Abstract
Limited availability of medical imaging datasets is a vital limitation when using “data hungry” deep learning to gain performance improvements. Dealing with the issue, transfer learning has become a de facto standard, where a pre-trained convolution neural network (CNN), typically on natural images (e.g., ImageNet), is finetuned on medical images. Meanwhile, pre-trained transformers, which are self-attention-based models, have become de facto standard in natural language processing (NLP) and state of the art in image classification due to their powerful transfer learning abilities. Inspired by the success of transformers in NLP and image classification, large-scale transformers (such as vision transformer) are trained on natural images. Based on these recent developments, this research aims to explore the efficacy of pre-trained natural image transformers for medical images. Specifically, we analyze pre-trained vision transformer on CheXpert and pediatric pneumonia dataset. We use CNN standard models including VGGNet and ResNet as baseline models. By examining the acquired representations and results, we discover that transfer learning from the pre-trained vision transformer shows improved results as compared to pre-trained CNN which demonstrates a greater transfer ability of the transformers in medical imaging.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10278-022-00666-z.
Keywords: Vision transformer, Chest X-rays, Transfer learning, Classification
Introduction
Chest radiography or chest X-ray (CXR) is one of the most frequently used medical imaging method for timely and accurate diagnosis of various chest and pulmonary diseases. CXRs are easily acquired, are cost-effective, and contain large amount of information about the region under study that make them useful for early screening and diagnosis [1, 2]. CXRs can be used to identify diseases such as Tuberculosis [3], pneumonia [4], cancer [5], cardiomegaly [6], etc. However, one of the major issues with CXRs is that for accurate diagnosis, they require careful interpretation by experienced radiologists which can take a lot of time and resources [7]. Furthermore, the interpretation of CXRs varies from radiologist to radiologist with large discrepancy rates reported [8]. The factors that affect the diagnosis accuracy of radiologists include large workload, negligence, lack of knowledge, and faulty reasoning, among other reasons [8].
In recent years, due to the increase in computational power and availability of large amounts of data, deep learning techniques have emerged as the state of the art in various image processing and computer vision applications [9–11]. Consequently, many studies have been carried out to aid radiologists using deep learning approaches, especially the convolutional neural networks (CNNs), for classification, localization, and segmentation of medical images [12–14]. Recently, however, the CNNs have been outperformed by the attention-based architecture known as Transformer [15, 16].
Transformer architecture has since become the state-of-the-art for natural language processing (NLP). Transformers are built on a self-attention-based mechanism that learns dependencies between input and output sequences without relying on recurrence. This allows transformer implementations to be easily parallelized and computationally efficient. Inspired by the success of transformers in NLP, [15] modified transformer architecture for computer vision, which they called Vision Transformer (ViT). [15] modified the original transformer such that it takes as input a sequence of fixed-size image patches that are treated similarly as words are treated in NLP application, and performed image classification. ViT, when trained from scratch, achieved lower performance as compared to convolution neural networks (CNNs). However, when ViT is pre-trained on a large dataset and transfer learning is performed to a smaller dataset, it outperformed the CNN architectures in many computer vision tasks such as object detection [17, 18], semantic segmentation [19], and image classification [15]. Although vision transformers have seen success on natural images, little work has been done in the medical imaging domain. CNN-based architectures are still commonly used for medical imaging and diagnostics [20] Compared to transformers, CNNs have some disadvantages such as convolution operations are difficult to capture global information [21], and CNNs are not able to capture long-range dependencies between different images that may be present in medical datasets [20].
To address the CNN issues, some authors have proposed transformer-based architectures for medical imaging and diagnostics [22–25]. [20] proposed a multi-modal medical imaging classification method. The authors combined CNN with transformer to learn both the low-level features and global features for effective image fusion and classification strategy. In [26], authors employed ViT architecture for COVID-19 classification using computed tomography (CT) scans and outperformed CNN-based DenseNet [27] architecture. In another study, [28] evaluated several deep learning architectures including DenseNet, EfficientNet, ResNet, and ViT for COVID-19 diagnosis using CT images and found ViT to outperform all other architectures. Although the methods discussed here have shown ViT to outperform CNN-based architectures, they fail to analyze the pretraining and finetuning aspects of transformers. Transformers require a large amount of training data to effectively exploit their capability [15]. However, in the medical domain, there is limited availability of large datasets [29, 30]. When trained on small datasets, ViT suffers from a lack of inductive bias that results in poor generalizability [15].
