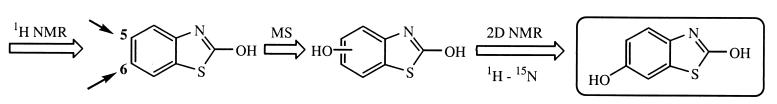
Abstract
The biodegradation of benzothiazole and 2-hydroxybenzothiazole by two strains of Rhodococcus was monitored by reversed phase high-pressure liquid chromatography and by 1H nuclear magnetic resonance (NMR). Both xenobiotics were biotransformed into a hydroxylated derivative of 2-hydroxybenzothiazole by these two strains. The chemical structure of this metabolite was determined by a new NMR methodology: long-range 1H-15N heteronuclear shift correlation without any previous 15N enrichment of the compound. This powerful NMR tool allowed us to assign the metabolite structure to 2,6-dihydroxybenzothiazole.
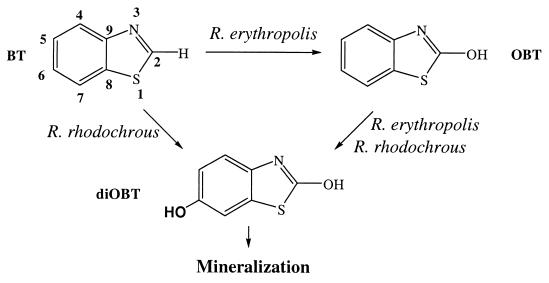
Benzothiazoles (BTs) are a family of xenobiotics widely used in a variety of industrial products and processes, such as pharmaceuticals, herbicides, fungicides, slimicides in the paper and pulp industry, antialgal agents, and, mainly, catalysts in the vulcanization process in rubber. BTs have been detected not only in industrial wastewater but also in various environmental compartments (15, 27) and are of concern for aquatic environment due to their limited biodegradability and potential toxicity (18). To date, only a few bacterial isolates have been shown to degrade some BTs as pure culture. These all belong to the Rhodococcus genus (8, 9, 17). However, the biodegradative pathways in these bacteria remain poorly known. De Wever et al. (8, 9) studied the biodegradation of BT and 2-hydroxybenzothiazole (OBT) by two Rhodococcus isolates (Rhodococcus erythropolis and Rhodococcus rhodochrous) by using high-pressure liquid chromatography (HPLC) with a UV detector and mass spectrometry. A common intermediate was detected and identified as dihydroxybenzothiazole (diOHBT), but its chemical structure could not be determined precisely (8). This prompted us to develop a new methodology in order to determine the structure of this metabolite, with our final goal being to use this tool more generally to study other benzothiazole derivatives.
Nuclear magnetic resonance (NMR) spectrometry is a powerful tool used to determine chemical structures and has been used, for example, to study the biodegradation of xenobiotics by microorganisms (7). However, most of the studies rely on specific enrichment of the compounds (e.g., 2H, 13C, 15N, and 19F). To overcome the difficulty of labeled-compound synthesis and also to increase the sensitivity of detection of metabolites, in situ 1H NMR has been recently developed that allows the study of molecules at natural abundance directly on the incubation medium without any previous purification step. This technique was successfully applied to study microbial metabolism (3) and, more specifically, to elucidate biodegradative pathways (2, 4–6, 16, 26). In this study we used 1H NMR to study the biodegradative pathway of BT and OBT by R. erythropolis and R. rhodochrous. Though this approach led to the characterization of some structural elements, it was not sufficient to assign unambiguously the position of the hydroxyl group on the benzene ring of the metabolite. To go further, we had to use long-range 1H-15N heteronuclear shift correlation by using GHMBC (gradient heteronuclear multiple-bond correlation) experiments at natural abundance. Although a large number of natural compounds and also pharmaceuticals and organic pollutants contain N atoms, only a few studies have been reported thus for that take advantage of the valuable information contained in 1H-15N scalar couplings. This can be easily explained by the difficulty of detecting the 15N nucleus because its sensitivity is very low due to a low natural abundance (0.37%) and a low gyromagnetic ratio. HMBC techniques (inverse-detection methods) have been used to measure long-range 1H-13C heteronuclear shift correlations (1) for the last 15 years but could not be applied to 1H-15N correlations at natural abundance until the 1995s, when spectrometers were equipped with gradients. Gradients greatly improved the sensitivity of detection for various reasons: (i) they suppress t1 noise and so improve the signal-to-noise ratio, (ii) they eliminate time-consuming phase cycling as a result of coherence pathways selection, and (iii) they can be used for solvent suppression. Other technical improvements were reached by using specific probes or new NMR sequences. More details are available in a recent and very interesting review (24) about the application of this technique to the determination of natural product structure, particularly of alkaloids. One- and two-dimensional inverse 1H-15N experiments have been used to detect 15N-enriched metabolites, mainly amino acids in mammalian (21, 28), insect (11, 12), and plant (25) cells. To our knowledge, this approach has never been applied in the field of biodegradation of xenobiotics by microorganisms. The present study is thus the first example of the determination of the structure of a metabolite issued from xenobiotic biodegradation using long-range 1H-15N heteronuclear shift correlation at natural abundance.
