Abstract
Based on nucleotide sequence homology with the Escherichia coli photolyase gene (phr), the phr sequence of Pseudomonas aeruginosa PAO1 was identified from the genome sequence, amplified by PCR, cloned, and shown to complement a known phr mutation following expression in Escherichia coli SY2. Stable, insertional phr mutants containing a tetracycline resistance gene cassette were constructed in P. aeruginosa PAO1 and P. syringae pv. syringae FF5 by homologous recombination and sucrose-mediated counterselection. These mutants showed a decrease in survival compared to the wild type of as much as 19-fold after irradiation at UV-B doses of 1,000 to 1,550 J m−2 followed by a recovery period under photoreactivating conditions. A phr uvrA mutant of P. aeruginosa PAO1 was markedly sensitive to UV-B irradiation exhibiting a decrease in survival of 6 orders of magnitude following a UV-B dose of 250 J m−2. Complementation of the phr mutations in P. aeruginosa PAO1 and P. syringae pv. syringae FF5 using the cloned phr gene from strain PAO1 resulted in a restoration of survival following UV-B irradiation and recovery under photoreactivating conditions. The UV-B survival of the phr mutants could also be complemented by the P. syringae mutagenic DNA repair determinant rulAB. Assays for increases in the frequency of spontaneous rifampin-resistant mutants in UV-B-irradiated strains containing rulAB indicated that significant UV-B mutability (up to a 51-fold increase compared to a nonirradiated control strain) occurred even in the wild-type PAO1 background in which rulAB only enhanced the UV-B survival by 2-fold under photoreactivating conditions. The frequency of occurrence of spontaneous nalidixic acid-resistant mutants in the PAO1 uvrA and uvrA phr backgrounds complemented with rulAB were 3.8 × 10−5 and 2.1 × 10−3, respectively, following a UV-B dose of 1,550 J m−2. The construction and characterization of phr mutants in the present study will facilitate the determination of the roles of light and dark repair systems in organisms exposed to solar radiation in their natural habitats.
Natural sunlight has a profound effect on much of the life on earth, both from the positive standpoint of the utilization of radiant energy for photochemical and photobiological processes and from the negative standpoint of the harmful effects of UV radiation (UVR) on organisms. The UV wavelengths which reach the earth's surface are classified as UV-A (320 to 400 nm) and UV-B (290 to 320 nm). The UV-A or far-UV wavelengths typically cause only indirect damage to cellular DNA through catalyzing the formation of chemical intermediates such as reactive oxygen species (10). In contrast, UV-B or near-UV radiation can cause direct DNA damage by inducing the formation of DNA photoproducts, of which the cyclobutane pyrimidine dimer (CPD) and the pyrimidine(6-4)pyrimidinone (6-4PP) are the most common (29). The accumulation of DNA photoproducts can be lethal to cells through the blockage of DNA replication and RNA transcription.
The effect of UV-B radiation on the ecology of microorganisms has been studied in aquatic systems and in the habitat of the plant leaf surface or phyllosphere. In marine environments, bacterioplankton are highly susceptible to UV-B, with results from field studies indicating that exposure to natural solar UVR can result in decreases in total cell density, reduction in amino acid uptake, increases in measurable CPDs, and significant inhibition of protein and DNA synthesis (3, 16, 26, 27). Solar UVRs also affect the population levels and species composition of bacteria recovered from the peanut phyllosphere at different times of day (39), and increased UV-B irradiation above ambient levels results in alterations of the relative abundance of certain fungal species in the oak phyllosphere (28). Examinations of the effects of UVR on individual bacterial isolates from marine or phyllosphere environments have shown a range of sensitivity levels among different species and among isolates within a species (2, 18, 39, 41). In one large-scale analysis, the majority of culturable isolates recovered from the peanut phyllosphere community were tolerant to relatively large doses of UV-C (254 nm) radiation in vitro (39).
Bacteria are particularly vulnerable to UVR damage because their small size limits effective cellular shading (13) and their unicellular nature places a paramount importance on the successful replication of the genome for growth to continue. Thus, the possession of mechanisms to repair UVR-induced DNA damage is an essential contributor to the ecological fitness of organisms that are regularly exposed to solar UVR. Bacteria have evolved four main mechanisms in the repair or damage tolerance of UVR-damaged DNA, including photoreactivation, nucleotide excision repair (NER), mutagenic DNA repair (MDR), and recombinational DNA repair (12). Photoreactivation in bacteria involves a single enzyme called photolyase which binds CPDs and, in a light-dependent step, monomerizes the CPD and dissociates from the repaired lesion (47). The photolyase enzyme contains two distinct chromophores, of which the first one (either 5,10-methenyl tetrahydrofolate or 8-hydroxy-5-deazaflavin) harvests light energy and transfers it to a reduced flavin chromophore that acts as the reaction center in CPD monomerization (22). CPD photolyases are widely but also sporadically distributed among eubacteria, archaea, and eukaryotes comprising two distinct classes: class I CPD photolyases are found in bacteria and lower eukaryotes, and class II CPD photolyases, with one exception, are found in higher eukaryotes (47).
Few ecological studies have delineated the importance of individual DNA repair or damage tolerance mechanisms in organisms for survival following exposure to solar UVR in their native habitats. An MDR system, encoded by the rulAB determinant of Pseudomonas syringae, confers a UVR tolerance phenotype that enables strains to maintain significantly larger populations in the bean phyllosphere following UV-B irradiation (40, 41). Spores of Bacillus subtilus were shown to require NER, spore photoproduct lyase (a photolyase), and an additional unknown mechanism for survival following exposure to solar radiation (45). The role of photoreactivation and dark repair has been inferred from field studies of CPD kinetics in bacterioplankton samples from the marine water column (16, 17). These studies showed that CPD levels followed a diel pattern with maximum damage present at late-afternoon samplings and subsequent reductions in CPDs during night-time hours (17). An additional study using artificial UV-B wavelengths and solar radiation also suggested the importance of exposure to photoreactivating wavelengths in bacterioplankton recovery following irradiation (19). While these studies suggest that both photoreactivation and dark repair are important processes in bacterioplankton, they have not resolved the importance of individual repair mechanisms nor have they accounted for the possibility of artifacts caused by differences in the relative abundance of particular organisms at each sampling time.