It has previously been shown that CNN-based architectures show improvement when they are pre-trained on large natural image datasets such as ImageNet [31] and finetuned on medical datasets [32]. Therefore, in this study, we explore the transfer learning capability of pre-trained transformers when finetuned on a medical dataset. In our work, we apply a pre-trained ViT on the CheXpert dataset [1] and show performance improvements over pre-trained CNN-based VGG-16 [33] and ResNet [9] architectures. We also analyze the impact of pretraining by comparing the pre-trained model with training from scratch and show that the pre-trained model has the advantage. The rest of the paper is structured as follows. The “Literature Review” section discusses the related work. The “Transformer Background” section discusses the background of transformer model. The “Methodology” section presents the proposed methodology. The “Experiment and Results” section discusses the results, and finally, the “Datasets” section concludes the paper.
Literature Review
Transfer Learning in Medical Imaging
With the advancement in the computer sciences and technology, transfer learning, which is primarily a substantial feature of deep learning, is now become indispensable to many applications as an integral part. It has been used by different fields of research in order to apply it in the field of radiology, training Inception, ResNet on retinal fundus images [34–37]. DenseNet, ResNet on chest x-rays [38, 39], and same are applied to ophthalmology. Besides this, the FDA have approved the related research on ophthalmology [40], with proper clinical arrangement [41]. [42] extracts characteristics from chest x-ray pictures using different neural network models pre-trained on ImageNet, then prepares five distinct models, analyzes their performance, and proposes an ensemble model that integrates outputs from all pre-trained models. Detection of Alzheimers disease in early stages is also its prominent application [43]. In 3D medical data, there are various transfer learning applications, such as [44] which create a Med3D network for 3D medical data classification and segmentation using a pre-trained ResNet-152. Other applications include the identification of skin cancer via photographs of dermatologist’s level [45] and the determination of the quality of human embryo for the IVF procedures [46]. [47] demonstrates that deep CNNs like inception-V3 trained on real-world radiographs can be utilized to transfer learning for fracture detection. The results were comparable to state-of-the-art for automated fracture diagnosis after training the model with a small sample set. [48] proposed Multi-view Convolutional Recurrent Neural Network (MVCRecNet), a deep learning approach that uses shape, size, and cross-slice changes in CT scan pictures to train model to identify lung cancer nodules from CT scan images. The model is given several viewpoints, allowing it to generalize better by learning robust characteristics. The datasets LIDC-IDRI and ELCAP were used in this study. [49] proposed Bayesian-based Convolutional Neural Network (B-CNN) takes advantage of model uncertainty and Bayesian confidence to increase TB detection and validation accuracy. The Montgomery and Shenzhen TB benchmark datasets were used to test the suggested methodology and it shows significant results in terms of TB identification accuracy, according to the findings.
Using the transfer learning methodology, [50] developed an extra layer of convolutional neural network blocks to integrate pre-trained ResNet and DenseNet models to establish higher performance above either model, and the suggested network was able to accurately classify lung diseases. [51] present their findings on the classification of histopathology images of oral cancer using various image classification models like Inception, ResNet, and MobileNet, concluding that transfer learning models perform well on histopathology. Despite the popularity and significance of transfer learning in the field of medical imaging, there has been little work done or research conducted in the relevant field. Even many common beliefs have been challenged by the latest research in this field of transfer learning in the area of natural image setting [52–56]. For instance, it has been shown in [53] that a transfer that has taken place between tasks that are similar in nature are not always resulted in the improvement of performance, and it has also been illustrated [55] that generalization of pre-trained features might be less than they are to be thought. In the medical imaging setting, many such open questions remain. As described above, in medical imaging, where present standard is taking an existing architecture that has been designed for natural image datasets like ImageNet, along with equivalent pre-trained weights, for example, ResNet and Inception, afterwards, the model is being finetuned on medical imaging data.