MATERIALS AND METHODS
Chemicals.
BT and OBT were purchased from Aldrich. Tetradeuterated sodium trimethylsilylpropionate (TSPd4) was purchased from Eurisotop (Saint Aubin, France).
Growth conditions.
R. erythropolis and R. rhodochrous were grown in 100-ml portions of Trypticase soy broth (bioMérieux, Marcy l'Etoile, France) in 500-ml Erlenmeyer flasks incubated at 30°C at 200 rpm. The cells were harvested after 20 h of culture.
Incubation with xenobiotic compounds.
Cells were centrifuged at 9,000 × g for 15 min at 5°C. The pellet was washed twice with Knapp buffer (K2HPO4, 1 g/liter; KH2PO4, 1 g/liter; FeCl3 4 mg/liter; MgSO4 · 7H2O, 40 mg/liter [pH 6.7]) and finally resuspended in this buffer (5 g of wet cells in 100 ml of buffer). The resting cells were incubated with 3 mM BT or OBT in 500-ml Erlenmeyer flasks at 30°C under agitation (200 rpm). The negative controls consisted of preparations incubated under the same conditions without a substrate or cells. Samples (1 ml) were taken every 30 min directly in the culture medium. They were centrifuged at 12,000 × g for 5 min and prepared for HPLC analysis or for 1H NMR analysis.
HPLC analyses.
HPLC analyses were performed using a Waters 600E chromatograph fitted with a reversed-phase column (Interchrom Nucléosil C18; 5 μm, 250 by 4.6 mm; Interchim) at room temperature. The mobile phase was acetonitrile-water (20/80, by volume), with a flow rate of 1 ml/min. Detection was performed with a Waters 486 UV detector set at 295 nm.
1H-NMR spectroscopy.
The preparation of NMR samples and the methods for performing a spectrum on a Bruker Avance 300 spectrometer at 298 K using 5-mm-diameter tubes have already been described (4), as was the method for quantification of the metabolites.
1H-15N GHMBC experiments.
The GHMBC experiments were performed on a Bruker Avance 300 spectrometer at 298 (±0.2) K. A 5-mm triple-tuned 1H-13C-15N probe equipped with a z-gradient coil was used. 1H and 15N 90° pulse lengths were 7.5 and 27 μs, respectively. Spectral widths were adjusted in both dimensions to encompass all 1H and 15N signals. Delay to allow nJNH correlations was set to 80 or 140 ms. The responses of 32 scans for each of 128 t1 increments were acquired. Zero-filling and phase-shifted sine window function in t1 and sine-squared window function in t2 were applied prior to two-dimensional Fourier transformation. A recycle delay of 1.5 s was used.
Ammonia and sulfate measurements.
Weatherburn's method (30) was used to assay for ammonia. To 100 μl of the sample was added successively 500 μl of solution A (phenol, 1 g; sodium nitroprusside [Na2(Fe(CN)5NO), 2H2O], 5 mg; distilled water, 100 ml) and 500 μl of solution B (NaOH, 0.5 g; sodium hypochlorite, 0.8 ml; distilled water, 100 ml). The mixture was stirred vigorously and put for 20 min in a water bath at 37°C. A blue coloration appeared, and the absorbance was measured at 625 nm. A standard curve was plotted using standard solutions of ammonium sulfate in Knapp buffer.
Sulfate was measured according to the method of Mainprize et al. (23). To 950 μl of the sample was added 50 μl of “developer solution” containing NaCl (7.5 g), ethanol (10 ml) concentrated HCl (3 ml), glycerol (5 ml), and distilled water (30 ml). BaCl2 (5 mg) was then added, and the solution mixed thoroughly for 1 min. The turbidity of the BaSO4 suspension, stabilized in the developer solution, was measured at 540 nm. A standard curve was plotted using standard solutions of ammonium sulfate in Knapp buffer.