Our long-term goal is to perform ecological studies in which the survival of and relative rates of DNA repair of organism derivatives with alterations in specific DNA repair pathways can be examined in microcosms and in the field. In the present study, we isolated the gene encoding photolyase (phr) from P. aeruginosa, a ubiquitous soil and aquatic organism, and utilized the gene to construct stable insertional phr mutants of P. aeruginosa and P. syringae. We then assessed the contribution of photoreactivation, NER, and MDR to survival and mutability following UV-B irradiation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains, plasmids, and specific oligonucleotides utilized which were relevant to this study are listed in Table 1. All bacterial strains were grown in Luria-Bertani (LB) medium (Difco) or King's medium B (KB) (23); E. coli strains and P. aeruginosa PAO1 were grown at 37°C, and P. syringae pv. syringae FF5 was grown at 28°C. Plasmid transfer from E. coli to Pseudomonas strains was accomplished by triparental mating using the helper plasmid pRK2013. Briefly, saline (0.85% NaCl) washed cells from 10-ml overnight cultures of donor, recipient, and helper strains were mixed in a 3:3:1 ratio, spotted onto LB agar (total volume, 70 μl)/ and incubated at 28°C for 8 h. The cells were scraped from the plates into 3 ml of saline, and the exconjugants were selected by plating on MG medium (20) or Pseudomonas Isolation Agar (Difco) supplemented with appropriate antibiotics. Oligonucleotides were purchased from Life Technologies (Gaithersburg, Md.). Antibiotics were added to the media in the following concentrations: ampicillin, 75 μg ml−1; carbenicillin, 200 μg ml−1; gentamicin, (GEN) 100 μg ml−1 for P. aeruginosa and 20 μg ml−1 for P. syringae; nalidixic acid, 100 μg ml−1; rifampin, 100 μg ml−1; and tetracycline (TET), 200 μg ml−1 for P. aeruginosa and 12.5 μg ml−1 for P. syringae.
TABLE 1.
Bacterial strains, plasmids, and oligonucleotide primers utilized in this study and their relevant characteristics or sequence
| Strain, plasmid, or primer | Relevant characteristics or sequencea | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH10B | Plasmid-free strain used for cloning | 14 |
| SY2 | Same as JM107, but Δphr::CmrΔuvrA::KmrΔrecA::Tcr | 46 |
| P. aeruginosa | ||
| PAO1 | Prototrophic, plasmid-free strain | A. M. Chakrabarty |
| GWS250 | Same as PAO1, but phr::Tcr | This study |
| UA11079 | Same as PAO1, but Rifr, uvrA::ΩHg (Hgr), plasmid-free strain | 31 |
| GWS251 | Same as UA11079, but phr::Tcr | This study |
| P. syringae pv. syringae | ||
| FF5 | Prototrophic, plasmid-free strain | 38 |
| GWS252 | Same as FF5, but phr::Tcr | This study |
| Plasmids | ||
| pBBR1MCS-3 | Source of Tcr determinant | 24 |
| pCR2.1 | Apr Kmr, direct cloning vector for PCR products | Invitrogen |
| pGem7zf- | Apr, cloning vector | Promega |
| pJB321 | Cbr, broad-host-range cloning vector | 4 |
| pJQ200SK | Gmr, sacB, suicide gene replacement vector | 30 |
| pRK2013 | Helper plasmid for triparental matings | 11 |
| pJJK17 | 3.8-kb rulAB as XbaI-BamHI in pJB321 | 21 |
| pJJK44 | 1.2-kb 5′ end and sequences upstream of phr as SacI-ClaI in pGem 7zf− | This study |
| pJJK45 | 1.2-kb 3′ end and sequences downstream of phr as KpnI-XbaI in pJJK44 | This study |
| pJJK46 | 1.4-kb Tcr determinant from pBBR1MCS-3 in pCR2.1 | This study |
| pJJK47 | 1.4-kb Tcr determinant from pJJK46 as BglII, blunt ended and ligated into SmaI site of pJJK45 | This study |
| pJJK48 | phr::Tcr cassette as SacI-XbaI in pJQ200SK | This study |
| pJJK55 | 2.4-kb phr gene as SacI-XbaI in pBluescript SK | This study |
| pJJK76 | 2.4-kb phr gene from pJJK55 as SacI-EcoRI in pJB321 | This study |
| pMS969 | Apr, contains phr from E. coli | 33 |
| Primers | ||
| phr 5′ SacI | 5′-GTACGAGCTCGCGCTCTAGGATTTTCC-3′ | This study |
| phr int ClaI | 5′-CCATCGATACCGCGCCGACTCCCAG-3′ | This study |
| phr int KpnI | 5′-GGGGTACCTCATCGACGGTGACC-3′ | This study |
| phr 3′ XbaI | 5′-GCTCTAGACTGGCCTCTGGCCAGGCC-3′ | This study |
| phr 3′ orf XbaI | 5′-GCTCTAGAGCGAGCCATTTGGCG-3′ | This study |
| 5′ BglII Tc | 5′-TCCAGATCTTCAGGTCGAGGTGGCCC-3′ | This study |
| 3′ BglII Tc | 5′-GGAAGATCTCAGGAACGCGGGCGCGCAC-3′ | This study |
Phenotype resistance (r) abbreviations are as follows: Ap, ampicillin; Cb, carbenicillin; Cm, chloramphenicol; Hg, mercury; Km, kanamycin; Rif, rifampin; Tc, tetracycline. For primer oligonucleotide sequences, the restriction sites incorporated in primers are underlined: AGATCT, BglII; ATCGAT, ClaI; GGTACC, KpnI; GAGCTC, SacI; TCTAGA, XbaI.
Molecular biology techniques.