Anyhow, there is a considerable difference between medical image diagnosis and ImageNet classification. The first prominent feature of medical imaging is that its tasks begin with a considerable large image of the region of interest in the body and afterwards to identify the pathologies, it uses local textures for variations. For instance, the small red spots or dots are the signs of diabetic retinopathy and microaneurysms in retinal fundus images [57], and the indication of pneumonia can be confirmed via chest x-rays by observing local white small opaque patches [1]. This is just the opposite of natural image as ImageNet, in which there has been a clear and transparent worldwide subject of image. Now there is an open and unanswered question like to what extent the ImageNet feature reuse is quite helpful for natural medical images.
Transformer for Medical Images
In the field of NLP self-attention models like transformers [16] are becoming very popular with time. The concept of self-attention is also tried in CNNs like for each query pixel, self-attention was only used in local neighborhoods instead of being global [58]. In [59] output of CNN is further processed by self-attention. The use of pre-trained transformers on a large corpus is widely used [60] and in medical field use of transformers on text is also well known like BioBERT [61], SciBERT [62], and ClinicalBERT [63]. It has become possible to train models of massive scales like 100B of parameters all this is due to computational efficiency and scalability of transformers such as massive models like generative pre-trained transformer (GPT-3) are state of the art in different NLP tasks [64].
Due to massive success of transformers in field of NLP, transformers are applied to computer vision tasks. Most recent transformers which are used for image classification include OpenAi Image GPT (iGPT) [65] which uses GPT for image generation and trained model on ImageNet but there are limitations in their model as it requires high computation power and low image quality, Google Vision transformer (Vit) [15] which uses original transformer architecture for image classification and converts images into patches and gives it to transformer, and Facebook Data efficient transformer (DeiT) [66] which also uses same architecture as of ViT and used knowledge distillation for better training of model in which CNN act as teacher model. Medical Transformer [23] proposed a novel transfer learning framework using transformer model. It models 3D volumetric images in the form of a sequence of 2D image slices. TransUNet [22] proposed a transformer-based U-Net architecture for medical image segmentation because CNN has limited capability while modeling long-range dependencies and transformers self-attention mechanism help in modeling better representation. Both CNN and transformer are used while modeling TransUNet. TransFuse [25] use transformer and CNN in parallel styles for medical image segmentation. Besides that, a BiFusion module is created to fuse features from both branches. Segtran [67] is a medical image segmentation system based on transformers. It contextualizes features by utilizing the limitless receptive fields of transformers. Segtran is able to see both the big picture and the minute details, resulting in excellent segmentation results. In image denoising [68] use a transformer-based neural network used to investigate long-range dependencies between low dose computed tomography (LDCT) pixels.
Transformer Background
Transformers are based on self-attention mechanism and already became a de facto standard in natural language processing (NLP) and state of the art in image classification and object detection. A key characteristic of transformers, that is well-adopted in NLP, is their effective transfer learning on downstream tasks.
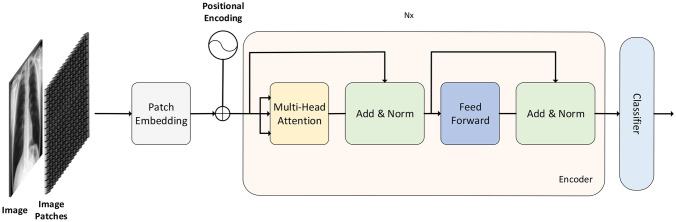
Transformer model used in image classification is based on original transformer [16] model which consists of two blocks encoder block and a decoder block. For image classification purpose only encoder part of transformer is used (Fig. 1). To feed input to the transformer model encoder part embedding is generated from patches of an image and positional encoding is attached to this patch embedding to keep the order of patches then these positional encoded patch embeddings are passed to encoder block. Encoder block consists of multi-head self-attention, layer normalization, feed-forward layers and then another layer normalization followed by feed-forward layer. To find the relation between each patch attention scores are computed using query, key and value matrices in self-attention layer. Multiple self-attentions scores are computed which acts as multi-head to get better representation and outputs of these heads are concatenated into one vector and input vector is added to it using skip connection and normalization is applied. After that output of this normalization is fed to feed-forward layer which is again added with previous layer output with the help of skip connections and normalization is applied. These skip connections allow the representation of different levels to interact with each other. Multiple encoder blocks can be stacked to gather in image transformer. At the end output of transformer encoder block is fed to classifier for classification purpose. Transformer output acts as image representation.