Isolation of the unknown metabolite.
For the isolation of the unknown metabolite, a resting-cell experiment with R. erythropolis was performed as described above but on a larger scale. R. erythropolis (45 g) was incubated in 900 ml of Knapp buffer with 3 mM OBT (nine Erlenmeyer flasks). The biodegradation kinetics was monitored by HPLC in order to stop the experiment at the maximum concentration in metabolite. After 2 h of incubation (metabolite concentration of 0.5 mM), the reaction mixture was centrifuged, and the supernatant was extracted with ethyl acetate for 24 h. The organic layer was dried on MgSO4, concentrated under vacuum, and purified over a silica gel column (eluent, ethyl acetate-chloroform [30/70]). The pure metabolite was obtained as a pale yellow solid (melting point of 219 to 221°C). In order to determine the structure of this compound, it was analyzed by mass spectroscopy (Electronic Impact) on a Hewlett-Packard MS 5989B spectrometer.
RESULTS AND DISCUSSION
In order to confirm the biodegradative pathways of BT and OBT by R. rhodochrous and R. erythropolis that were described previously (8) and to demonstrate the formation of other metabolites that were not detected by HPLC with a monochromatic UV detector, new assays of biodegradation were performed using 1H NMR spectroscopy. The biodegradation conditions were slightly modified; in particular, the xenobiotic concentration was increased (3 mM instead of 1 mM). The samples were analyzed in parallel by inverse-phase HPLC and by 1H NMR as previously described (4).
Biodegradation of BT and OBT by R. erythropolis.
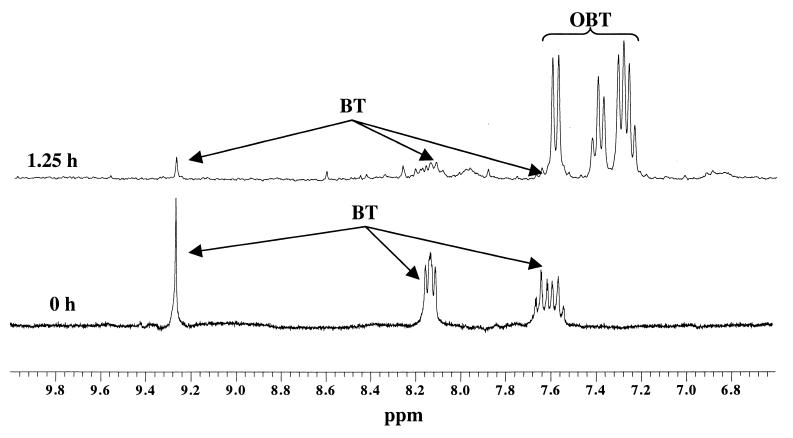
The biodegradation of BT (3 mM) by resting cells of R. erythropolis was monitored by both analytical methods used, i.e., HPLC and 1H NMR. After 30 min of incubation, a first metabolite was detected. Two 1H NMR spectra recorded after 0 and 1.25 h of incubation are shown in Fig. 1. By studying the 1H NMR spectra obtained at different times, we could observe the signals of BT (7.57 [t], 7.65 [t], 8.14 [2xd], and 9.28 [s]) decreasing and new signals appearing in the aromatic region, i.e., two triplets at 7.24 and 7.38 ppm and two doublets at 7.29 and 7.58 ppm. They were assigned to OBT by adding the commercially available compound to the sample. This was also confirmed by HPLC (coelution of the OBT standard). After BT exhaustion, another metabolite appeared on the HPLC chromatogram. Its retention time (7 min) was shorter than that of OBT (25 min), indicating a higher polarity of this compound. Using 1H NMR, we could observe the disappearance of OBT signals. However, no signal corresponding to the new metabolite observed by HPLC could be detected due to the lower sensitivity of NMR.
FIG. 1.
1H NMR spectra of the samples taken after 0 and 1.25 h of incubation of BT (3 mM) with resting cells of R. erythropolis.