Plasmid isolation from P. syringae was accomplished using the technique of Crosa and Falkow (7). Restriction digestions, isolation of DNA fragments from agarose gels, PCR amplifications, ligations, and Southern transfer to nylon membranes were performed using standard techniques (32). Genomic DNA from Pseudomonas strains was isolated using a genomic DNA isolation kit (Qiagen, Valencia, Calif.). DNA fragments used as probes were labeled with digoxigenin-11-dUTP (Genius kit; Boehringer Mannheim, Indianapolis, Ind.) according to the instructions of the manufacturer. Hybridizations at 65°C followed by high-stringency washes were performed as described previously (38).
Construction of photolyase-deficient mutants of P. aeruginosa PAO1 and P. syringae pv. syringae FF5.
The putative photolyase gene sequence from P. aeruginosa PAO1 was identified by performing a BLASTx search of the 15 March 1999 sequence release of the P. aeruginosa genome sequencing project, which was available at (http://www.pseudomonas.com), using the known sequence of the E. coli phr gene (33). A single region of significant sequence homology was detected, suggesting the presence of a phr gene of 1,446 bp within the PAO1 genome. The intact putative PAO1 phr gene was amplified with the oligonucleotide primers phr 5′ SacI and phr 3′ orf XbaI, excised with the appropriate enzymes, and ligated into pJB321, creating the plasmid pJJK76. A strategy was also designed to amplify the putative phr sequence, including approximately 900 bp of flanking sequence, in two discontinuous segments. The primers phr 5′ SacI and phr int ClaI and phr int KpnI and phr 3′ XbaI were utilized to amplify the 5′ and 3′ regions as 1.2-kb fragments, respectively. The amplified fragments were digested with the appropriate restriction enzymes and sequentially ligated into pGem 7zf−, creating the plasmids pJJK44 and pJJK45. These cloning steps resulted in the deletion of approximately 57% of the phr coding sequence. With the fragments ligated in this manner, a SmaI site was maintained within the pGem7zf− polylinker between the ClaI and KpnI sites. This site was used to insert a tetracycline resistance (Tcr) determinant (amplified from pBBR1MCS-3 using the primers Tc 5′ BglII and Tc 3′ BglII) that was excised from pJJK46 with BglII, and the ends were made blunt with Klenow fragment prior to ligation, creating pJJK47, and thereby generating an insertional mutation within phr. The entire cassette consisting of flanking sequences, phr sequences, and Tcr determinant was excised with SacI and XbaI and ligated into the suicide gene replacement vector pJQ200SK creating pJJK48. pJJK48 was transferred into P. aeruginosa PAO1, P. aeruginosa UA11079, and P. syringae pv. syringae FF5 by triparental mating. Following transfer, those recipient cells in which a plasmid integration event had occurred were selected on MG supplemented with GEN. Several isolated Gmr colonies were then cultured in LB broth containing TET. After 2 days of incubation, 0.1-ml samples were plated onto LB containing TET and 5% sucrose to counterselect against the sacB gene encoded on pJQ200SK. The sucrose-resistant (Sucr) colonies recovered were subsequently tested for sensitivity to GEN as a phenotypic assay for loss of the vector sequences. Confirmation of loss of the vector sequences and the insertion of the Tcr cassette within the phr gene was done using Southern hybridization analysis of SacI- or XbaI-digested genomic DNA from the relevant strains using the PAO1 phr coding sequence from pJJK76 and the Tcr determinant from pJJK46 as probes.
UV-B irradiation and MDR assays.
Bacterial strains were grown to late-log phase in LB medium containing the appropriate antibiotics; 1 ml of the cultures were pelleted, washed with an equal volume of sterile saline solution, and resuspended in an equal volume of saline. For assays involving E. coli SY2/pMS969 and SY2/pJJK76, 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the culture medium prior to inoculation, since the phr gene in pMS969 is under the regulatory control of the vector tac promoter and the phr gene in pJJK76 is under the regulatory control of the pJB321 vector lac promoter. It should be noted that E. coli SY2 is a derivative of JM107 which contains the lacIq gene on the F′ plasmid; pMS969, but not pJJK76, also encodes the lacIq gene. Cell suspensions were then diluted with an additional 9 ml of saline, placed in a sterile glass petri dish, and irradiated with UV-B wavelengths (peak at 302 nm) using an XX-15M model UV-B lamp (UVP Products, San Gabriel, Calif.) filtered through cellulose diacetate (Kodacel; Eastman Kodak, Rochester, N.Y.) to eliminate any UV-C wavelengths (<290 nm) given off. The UV-B lamp was turned on 15 min prior to use to allow for stabilization of the UVR output. The energy output of the XX-15M lamp was monitored with a UV-X radiometer (UVP Products) and determined to be 4.0 J m−2 s−1. Cell suspensions were mixed continuously while receiving the UV-B dose to eliminate survival as a result of shading. For the light repair assays, irradiated cells were maintained in covered glass petri dishes and illuminated for 1 h under white fluorescent lamps at 25°C prior to enumeration by dilution plating. Light intensity was measured with an LI-190SA quantum sensor (Licor, Lincoln, Nebr.) and was approximately 120 μmol s−1 m−2 (i.e., 7.5 × 1019 photons s−1 m−2). For the dark repair assays, irradiated cells were plated under conditions in which the UV-B lamp provided the only source of illumination.
For the MDR assays, 0.1 ml of untreated cells and cells from all UV-B treatments were diluted in 0.9 ml of sterile saline and added to 1 ml of 2× LB broth. Following overnight incubation in total darkness, appropriate dilutions of cell suspensions were plated on LB medium, LB medium containing rifampin, or LB medium containing nalidixic acid. The mutation frequency to rifampin resistance (Rifr) or nalidixic acid resistance (Nalr) was calculated as the number of Rifr or Nalr mutants per 108 cells.
RESULTS
Cloning of the P. aeruginosa PAO1 photolyase gene.