Fig. 1.
Image transformer
Methodology
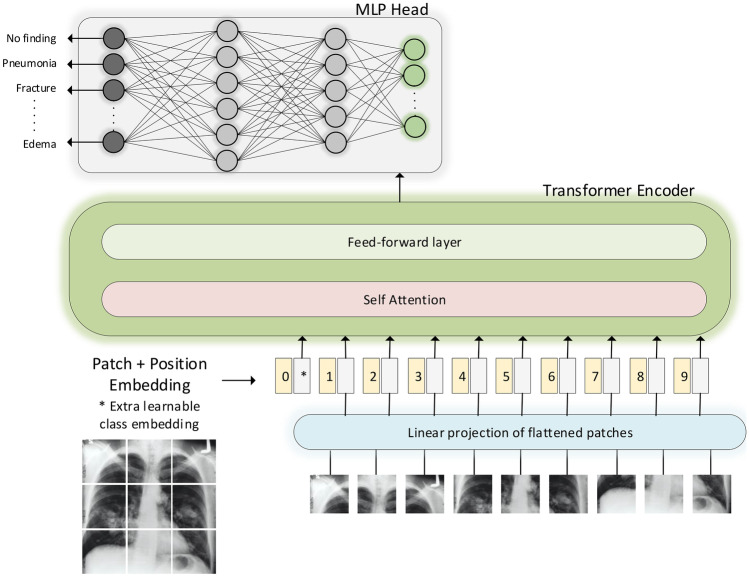
To evaluate the transfer learning performance of ViT from natural images to medical images, we train a standard ViT model both from random initialization and doing transfer learning from ImageNet [31] dataset. The ViT model we are using is closely related to original Transformer [16] and inspired from [15]. The overview of the model is shown in Fig. 2 which is used for medical image classification. The input images are reshaped into fixed-size 2D patches which are flattened and combined with position embeddings before feeding them to the ViT in a sequence. The transformer encoder consists of repeated blocks that each contains normalization, multi-head attention, and multi-layer perceptron (MLP) layers. The output of the encoder blocks is connected to a classification head that consists of MLP that maps the encoded feature vector to one of the output classes. We compare the transfer learning performance of ViT with CNN-based architectures including VGG-16 [33] and ResNet-50 [9]. These CNNs are also trained by doing transfer learning from ImageNet [31]. The performance evaluation of ViT and CNNs is performed based on evaluation metrics including accuracy, precision, recall, and F1-score.
Fig. 2.
Vision transformer
Transformer Encoder
The encoder block in vision transformer takes radiograph scans, sliced into patches of size 16 16. The patches are represented using a patch feature matrix X after adding positional encoding. The purpose of positional encoding is to preserve spatial structure of radiograph scans. The dependencies between patches are modeled by using a self-attention mechanism which work based on three embeddings: Query (Q), Key (K), and Value (V), defined as follows:
| 1 |
| 2 |
| 3 |
, and are used to project patch features onto embeddings Q, K, and V respectively. The ViT encoder pipeline mainly works in two steps: self-attention and attention-based feature weighting. The self-attention mechanism is used to model the dependencies between patches. The self-attention pipeline works as follows: In the first step, a similarity between patches embeddings is computed by taking a dot product between Q and K as follows:
| 4 |
The scores are then scaled down by dividing by the square root of the Q and K dimension. This allows for more stable gradients as multiplying values can have explosive effects:
| 5 |
The softmax layer is used to convert similarity score between Q and K into a probability distribution. As a result, the model may be more certain about which patch to pay attention to.