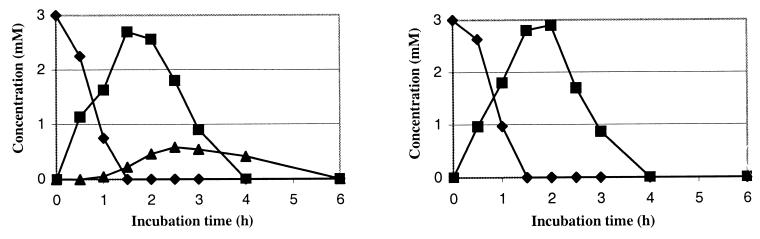
The use of standard curves for the identified compounds in HPLC and the addition of a reference in the sample (TSPd4 solution of known concentration in D2O) allowed the quantification of each compound. (For the metabolite, the quantitative curves, for both methods, were plotted only after its isolation and the determination of its chemical structure.) Figure 2 shows the time courses of the concentration of BT and its metabolites with R. erythropolis, as analyzed by HPLC and 1H NMR. The results obtained by both techniques are very similar. Both methods are quantitative.
FIG. 2.
Time courses of the concentration of BT (⧫), OBT (■), and the unknown metabolite (▴) during the biodegradation of BT (3 mM) by resting cells of R. erythropolis. A comparison of the results obtained by HPLC (left) and by 1H NMR (right) is shown. Similar results were obtained in three independent experiments.
With this strain, BT was quantitatively transformed into OBT, which was then converted to an unknown, more-polar metabolite. This one, whose highest concentration reached 0.6 mM after 2 h 30 min, was then degraded in its turn within 6 h of incubation.
The degradation of OBT (3 mM) by R. erythropolis was also studied by both analytical methods: HPLC and 1H NMR (data not shown). OBT was completely degraded within 5 h. The sole metabolite, observed only by HPLC, presented the same retention time (7 min) as the compound formed during the biodegradation of BT. Coinjection of samples issued from both of these biodegradations showed only one peak on the HPLC chromatogram whatever the elution conditions. The maximum concentration in metabolite (0.54 mM) was obtained after 3 h of incubation.
Since the metabolite has completely disappeared in its turn within 6 h, we wanted to know if mineralization had occurred. Preliminary assays to determine the production of ammonia and sulfate were carried out using spectrophotometric means according to the methods of Weatherburn (30) and Mainprize et al. (23), respectively. The stoichiometry for the degradation of OBT and ammonia and sulpfate production obtained was 1/0.6/0.4 (theoretical stoichiometry = 1/1/1). A very similar ratio was obtained during BT degradation. The values obtained are lower than those expected for a complete mineralization, but this is generally the case in such experiments (23), in particular because nitrogen and sulfur can be either assimilated by the cells or discharged in a different form than NH4+ and SO42− (10).
The BT and OBT degradation pathways are closely related and go through the same unknown, more-polar metabolite. In both cases, the medium turned black.
Biodegradation of BT and OBT by R. rhodochrous.
The kinetics of BT and OBT biodegradation observed with R. rhodochrous showed a different behavior for this strain. No OBT was detected during the BT biodegradation whatever analytical method was used. Only the unknown metabolite was observed. Some hypotheses can be proposed: either the life span of OBT in this case is too short (its rate of biodegradation being very high), its concentration remains too low to be detected even by HPLC, or R. rhodochrous follows a different pathway, converting BT directly into the unknown metabolite. Several experiments of HPLC coinjection of different samples obtained with both strains and/or with both xenobiotics (BT and OBT) gave evidence that the metabolite observed was always the same one.
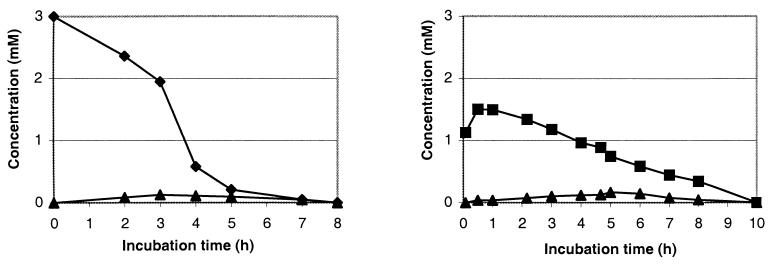
The time courses of BT, OBT, and their metabolite concentrations measured by HPLC are presented in Fig. 3. The biodegradative rates of BT and OBT with R. rhodochrous were much slower than those observed with the other strain: the complete disappearance of the xenobiotic was observed after 8 h of incubation for BT (1.5 h for R. erythropolis) and after 10 h for OBT (5 h for R. erythropolis). Moreover, with this strain the metabolite concentration remained very low throughout the biodegradation (<0.2 mM).