A search of the in-progress P. aeruginosa genome sequence (15 March, 1999 update) using the program BLASTx (1) revealed the presence of a single open reading frame (ORF) with significant sequence similarity with the E. coli phr sequence. A recent, additional search of the completed P. aeruginosa genome sequence confirmed that only a single sequence showed homology with E. coli phr and indicated that this putative phr sequence was assigned gene number PA4660 (6, 37). A multiple sequence alignment of the PAO1 phr sequence with other known phr sequences in the GenBank database indicated that the PAO1 phr sequence was most similar to that of class I photolyases (data not shown). The PAO1 phr gene appears to be expressed as part of an operon, since immediately upstream are three additional genes, PA4657 to PA4659, that would be transcribed in the same direction as phr (Table 2). There are 213 bp of noncoding sequence between genes PA4657 and PA4658 and 414 bp of noncoding sequence upstream of gene PA4657, where promoter sequences regulating the expression of phr could be located. Genes PA4656 and PA4661 (immediately downstream of phr) are both transcribed from the opposite DNA strand (37).
TABLE 2.
Description of ORFs adjacent to the P. aeruginosa PAO1 phr gene
| Gene | Location (bp)a | Closest homologb (GenBank accession no.) |
|---|---|---|
| phr | 5,227,887–5,229,332 | Photolyase from E. coli (AAC73802) |
| PA4659 | 5,226,991–5,227,890 | Transcriptional regulator of the MerR family from Vibrio cholerae (AAF95970) |
| PA4658 | 5,226,025–5,226,981 | Photoreactivation-associated protein from Methanothermobacter thermoautotrophicus (AAB85401) |
| PA4657 | 5,224,889–5,225,872 | Hypothetical protein from Synechocystis sp. (D90906) |
The gene location is taken from the complete genome sequence of PAO1 available at http://www.pseudomonas.com.
The closest homologs were identified in the present study through the use of BLASTp (1) searches.
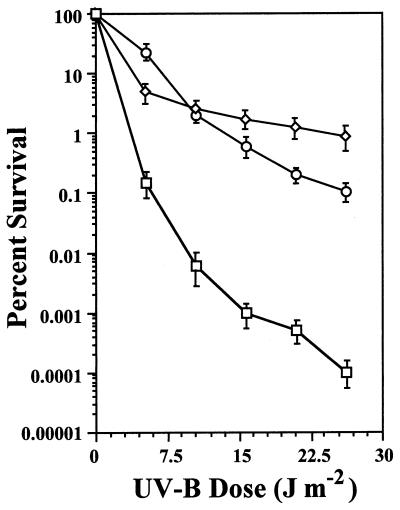
The putative phr ORF and approximately 0.9 kb of upstream and downstream flanking DNA was amplified from a preparation of P. aeruginosa PAO1 genomic DNA and ligated into pJB321 creating the plasmid pJJK76. To show that the sequence we had cloned contained the functional PAO1 photolyase gene, we transformed the E. coli phr mutant SY2 with either the cloned PAO1 phr gene on pJJK76 or the cloned E. coli phr gene on pMS969. Strain SY2 also carries mutant recA and uvrA alleles and is thus highly sensitive to UVR. Following irradiation with UV-B and exposure to visible light, the survival of E. coli SY2/pJJK76 and SY2/pMS969 was significantly increased (Fig. 1). The survival increase of SY2/pJJK76 was as great as 8,500-fold at the 26 J m−2 dose (Fig. 1). These initial results indicated that pJJK76 contained an active photolyase gene from P. aeruginosa PAO1. The slightly increased survival of SY2/pJJK76 relative to SY2/pMS969 at higher UV-B dose levels could be due to copy number differences between the respective plasmids, differential expression of the respective phr genes (possibly due to the presence of lacIq on pMS969), or to an increased efficiency of P. aeruginosa photolyase relative to E. coli photolyase.
FIG. 1.
Survival of E. coli SY2/pJB321 (phr) (□), SY2/pMS969 (phr/E. coli phr+) (○), and SY2/pJJK76 (phr/P. aeruginosa phr+) (◊) after UV-B irradiation. Each datum point represents the mean (± the standard error of the mean) from three replicate experiments.
Construction of photolyase knockout mutants of P. aeruginosa PAO1, P. aeruginosa UA11079, and P. syringae pv. syringae FF5.
We utilized a PCR strategy to amplify 5′ and 3′ sections of the PAO1 phr gene along with approximately 900 bp of the flanking sequences on each end. Briefly, mutants were constructed via homologous recombination using pJJK48, a construct in the gene replacement vector pJQ200SK which contains a Tcr cassette inserted within phr. Following matings of pJJK48 with P. aeruginosa PAO1, UA11079, and P. syringae pv. syringae FF5, Gmr Tcr colonies were obtained, suggesting that the plasmid had recombined with the recipient strains at the phr locus. Following this initial recombination event, counterselection using TET and 5% sucrose was utilized, resulting in the selection of Sucr colonies which had undergone a second homologous recombination event generating the mutant strains GWS250 (PAO1 phr::Tc), GWS251 (UA11079 phr::Tc), and GWS252 (FF5 phr::Tc). The replacement of the wild-type phr sequence with the phr::Tc sequence in each strain was confirmed by separate hybridizations of genomic DNA digested with SacI or XbaI with the PAO1 phr sequence from pJJK76 and with the Tcr cassette from pJJK46 (data not shown).
UV-B sensitivity analysis of photolyase knockout mutants.
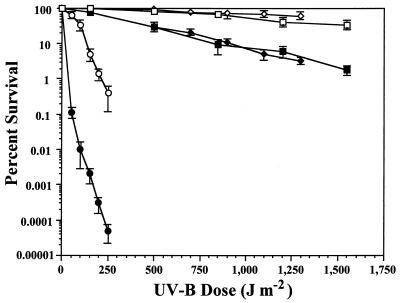
The survival of P. aeruginosa GWS250 was reduced by as much as 7-fold following UV-B doses up to 1,200 J m−2 and was reduced by 19-fold following a dose of 1,550 J m−2 compared to the survival of the wild-type PAO1 (Fig. 2). Similar results were observed in the comparison of P. syringae pv. syringae GWS252 and the FF5 wild-type (Fig. 2). We also observed the UV-B sensitivity of the PAO1 uvrA strain UA11079, and the PAO1 uvrA phr strain GWS251 constructed in this study. The maximum UV-B dose administered to these strains was 250 J m−2, approximately six times less than that of the wild-type or PAO1 phr backgrounds. The P. aeruginosa strain UA11079 was markedly reduced in survival, compared to the phr strain GWS250, following UV-B doses of as low as 150 to 250 J m−2 with recovery under photoreactivating conditions (Fig. 2). The P. aeruginosa uvrA phr double-mutant strain GWS251 was further reduced in UV-B survival with a difference of as high as 8,000-fold observed following the 250-J m−2 dose (Fig. 2). Complementation of the mutant strains GWS250, GWS251, and GWS252 with the cloned P. aeruginosa phr gene on pJJK76 restored the survival of strains to wild-type levels (data not shown).