| 6 |
The objective of attention-based feature weighting step aims to weight chest embeddings V based on self-attention scores computed in the previous step.
| 7 |
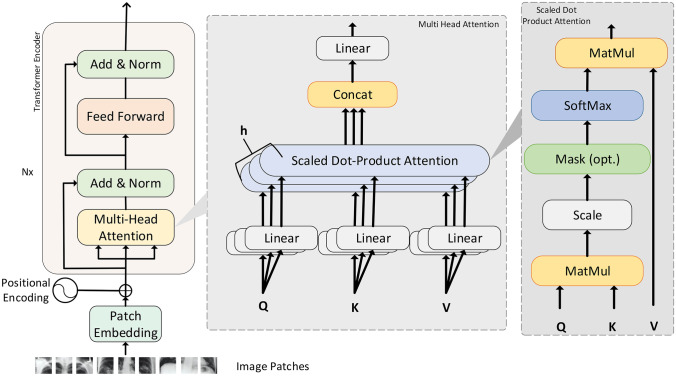
Attention scores as shown in above Eq. (7) is calculated as illustrated in Fig. 3. Where MatMul stands for matrix multiplication. concat is an abbreviation for concatenation.
Fig. 3.
Transformer encoder block with multi-head attention and scaled dot product attention
Multi-headed Attention
In multi-headed attention, each attention mechanism acts as a head, and each head learns something distinct, resulting in a better representation power for the encoder model. Before applying self-attention, query, key, and value are divided into N vectors to make this a multi-headed attention calculation as shown in Fig. 3. After that, the divided vectors go through the self-attention process one by one. Each step of self-awareness is referred to as a head. Before passing through the final linear layer, each head produces an output vector that is concatenated into a single vector.
Evaluation Metrics
To evaluate the performance of system model different evaluation metrics like Accuracy, Precision, Recall, and F1 score are used.
Accuracy
It is simply a ratio between correct predictions and total number of predication. It measures how many times a model correctly predicts label.
| 8 |
Precision
It measures how many times a model correctly predicts positive out of all positive prediction made by model.
| 9 |
Recall / Sensitivity
It measures how many times a model correctly predicts a label positive from an overall positive class.
| 10 |
F1 Score
It’s a combination of precision and recall and balance both. A perfect model have F1 score 1 and worst have 0. Better F1 score tells that model give low false positives and false negatives
| 11 |
Experiments and Results
The following section discusses the datasets used for experimentations and presents the results and discussion.
Datasets
For evaluation of proposed methodology 2 datasets have been used CheXpert [1] and Pediatric pneumonia dataset [69].
CheXpert
CheXpert [1] is a massive public dataset of 224,316 chest radiographs from 65,240 patients for chest radiograph analysis. CheXpert data is compiled from examinations performed at Stanford Hospital in both inpatient and outpatient settings between October 2002 and July 2017, as well as the radiology reports that accompanied them. CheXpert consists of chest X-rays of different sizes which are then resized to 224 224. Each of these X-rays is labelled into 14 observations No finding, Enlarged Cardiom, Cardiomegaly, Lung Lesion, Lung Opacity, Edema, Consolidation, Pneumonia, Atelectasis, Pneumothorax, Pleural Other, Fracture and Support Devices as positive, negative or uncertain. Distribution of CheXpert instances across 14 observations is shown in Table 1. We have decided to replace all uncertain labels with positive labels as it is also feasible in a real-world scenario, as if a patient gets a false negative result, the patient will accept it as compared to a false positive, then he or she is more likely to get a second opinion which will then clear the classification. Samples from CheXpert dataset are shown in Fig. 4
Table 1.