FIG. 3.
Biodegradation of 3 mM BT (left) or 3 mM OBT (right) by R. rhodochrous. Symbols: ⧫, BT; ■, OBT; ▴, unknown metabolite.
The stoichiometry for the degradation of OBT and ammonia and sulfate production obtained was 1/0.3/0.3 (theoretical stoichiometry = 1/1/1). A very similar ratio was obtained during BT degradation. As observed previously with R. erythropolis, the values obtained are lower than those expected for a complete mineralization.
Determination of the chemical structure of the unknown metabolite.
In order to determine the chemical structure of the unknown metabolite obtained during the BT and OBT degradation, a quantitative assay was carried out with R. erythropolis, this strain giving the highest concentration in metabolite. R. erythropolis (45 g) was incubated in 900 ml of Knapp buffer with 3 mM OBT. The biodegradation kinetics was monitored by HPLC in order to stop the experiment at the maximum concentration in metabolite (0.5 mM after 2 h of incubation). Extraction of the supernatant with ethyl acetate, followed by purification over a silica gel column, yielded the pure metabolite as a pale yellow solid.
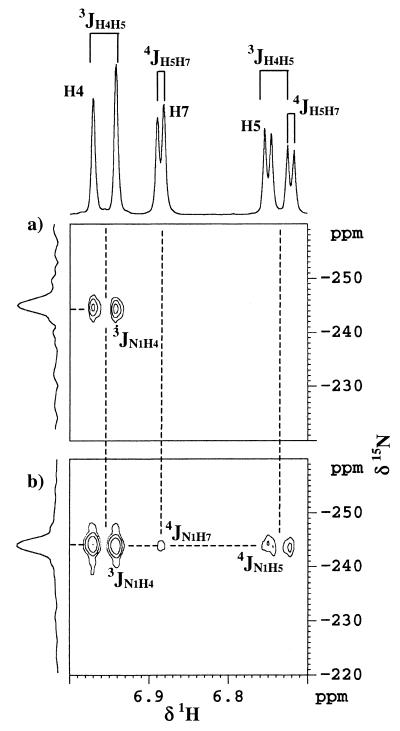
We checked that the isolated product corresponded to the unknown metabolite observed previously by coelution with a sample taken during the degradation of OBT and also by thin-layer chromatography. The purified product was first analyzed by 1H NMR in CD3OD. Only three signals are visible in the aromatic region, each one corresponding to an equivalent number of protons (Fig. 4). So a substituent was present on the aromatic ring. By analyzing the coupling constants, we could deduce that the substituent was either at position 5 or position 6 (Fig. 5). Indeed, the doublet at 6.97 ppm presented a coupling constant of 8.5 Hz (3J), meaning that this proton was coupled with another proton away from three bonds. The same coupling constant was observed on the signal at 6.74 ppm (dd). This proton was then not only coupled with the doublet at 6.97 ppm but also coupled with the proton at 6.89 ppm, with a small coupling constant (4J = 2.6 Hz) corresponding to a four-bond coupling. All of this information showed that the substituent was either at position 5 or position 6. However, the assignment of protons 4 and 7 was not possible because of the difficulty of knowing the electronic effects of the nitrogen and the sulfur atoms on these protons. Neither a semiempirical approach nor ab initio calculations are precise enough to determine nuclear shielding constant within 0.5 ppm (14).
FIG. 4.
1H-15N GHMBC spectra of OBT metabolite recorded in CD3OD; the experiments were set up for assumed 3J (delay = 80 ms) (a) or 4J (delay = 140 ms) (b) long-range couplings.
FIG. 5.
Sequence of experiments to determine the chemical structure of the unknown metabolite.
Another method to get information on the metabolite structure is mass spectrometry (Fig. 5). The mass spectrum of the isolated compound presented a peak with a mass/charge ratio of 167, differing from the molecular weight of OBT by 16. Thus, the metabolite formed during the biodegradation of BT and OBT is a hydroxylated derivative of the substrate OBT.