FIG. 2.
Survival of P. aeruginosa PAO1 (□), GWS250 (PAO1 phr) (■), UA11079 (PAO1 uvrA) (○), and GWS251 (PAO1 uvrA phr) (●) and P. syringae pv. syringae FF5 (◊) and GWS252 (FF5 phr) (⧫) after UV-B irradiation. Each datum point represents the mean (± the standard error of the mean) from three replicate experiments.
Examination of the role of photoreactivation, NER, and MDR in the UV-B survival of P. aeruginosa under photoreactivating conditions.
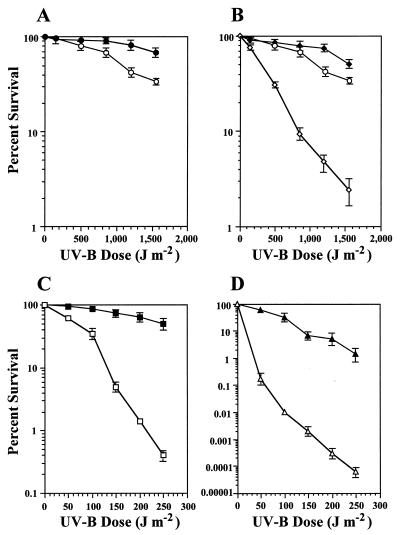
The cloned rulAB determinant on pJJK17 conferred a twofold increase in survival to the wild-type PAO1 strain following the higher UV-B doses (1,200 and 1,550 J m−2) administered (Fig. 3A). This is in contrast to previous experiments performed under dark conditions, as assessments of bacterial MDR are normally done, in which pJJK17 increased the survival of PAO1 approximately 20-fold following a UV-B dose of 1,550 J m−2 (21). In the PAO1::phr background (GWS250), the rulAB determinant conferred a large increase in strain survival (Fig. 3B; for comparison, the UV-B survival curve of the wild-type PAO1 background is also shown). Likewise for the PAO1::uvrA mutant (UA11079) and the PAO1::uvrA phr double mutant (GWS251), the rulAB determinant increased the UV-B survival, only at much larger magnitudes (Fig. 3C and D). In the case of the PAO1::uvrA mutant, the survival increase was 125-fold following the 250 J m−2 dose (Fig. 3C), while a phenomenally large increase in UV-B survival of up to 23,333-fold at 250 J m−2 was observed in the PAO1::uvrA phr mutant containing rulAB (Fig. 3D).
FIG. 3.
Survival of P. aeruginosa PAO1/pJB321 (○) and PAO1/pJJK17 (rulAB+) (●) (A), GWS250/pJB321 (PAO1 phr) (◊), PAO1/pJB321 (○), and GWS250/pJJK17 (PAO1 phr rulAB+) (⧫) (B), UA11079/pJB321 (PAO1 uvrA) (□) and UA11079/pJJK17 (PAO1 uvrA rulAB+) (■) (C), and GWS251/pJB321 (PAO1 uvrA phr) (▵) and GWS251/pJJK17 (PAO1 uvrA phr rulAB+) (▴) (D) after UV-B irradiation. Each datum point represents the mean (± the standard error of the mean) from three replicate experiments.
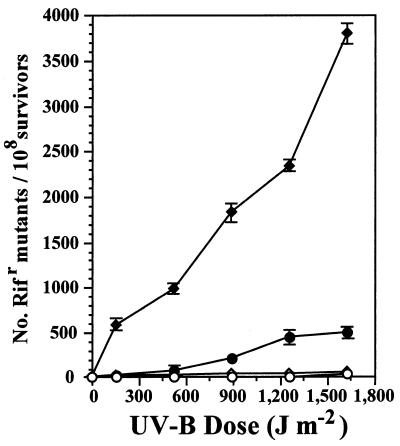
The by-product of MDR, an increase in cellular mutation frequency, can be assayed by examining the increase in levels of occurrence of spontaneous mutants following irradiation. We examined increases in the frequency of Rifr mutants, or Nalr mutants in the case of strains UA11079 and GWS251, which were already resistant to rifampin. Under photoreactivating conditions, when the increase in survival of PAO1/pJJK17 was only 2-fold or less, relatively large increases in the frequency of Rifr mutants (as much as 51-fold at the 1,200 J m−2 dose compared to a nonirradiated control) were observed (Fig. 4). The presence of pJJK17 in the PAO1::phr background (GWS250) resulted in larger increases in the frequency of Rifr mutants observed (Fig. 4; ranging from approximately 23-fold at the 150 J m−2 dose to 73-fold at the 1,550 J m−2 dose).
FIG. 4.
Analysis of rulAB-mediated MDR in P. aeruginosa PAO1/pJB321 (○), PAO1/pJJK17 (rulAB+) (●), GWS250/pJB321 (PAO1 phr) (◊), and GWS250/pJJK17 (PAO1 phr rulAB+) (⧫). Rifs strains were irradiated with different doses of UV-B, samples were removed to initiate cultures which were incubated in LB medium for 18 h, and the number of Rifr colonies was determined. The number of spontaneous mutations conferring Rifr in the absence of UV-B irradiation has been subtracted. Each datum point represents the mean (± the standard error of the mean) from three replicate experiments.
