Spatarelu CP, Zhang H, Trung Nguyen D, Han X, Liu R, Guo Q, Notbohm J, Fan J, Liu L, Chen Z. Biomechanics of collective cell migration in cancer progression: experimental and computational methods. ACS Biomater Sci Eng. 2019;5:3766–3787. doi: 10.1021/acsbiomaterials.8b01428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sengupta D, Bhattacharya G, Ganguli S, Sengupta M. Structural insights and evaluation of the potential impact of missense variants on the interactions of SLIT2 with ROBO1/4 in cancer progression. Sci Rep. 2020;10:21909. doi: 10.1038/s41598-020-78882-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kotini M, Barriga EH, Leslie J, Gentzel M, Rauschenberger V, Schambony A, Mayor R. Gap junction protein Connexin-43 is a direct transcriptional regulator of N-cadherin in vivo. Nat Commun. 2018;9:3846. doi: 10.1038/s41467-018-06368-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Menter DG, Dubois RN. Prostaglandins in cancer cell adhesion, migration, and invasion. Int J Cell Biol. 2012;2012:723419. doi: 10.1155/2012/723419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kashani AS, Packirisamy M. Cancer cells optimize elasticity for efficient migration. R Soc Open Sci. 2020;7:200747. doi: 10.1098/rsos.200747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Thiele JA, Bethel K, Králíčková M, Kuhn P. Circulating tumor cells: fluid surrogates of solid tumors. Annu Rev Pathol. 2017;12:419–447. doi: 10.1146/annurev-pathol-052016-100256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hwang WL, Pleskow HM, Miyamoto DT. Molecular analysis of circulating tumors cells: Biomarkers beyond enumeration. Adv Drug Deliv Rev. 2018;125:122–131. doi: 10.1016/j.addr.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 118.Sun YF, Yang XR, Zhou J, Qiu SJ, Fan J, Xu Y. Circulating tumor cells: advances in detection methods, biological issues, and clinical relevance. J Cancer Res Clin Oncol. 2011;137:1151–1173. doi: 10.1007/s00432-011-0988-y. [DOI] [PubMed] [Google Scholar]
- 119.Costa JL, Schmitt FC. Liquid biopsy: a new tool in oncology. Acta Cytol. 2019;63:448. doi: 10.1159/000501355. [DOI] [PubMed] [Google Scholar]
- 120.Moon DH, Lindsay DP, Hong S, Wang AZ. Clinical indications for, and the future of, circulating tumor cells. Adv Drug Deliv Rev. 2018;125:143–150. doi: 10.1016/j.addr.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 121.Masuda T, Hayashi N, Iguchi T, Ito S, Eguchi H, Mimori K. Clinical and biological significance of circulating tumor cells in cancer. Mol Oncol. 2016;10:408–417. doi: 10.1016/j.molonc.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Abalde-Cela S, Piairo P, Diéguez L. The significance of circulating tumour cells in the clinic. Acta Cytol. 2019;63:466–478. doi: 10.1159/000495417. [DOI] [PubMed] [Google Scholar]
- 123.Fernandes Marques J, Pereira Reis J, Fernandes G, Hespanhol V, Machado JC, Costa JL. Circulating tumor DNA: a step into the future of cancer management. Acta Cytol. 2019;63:456–465. doi: 10.1159/000492917. [DOI] [PubMed] [Google Scholar]
- 124.Li J, Liu R, Huang C, Chen S, Xu M. The introduction and clinical application of cell-free tumor DNA. Methods Mol Biol. 2018;1754:45–65. doi: 10.1007/978-1-4939-7717-8_4. [DOI] [PubMed] [Google Scholar]