To assign unambiguously the position of the hydroxyl group on the benzene ring of the metabolite, long-range 1H-15N heteronuclear shift correlation and, more precisely GHMBC were used. Among the new sequencing methods available recently (24), the choice of the GHMBC experiment (20) was guided by the ease in setting up this kind of experiment and by its high distribution on most commercial spectrometers. In the scheme of the pulse program (for details, see reference 24) a variable delay is included between the two first 90° 15N pulses. This evolution period allows the system to be converted into zero and double quantum coherence and is proportional to 1/(2 × nJ15N-1H). Thus, this delay must be optimized in order to select 2J, 3J, or 4J couplings. 2J and 3J are generally in the range of 5 to 10 Hz, while only few precise data are available for 4J. We can only estimate that such values are in the range of 0.5 to 4 Hz (24). Another point of interest is that the desired coupling constant can be overestimated (until 30%) without important loss of information. Usually, the 4JNH are difficult to observe in nonaromatic systems since the corresponding coupling could be very weak. In our case, the delay to allow nJNH correlations was set to 80 and 140 ms in order to select assumed 3JNH (6.2 Hz) and 4JNH (3.5 Hz), respectively.
As chemical shift is concerned, since there is not a single chemical shift referencing, we used CH3NO2 as an external reference (0 ppm) and more precisely enriched urea (for practical convenience) calibrated at −306 ppm from nitromethane (13). The chemical shifts are reported negative upfield from CH3NO2 resonance.
Figure 4a represents the 1H-15N GHMBC recorded with an 80-ms evolution period. On this spectrum, only one correlation can be observed with the doublet at 6.97 ppm. Thus, this proton can be assigned to H4, since no other 3J can be observed in this compound. In Fig. 4b, the delay fixed at 140 ms (while other conditions remain the same) allows the observation of a 4J coupling between nitrogen and the doublet of doublets resonating at 6.73 ppm. This proton can be assigned to H5 because of its 8.5-Hz (3J) coupling constant with H4. No doubt exists concerning the attribution of the substituent in position 6. So the unknown metabolite is 2,6-dihydroxybenzothiazole (diOBT), corresponding to a hydroxylation in position 6 of OBT (Fig. 5).
The same sequence of experiments (Fig. 5) was carried out starting from a quantitative assay with R. erythropolis and led to an identical structure for the metabolite, observed during the biodegradation of BT and OBT with this strain.
The case of structural determination solved in these experiments using 1H-15N GHMBC is quite ideal for two reasons: (i) 4JNH could be observed easily (the coupling constant seems to be relatively important) without looking for several delays and (ii) no 2J couplings exist in this compound, which avoids confusion between two- and three-bond correlations (indeed, 2J and 3J are usually of the same order).
Even if it would be the case, some solutions have been developed recently. Concerning the observation of 4JNH, ACCORD-HMBC (29) and IMPEACH-MBC (19) offer the capability of surveying broad ranges of potential long-range couplings in a single experiment. The second point mentioned above could be circumvented by the use of the 2J,3J-HMBC experiment (22), which is capable of differentiating two-bond from three-bond couplings. However, this sequence was checked only for 1H-13C correlations.
One last point is the development of cold metal or superconducting coil NMR probes, which allow the detection of smaller quantities with an increased signal-to-noise ratio. This technical aspect will certainly be one of the most important improvements of the 15N detection method in the future.
In conclusion, the complementarity of a classical analytical method such as HPLC equipped with a UV detector, which has a great sensitivity, and 1H NMR carried out directly on the culture medium without any purification allowed us to confirm structural information previously obtained by De Wever et al. (8). With R. erythropolis, BT is first oxidized into OBT, which is further transformed into diOBT (Fig. 6); the same metabolite is obtained when OBT is incubated with the cells. With R. rhodochrous, the biodegradative pathways of BT and OBT are very similar, except that OBT was not detected during the BT degradation. The determination of the exact chemical structure of the metabolite formed was possible by using long-range 1H-15N heteronuclear shift correlation at natural abundance. The hydroxyl group was shown to be at the C6 position on the benzene ring (Fig. 6). diOBT is a common intermediate to OBT and BT degradation by both strains of Rhodococcus. This metabolite was then degraded quite rapidly. This result was confirmed by direct incubation of diOBT (1.7 mM) with resting cells of R. rhodochrous. It was completely degraded within 4 h of incubation, and no other metabolite was detected. However, further experiments are still needed in order to understand which mechanism underlies this initial oxidation.
FIG. 6.
Biodegradative pathways of BT and OBT by two strains of Rhodococcus.