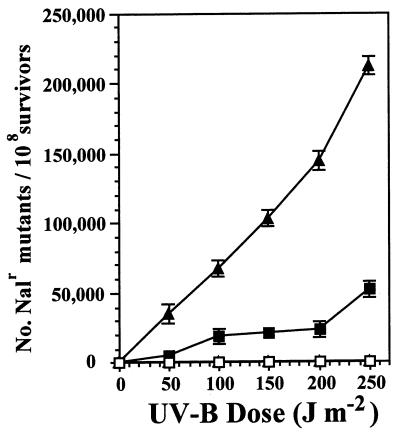
The frequency of occurrence of Nalr mutants was assayed in the PAO1 uvrA and uvrA phr backgrounds because the PAO1 uvrA strain UA11079 was already Rifr. By comparison, Nalr mutants occurred at a frequency approximately sevenfold greater than that of Rifr mutants in strain PAO1/pJJK17 following UV-B doses over the range utilized in this study (data not shown). The significant increase in the UV-B survival of strains UA11079 and GWS251 conferred by pJJK17 (Fig. 3C and D) was accompanied by a corresponding increase in the frequency of Nalr mutants (Fig. 5). The overall frequency of occurrence of Nalr mutants observed in strain GWS251 was as high as 2.1 × 10−3 at the 250-J m−2 dose (Fig. 5). The trajectory of the increase in Nalr mutants with higher UV-B doses was smaller in the UA11079/pJJK17 combination as compared with the GWS251/pJJK17 combination. These data were correlated with the UV-B sensitivity data of these strains that showed a smaller increase in UV-B sensitivity in UA11079/pJJK17 (Fig. 3C and D).
FIG. 5.
Analysis of rulAB-mediated MDR in P. aeruginosa UA11079/pJB321 (PAO1 uvrA) (□), UA11079/pJJK17 (PAO1 uvrA rulAB+) (■), GWS251/pJB321 (PAO1 uvrA phr) (▵), and GWS251/pJJK17 (PAO1 uvrA phr rulAB+) (▴). Nals strains were irradiated with different doses of UV-B, samples were removed to initiate cultures which were incubated in LB medium for 18 h, and the number of Nalr colonies was determined. The number of spontaneous mutations conferring Nalr in the absence of UV-B irradiation has been subtracted. Each datum point represents the mean (± the standard error of the mean) from three replicate experiments.
DISCUSSION
Photoreactivation is thought to be an important component of the bacterial arsenal in the repair or reversal of UV-B-mediated DNA damage; however, relevant data generated using defined mutants are lacking, thus precluding comparative examinations of the relative importance of photoreactivation and other DNA repair mechanisms to cell survival. Our analyses of the UV-B survival of photolyase-deficient mutants of P. aeruginosa and P. syringae demonstrated the importance of this DNA repair mechanism and also indicated that other dark repair mechanisms, such as NER and MDR, are active contributors to the overall cellular repair effort even when repair is ongoing under photoreactivating conditions.
Our work confirms the importance of NER since the P. aeruginosa uvrA mutant UA11079 was greatly increased in UV-B sensitivity compared to the phr mutant GWS250. Most photolyase enzymes are specific for pyrimidine dimers and are incapable of reversing other DNA lesions (47). In contrast, NER actively repairs CPDs and 6-4PPs, the two most common lesions caused by UV-B irradiation (29). The sharp increase in UV-B sensitivity of the P. aeruginosa uvrA phr double-mutant GWS251 is comparable to results obtained with uvr phr mutants of E. coli and of the yeast Saccharomyces cerevisiae (33, 4). These observations are attributed to the cooperative action of photolyase and the excision repair proteins. In these organisms, the photolyase enzyme binds to CPDs even in the absence of photoreactivating light, and the enzyme-bound complex enhances CPD recognition by the UvrABC excision nuclease (33, 34). Although not investigated in the present study, our results suggest that the photolyase and Uvr proteins of P. aeruginosa act in a similar cooperative fashion.
The ability of the rulAB determinant to restore the survival of the P. aeruginosa phr and uvrA mutants and to increase the survival of the phr uvrA mutants by over 23,000-fold illustrates the ability of an MDR system to effect large-scale DNA repair. The associated hypermutability of Uvr− MDR+ strains has been demonstrated previously in E. coli under low-UVR fluence conditions (42). Bacterial MDR systems present an interesting problem in the ecology of microorganisms. Some of these systems, such as rulAB, confer UVR tolerance and also elevate the cellular mutation rate. In other enterobacterial systems such as umuDC, the effect on the cellular mutation rate is substantial, while the contribution to UVR tolerance is less clear (43). It has alternately been argued that this elevation in mutation rate would be deleterious to cells (5, 44) or could be a source of genetic change that would be beneficial to cells inhabiting changing environments (9, 35). Our data indicate that, even in wild-type PAO1, the rulAB determinant confers a small increase in UV-B survival under photoreactivating conditions that is accompanied by a relatively larger increase in the occurrence of Rifr mutants. Thus, in nature, UVR-induced mutability could consistently occur in organisms expressing rulAB or similar MDR systems following DNA damage accumulation under light or dark conditions.
The availability of phr mutants of P. aeruginosa and P. syringae will facilitate the determination of the ecological importance of photoreactivation and MDR under field conditions. Under dark repair conditions, P. aeruginosa is characterized as relatively UV sensitive (8). Plasmid-encoded MDR determinants have been previously described in native P. aeruginosa isolates (25, 36), but the distribution of these determinants has not been systematically analyzed. In P. syringae, we have previously shown that the phenotype of tolerance to UVR conferred by rulAB is required for the maintenance of populations in their phyllosphere habitat (41). Phyllosphere population size is correlated with disease incidence in P. syringae, and phyllosphere populations represent an inoculum source in terms of dissemination of the bacterium in the environment (15). We have also shown that rulAB-mediated MDR is readily detectable in established phyllosphere populations of P. syringae (21). The availability of the phr mutant P. syringae pv. syringae GWS252 will also enable us to examine the contribution of phr and rulAB to solar UVR survival and mutability under field conditions.