Distribution of CheXpert images across different classes
| Pathology | Positive | Uncertain | Negative |
|---|---|---|---|
| No Finding | 16627 | 0 | 171014 |
| Enlarged Cardiom | 9020 | 10148 | 168473 |
| Cardiomengaly | 23002 | 6597 | 158042 |
| Lung Lesion | 6856 | 1071 | 179717 |
| Lung Opacity | 92669 | 4341 | 90631 |
| Edema | 48905 | 11571 | 127165 |
| Consolidation | 12730 | 23976 | 150935 |
| Pneumonia | 4576 | 15658 | 167407 |
| Atelectasis | 29333 | 29377 | 128931 |
| Pneumothorax | 17313 | 2663 | 167665 |
| Pleural Effusioin | 75696 | 9419 | 102526 |
| Pleural Other | 2441 | 1771 | 183429 |
| Fracture | 7270 | 484 | 179887 |
| Support Devices | 105831 | 898 | 80912 |
Fig. 4.
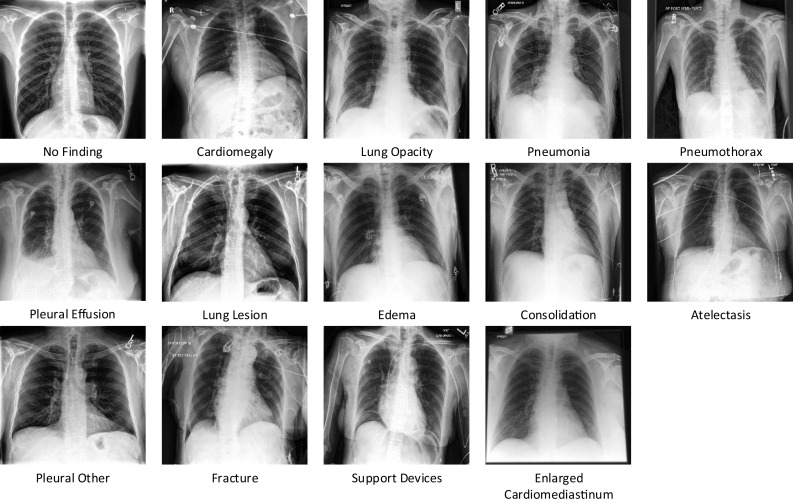
CheXpert samples
Pediatric Pneumonia Dataset
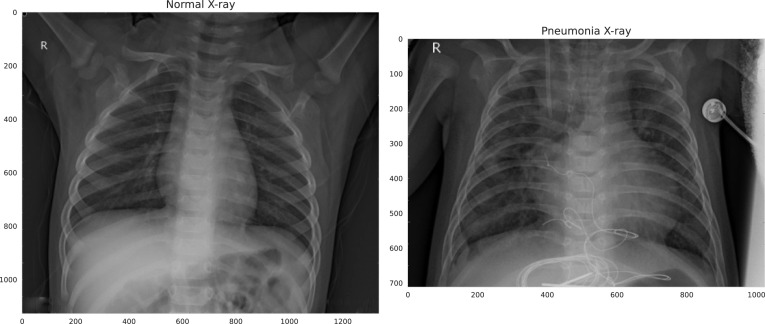
Pneumonia, which outnumbers all other infectious diseases, is the greatest cause of death among babies [70]. Anterior-posterior chest X-ray pictures were chosen from retrospective cohorts of pediatric patients aged one to five years old at Guangzhou Women and Children’s Medical Center in Guangzhou for the labelled Chest X-Ray Images for classification dataset [69]. There are 5863 chest x-ray images in this collection, divided into two classes: normal and pneumonia. Sample images from both classes are shown in Fig. 5
Fig. 5.
Pediatric chest X-ray sample
Network Training
For all networks, we use the transfer learning technique. As demonstrated in Fig. 6, many tactics are employed for this purpose. In some scenarios, the entire model is trained after being initialized with pre-trained weights. Freezing a few to several layers of the model is another strategy. The pre-trained model is loaded in our scenario, then the pre-trained model’s classification head is removed and replaced with a new head based on dataset classes, the network parameters are frozen, and the model is trained. Finally, the model is finetuned. Table 2 shows the training, validation, and test sets for both the CheXpert and Pediatric pneumonia datasets.
Fig. 6.