The major interest of our work is to show that long-range 1H-15N heteronuclear shift correlation can be used to study metabolites without any previous 15N enrichment of the starting xenobiotic (at natural abundance). Xenobiotics that contain N nuclei are numerous, so this tool could be developed in the future in the environmental field.
REFERENCES
- 1.Bax A, Summers M F. 1H and 13C assignments from sensitivity-enhanced detection of heteronuclear multiple-bond connectivity by 2D multiple quantum NMR. J Am Chem Soc. 1986;108:2093–2098. [Google Scholar]
- 2.Besse P, Combourieu B, Poupin P, Sancelme M, Truffaut N, Veschambre H, Delort A M. Degradation of morpholine and thiomorpholine by an environmental Mycobacterium involves a cytochrome P450. Direct evidence of the intermediates by in situ 1H-NMR. J Mol Biocatal B Enzymol. 1998;5:403–409. [Google Scholar]
- 3.Brecker L, Ribbons D W. Biotransformations monitored in situ by proton nuclear magnetic resonance spectroscopy. Trends Biochem. 2000;18:197–202. doi: 10.1016/s0167-7799(00)01425-6. [DOI] [PubMed] [Google Scholar]
- 4.Combourieu B, Besse P, Sancelme M, Veschambre H, Delort A M, Poupin P, Truffaut N. Morpholine degradation pathway of Mycobacterium aurum MO1: direct evidence of intermediates by in situ 1H nuclear magnetic resonance. Appl Environ Microbiol. 1998;64:153–158. doi: 10.1128/aem.64.1.153-158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Combourieu B, Poupin P, Besse P, Sancelme M, Veschambre H, Truffaut N, Delort A M. Thiomorpholine and morpholine oxidation by a cytochrome P450 in Mycobacterium aurum MO1. Evidence of the intermediates by in situ 1H NMR. Biodegradation. 1998;9:433–442. doi: 10.1023/a:1008321610465. [DOI] [PubMed] [Google Scholar]
- 6.Combourieu B, Besse P, Sancelme M, Godin J P, Monteil A, Veschambre H, Delort A M. Common degradative pathways of morpholine, thiomorpholine, and piperidine by Mycobacterium aurum MO1: evidence from 1H-nuclear magnetic resonance and ionspray mass spectrometry performed directly on the incubation medium. Appl Environ Microbiol. 2000;66:3187–3193. doi: 10.1128/aem.66.8.3187-3193.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delort A M, Combourieu B. Microbial degradation of xenobiotics. In: Barbotin J N, Portais J C, editors. NMR in microbiology: theory and applications. London, United Kingdom: Horizon Scientific Press; 2000. pp. 411–430. [Google Scholar]
- 8.De Wever H, Vereecken K, Stolz A, Verachtert H. Initial transformations in the biodegradation of benzothiazoles by Rhodococcus isolates. Appl Environ Microbiol. 1998;64:3270–3274. doi: 10.1128/aem.64.9.3270-3274.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Wever H, de Cort S, Noots I, Verachtert H. Isolation and characterization of Rhodococcus rhodochrous for the degradation of the wastewater component 2-hydroxybenzothiazole. Appl Microbiol Biotechnol. 1997;47:458–461. [Google Scholar]
- 10.Doelle H W. Microbial metabolism. 2nd ed. New York, N.Y: Academic Press, Inc.; 1975. [Google Scholar]
- 11.Doverskog M, Jacobsson U, Chapman B E, Kuchel P W, Häggström L. Determination of NADH-dependent glutamate synthase (GOGAT) in Spodoptera frugiperda (Sf9) insect cells by selective 1H/15N NMR in vitro assay. J Biotechnol. 2000;79:87–97. doi: 10.1016/s0168-1656(00)00215-7. [DOI] [PubMed] [Google Scholar]
- 12.Drews M, Doverskog M, Öhman L, Chapman B E, Jacobsson U, Kuchel P W, Häggström L. Pathways of glutamine metabolism in Spodoptera frugiperda (Sf9) insect cells: evidence for the presence of the nitrogen assimilation system, and a metabolic switch by 1H/15N NMR. J Biotechnol. 2000;78:23–37. doi: 10.1016/s0168-1656(99)00231-x. [DOI] [PubMed] [Google Scholar]