The difficulty with the currently available data on the importance of dark repair processes in organisms exposed to solar radiation (e.g., references 16 and 17) is that the contribution of photoreactivation to repair when organisms are sampled is unknown. Since the spectrum of wavelengths of solar radiation concurrently contains UV-B damaging wavelengths and photoreactivating wavelengths, sampling organisms after solar radiation exposure and placing them under dark conditions would still most likely result in the occurrence of some level of photoreactivation. Studies with genetically defined phr mutants would eliminate this problem and facilitate investigations into other ecological questions concerning photoreactivation, such as if photoreactivation enables P. syringae strains to colonize a larger sunlight-exposed surface area in the phyllosphere or if there are any alterations in the relative expression of damage-inducible genes such as recA and rulAB in wild-type and photolyase-deficient backgrounds.
ACKNOWLEDGMENTS
We thank the Pseudomonas Genome Project for making preliminary data of the P. aeruginosa PAO1 genome sequencing project available to the scientific community and Jordi Barbe, Akira Yasui, and the E. coli Genetic Stock Center for strains and plasmids.
This work was supported by the U.S. Department of Agriculture (NRICGP 9702832 and NRICGP 1999-02516) and the Texas Agricultural Experiment Station.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Arrieta J M, Weinbauer M G, Herndl G J. Interspecific variability in sensitivity to UV radiation and subsequent recovery in selected isolates of marine bacteria. Appl Environ Microbiol. 2000;66:1468–1473. doi: 10.1128/aem.66.4.1468-1473.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey C A, Neihof R A, Tabor P S. Inhibitory effect of solar radiation on amino acid uptake in Chesapeake Bay bacteria. Appl Environ Microbiol. 1983;46:44–49. doi: 10.1128/aem.46.1.44-49.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blatny J M, Brautaset T, Winther-Larsen H C, Haugan K, Valla S. Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl Environ Microbiol. 1997;63:370–379. doi: 10.1128/aem.63.2.370-379.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bridges B A. DNA repair: getting past a lesion—at a cost. Curr Biol. 1998;8:R886–R888. doi: 10.1016/s0960-9822(07)00552-0. [DOI] [PubMed] [Google Scholar]
- 6.Croft L, Beatson S A, Whitchurch C B, Huang B, Blakely R L, Mattick J S. An interactive web-based Pseudomonas aeruginosagenome database: discovery of new genes, pathways and structures. Microbiology. 2000;146:2365–2373. doi: 10.1099/00221287-146-10-2351. [DOI] [PubMed] [Google Scholar]
- 7.Crosa J H, Falkow S. Plasmids. In: Gerhardt P, Murray R G E, Costilow R N, Nester E W, Wood W A, Krieg N R, Phillips G B, editors. Manual of methods for general bacteriology. Washington, D.C.: ASM Press; 1981. pp. 266–282. [Google Scholar]
- 8.Degiorgi C F, Fernandez R O, Pizarro R A. Ultraviolet-B lethal damage on Pseudomonas aeruginosa. Curr Microbiol. 1996;33:141–146. doi: 10.1007/s002849900090. [DOI] [PubMed] [Google Scholar]
- 9.Echols H. SOS functions, cancer, and inducible evolution. Cell. 1981;25:1–2. doi: 10.1016/0092-8674(81)90223-3. [DOI] [PubMed] [Google Scholar]
- 10.Eisenstark A. Bacterial genes involved in response to near-ultraviolet radiation. Adv Genet. 1989;26:99–147. doi: 10.1016/s0065-2660(08)60224-2. [DOI] [PubMed] [Google Scholar]
- 11.Figurski D H, Helinski D R. Replication of an origin containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;79:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 13.Garcia-Pichel F. A model for internal self-shading in planktonic organisms and its implications for the usefulness of ultraviolet sunscreen. Limnol Oceanogr. 1994;39:1704–1717. [Google Scholar]
- 14.Grant S G, Jessee J, Bloom F R, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia-colimethylation-restriction mutants. Proc Natl Acad Sci USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirano S S, Upper C D. Population biology and epidemiology of Pseudomonas syringae. Annu Rev Phytopathol. 1990;28:155–177. [Google Scholar]
- 16.Jeffrey W H, Aas P, Lyons M M, Coffin R B, Pledger R J, Mitchell D L. Ambient solar radiation-induced photodamage in marine bacterioplankton. Photochem Photobiol. 1996;64:419–427. [Google Scholar]
- 17.Jeffrey W H, Pledger R J, Aas P, Hager S, Coffin R B, VonHaven R, Mitchell D L. Diel and depth profiles of DNA photodamage in bacterioplankton exposed to ambient solar ultraviolet radiation. Mar Ecol Prog Ser. 1996;137:283–291. [Google Scholar]
- 18.Joux F, Jeffrey W H, Lebaron P, Mitchell D L. Marine bacterial isolates display diverse responses to UV-B radiation. Appl Environ Microbiol. 1999;65:3820–3827. doi: 10.1128/aem.65.9.3820-3827.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaiser E, Herndl G J. Rapid recovery of marine bacterioplankton activity after inhibition of UV radiation in coastal waters. Appl Environ Microbiol. 1997;63:4026–4031. doi: 10.1128/aem.63.10.4026-4031.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keane P J, Kerr A, New P B. Crown gall of stone fruit. II. Identification and nomenclature of Agrobacteriumisolates. Aust J Biol Sci. 1970;23:585–595. [Google Scholar]
- 21.Kim J-J, Sundin G W. Regulation of the rulAB mutagenic DNA repair operon of Pseudomonas syringae by UV-B (290 to 320 nanometers) radiation and analysis of rulAB-mediated mutability in vitro and in planta. J Bacteriol. 2000;182:6137–6144. doi: 10.1128/jb.182.21.6137-6144.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim S-T, Sancar A. Photochemistry, photophysics, and mechanism of pyrimidine dimer repair by DNA photolyase. Photochem Photobiol. 1993;57:895–904. doi: 10.1111/j.1751-1097.1993.tb09232.x. [DOI] [PubMed] [Google Scholar]
- 23.King E O, Ward N K, Raney D E. Two simple media for the detection of pyocyanin and fluorescein. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 24.Kovach M E, Elzer P H, Hill D S, Robertson G T, Farris M A, Roop R M, Peterson K M. Four new derivatives of the broad-host-range cloning vector pBBR1mcs, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 25.Lehrbach P R, Kung A H C, Lee B T O. R plasmids which alter ultraviolet light-sensitivity and enhance ultraviolet light-induced mutability in Pseudomonas aeruginosa. J Gen Microbiol. 1978;108:119–123. doi: 10.1099/00221287-108-1-119. [DOI] [PubMed] [Google Scholar]
- 26.Lyons M M, Aas P, Pakulski J D, Van Waasbergen L, Mitchell D L, Miller R V, Jeffrey W H. Ultraviolet radiation induced DNA damage in coral reef microbial communities. Mar Biol. 1998;130:537–543. [Google Scholar]
- 27.Mullar-Niklas G, Heissenberger A, Puskeric S, Herndl G J. Ultraviolet-B radiation and bacterial metabolism in coastal waters. Aquat Microb Ecol. 1995;9:111–116. [Google Scholar]
- 28.Newsham K K, Low M N R, McLeod A R, Greenslade P D, Emmett B A. Ultraviolet-B radiation influences the abundance and distribution of phylloplane fungi on pedunculate oak (Quercus robur) New Phytol. 1997;136:287–297. [Google Scholar]
- 29.Pfeifer G P. Formation and processing of UV photoproducts: effects of DNA sequence and chromatin environment. Photochem Photobiol. 1997;65:270–283. doi: 10.1111/j.1751-1097.1997.tb08560.x. [DOI] [PubMed] [Google Scholar]
- 30.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 31.Rivera E, Vila L, Barbe J. Expression of the Pseudomonas aeruginosa uvrAgene is constitutive. Mutat Res. 1997;377:149–155. doi: 10.1016/s0027-5107(97)00061-4. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Sancar A, Smith F W, Sancar G B. Purification of Escherichia coliDNA photolyase. J Biol Chem. 1984;259:6028–6032. [PubMed] [Google Scholar]
- 34.Sancar G, Smith F W. Interactions between yeast photolyase and nucleotide excision repair proteins in Saccharomyces cerevisiae and Escherichia coli. Mol Cell Biol. 1989;9:4767–4776. doi: 10.1128/mcb.9.11.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sedgwick S G, Goodwin P A. Differences in mutagenic and recombinational DNA repair in enterobacteria. Proc Natl Acad Sci USA. 1985;82:4172–4176. doi: 10.1073/pnas.82.12.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stokes H W, Krishnapillai V. Prevalence of Pseudomonas aeruginosaFP plasmids which enhance spontaneous and UV-induced mutagenesis. Mutat Res. 1978;50:19–28. doi: 10.1016/0027-5107(78)90056-8. [DOI] [PubMed] [Google Scholar]
- 37.Stover C K, Pham X Q, Erwin A L, Mizoguchi S D, Warrener P, Hickey M J, Brinkman F S L, Hufnagle W O, Kowalik D J, Lagrou M, Garber R L, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody L L, Coulter S N, Folger K R, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong G K-S, Wu Z, Paulsen I T, Reizer J, Saier M H, Hancock R E W, Lory S, Olson M V. Complete genome sequence of Pseudomonas aeruginosaPAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 38.Sundin G W, Bender C L. Ecological and genetic analysis of copper and streptomycin resistance in Pseudomonas syringaepv. syringae. Appl Environ Microbiol. 1993;59:1018–1024. doi: 10.1128/aem.59.4.1018-1024.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sundin G W, Jacobs J L. Ultraviolet radiation (UVR) sensitivity analysis and UVR survival strategies of a bacterial community from the phyllosphere of field-grown peanut (Arachis hypogeaeL.) Microb Ecol. 1999;38:27–38. doi: 10.1007/s002489900152. [DOI] [PubMed] [Google Scholar]
- 40.Sundin G W, Kidambi S P, Ullrich M, Bender C L. Resistance to ultraviolet light in Pseudomonas syringae: sequence and functional analysis of the plasmid-encoded rulABgenes. Gene. 1996;177:77–81. doi: 10.1016/0378-1119(96)00273-9. [DOI] [PubMed] [Google Scholar]
- 41.Sundin G W, Murillo J. Functional analysis of the Pseudomonas syringae rulAB determinant in tolerance to ultraviolet B (290–320 nm) radiation and distribution of rulAB among P. syringaepathovars. Environ Microbiol. 1999;1:75–87. doi: 10.1046/j.1462-2920.1999.00008.x. [DOI] [PubMed] [Google Scholar]
- 42.Witkin E M. Ultraviolet mutagenesis and inducible DNA repair in Escherichia coli. Bacteriol Rev. 1976;40:869–907. doi: 10.1128/br.40.4.869-907.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woodgate R. Construction of a umuDC operon substitution mutation in Escherichia coli. Mutat Res. 1992;281:221–225. doi: 10.1016/0165-7992(92)90012-7. [DOI] [PubMed] [Google Scholar]
- 44.Woodgate R, Sedgwick S G. Mutagenesis induced by bacterial UmuDC proteins and their plasmid homologues. Mol Microbiol. 1992;6:2213–2218. doi: 10.1111/j.1365-2958.1992.tb01397.x. [DOI] [PubMed] [Google Scholar]
- 45.Xue Y, Nicholson W L. The two major spore DNA repair pathways, nucleotide excision repair and spore photoproduct lyase, are sufficient for the resistance of Bacillus subtilusspores to artificial UV-C and UV-B, but not to solar radiation. Appl Environ Microbiol. 1996;62:2221–2227. doi: 10.1128/aem.62.7.2221-2227.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yasuhira S, Yasui A. Visible light-inducible photolyase gene from the goldfish Carassius auratus. J Biol Chem. 1992;267:25644–25647. [PubMed] [Google Scholar]
- 47.Yasui A, Eker A P M. DNA photolyases. In: Nickoloff J A, Hoekstra M F, editors. DNA damage and repair. 2. DNA repair in higher eukaryotes. Totowa, N.J: Humana Press, Inc.; 1998. pp. 9–32. [Google Scholar]