- 13.Farrow N A, Kanamori K, Ross B D, Parivar F. A 15N-NMR study of cerebral, hepatic and renal nitrogen metabolism in hyperammonaemic rats. Biochem J. 1990;270:473–481. doi: 10.1042/bj2700473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukui H. Theory and calculation of nuclear shielding constants. Progr NMR Spectrosc. 1997;31:317–342. [Google Scholar]
- 15.Fiehn O, Reemtsma T, Jekel M. Extraction and analysis of various benzothiazoles from industrial wastewater. Anal Chim Acta. 1994;295:297–305. [Google Scholar]
- 16.Gaines G L, III, Smith L, Neidle E L. Novel nuclear magnetic resonance spectroscopy methods demonstrate preferential carbon source utilization by Acinetobacter calcoaceticus. J Bacteriol. 1996;178:6833–6841. doi: 10.1128/jb.178.23.6833-6841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaja M A, Knapp J S. The microbial degradation of benzothiazoles. J Appl Microbiol. 1997;83:327–334. [Google Scholar]
- 18.Gold L S, Slone T H, Stern B R, Bernstein L. Comparison of target organs of carcinogenicity for mutagenic and non-mutagenic chemicals. Mutat Res. 1993;286:75–100. doi: 10.1016/0027-5107(93)90004-y. [DOI] [PubMed] [Google Scholar]
- 19.Hadden C E, Martin G E, Krishnamurthy V V. Improved performance accordion heteronuclear multiple-bond correlation spectroscopy—IMPEACH-MBC. J Magn Reson. 1999;140:274–280. doi: 10.1006/jmre.1999.1840. [DOI] [PubMed] [Google Scholar]
- 20.Hurd R E, John B K. Gradient-enhanced proton detected heteronuclear multiple quantum spectroscopy. J Magn Reson. 1991;91:648–653. [Google Scholar]
- 21.Kanamori K, Ross B D, Parivar F. Selective observation of biologically important 15N-labelled metabolites in isolated rat brain and liver by 1H-detected multiple-quantum-coherence spectroscopy. J Magn Reson. 1991;93:319–328. [Google Scholar]
- 22.Krishnamurthy V V, Russel D J, Hadden C E, Martin G E. 2J, 3J-HMBC: a new long-range heteronuclear shift correlation technique capable of differentiating 2JCH from 3JCH correlations to protonated carbons. J Magn Reson. 2000;146:232–239. doi: 10.1006/jmre.2000.2141. [DOI] [PubMed] [Google Scholar]
- 23.Mainprize J, Knapp J S, Callely A G. The fate of benzothiazole-2-sulphonic acid in biologically treated industrial effluents. J Appl Bacteriol. 1976;40:285–291. doi: 10.1111/j.1365-2672.1976.tb04176.x. [DOI] [PubMed] [Google Scholar]
- 24.Martin G E, Hadden C E. Long-range 1H-15N heteronuclear shift correlation at natural abundance. J Nat Prod. 2000;63:543–585. doi: 10.1021/np9903191. [DOI] [PubMed] [Google Scholar]
- 25.Mesnard F, Azaroual N, Marty D, Fliniaux M-A, Robins R J, Vermeersch G, Monti J-P. Use of 15N reverse gradient two-dimensional nuclear magnetic resonance spectroscopy to follow metabolic activity in Nicotiana plumbaginifolia cell-suspension cultures. Planta. 2000;210:446–453. doi: 10.1007/pl00022919. [DOI] [PubMed] [Google Scholar]
- 26.Poupin P, Truffaut N, Combourieu B, Besse P, Sancelme M, Veschambre H, Delort A-M. Degradation of morpholine by an environmental strain of Mycobacterium involves a cytochrome P450. Appl Environ Microbiol. 1998;64:159–165. doi: 10.1128/aem.64.1.159-165.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reemtsma T, Fiehn O, Kalnowski G, Jekel M. Microbial transformations and biological effects of fungicide derived-benzothiazoles determined in industrial wastewater. Environ Sci Technol. 1995;29:478–485. doi: 10.1021/es00002a025. [DOI] [PubMed] [Google Scholar]
- 28.Street J C, Delort A M, Bradock P S H, Brindle K M. A 1H/15N NMR study of nitrogen metabolism in cultured mammalian cells. Biochem J. 1993;291:485–492. doi: 10.1042/bj2910485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner R, Berger S. ACCORD-HMBC: a superior technique for structural elucidation. Magn Reson Chem. 1998;36:S44–S48. [Google Scholar]
- 30.Weatherburn M W. Phenol-hypochlorite reaction for determination of ammonia. Anal Chem. 1967;39:971–974. [Google Scholar]