Transfer learning strategies
Table 2.
CheXpert and pediatric pneumonia dataset split for training, validation and testing
| Dataset | Training | Validation | Test | Sum |
|---|---|---|---|---|
| CheXpert | 152984 | 19124 | 19123 | 191231 |
| Pediatric Pneumonia | 5216 | 16 | 624 | 5856 |
The ADAM optimizer is used to optimize all of the networks. 0.0001 and 32 are the learning rate and batch size, respectively. A patience of 10 epochs is chosen as the stopping condition. On a GPU-based desktop machine with 128 GB RAM, Nvidia TitanX Pascal (12 GB VRAM), and a ten-core Intel Xeon processor, we train networks.
Experiments on Pediatric Pneumonia Dataset
Pediatric pneumonia is a binary classification task in which X-rays images are classified as normal or pneumonia. Through experimentation on a dataset of pediatric pneumonia patients [70] it is revealed that pre-trained transformer transfer learning performs better as compared to other state-of-the-art CNN-based vision models. Performance comparison of vision transformer and other CNN-based deep learning models is shown in Table 3
Table 3.
Performance comparison on pediatric pneumonia dataset
| Model | Precision | Recall | F1-score | AUC | Accuracy |
|---|---|---|---|---|---|
| ResNet-50 | 0.8 | 0.72 | 0.73 | 0.72 | 0.78 |
| Inception-V3 | 0.86 | 0.79 | 0.81 | 0.79 | 0.83 |
| VGG-16 | 0.88 | 0.83 | 0.85 | 0.89 | 0.82 |
| Vision Transformer | 0.89 | 0.84 | 0.86 | 0.87 | 0.87 |
ResNet-50
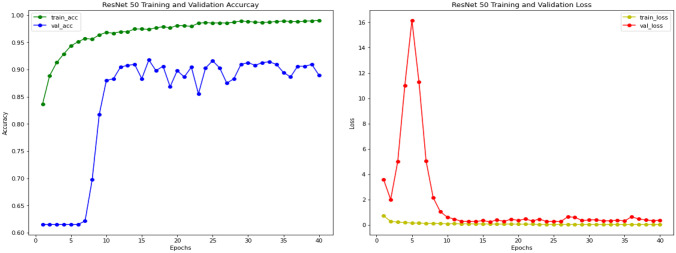
Experimental results of ResNet-50 on Pediatric pneumonia are done using pre-trained ResNet-50 base model. ResNet model uses residual connections or skip connection for learning representation. On top of that base model an additional classifier is added to classify images of chest X-rays into normal and pneumonia classes. Training and validation accuracy curves of ResNet model can be seen in Fig. 8; accuracy and loss are computed on 30 epochs of training.
Fig. 8.
ResNet-50 training and validation accuracy and loss
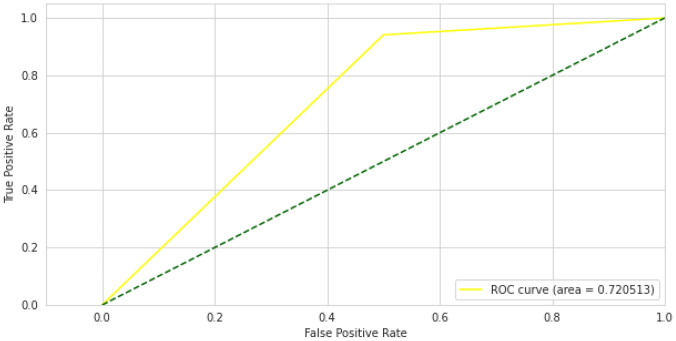
After training and validation of ResNet-50 model we have computed receiver operating characteristic curve or ROC for ResNet-50 model and it shows performance above 0.5 and area under ROC curve which is known as AUC is 0.72 as shown in Fig. 9.
Fig. 9.
ResNet-50 ROC
Confusion matrix for ResNet-50 is also computed for better understanding of results and shows the number of TP, FP, TN and FN for classifying X-ray images into normal and pneumonia classes as shown in Fig. 10
Fig. 10.
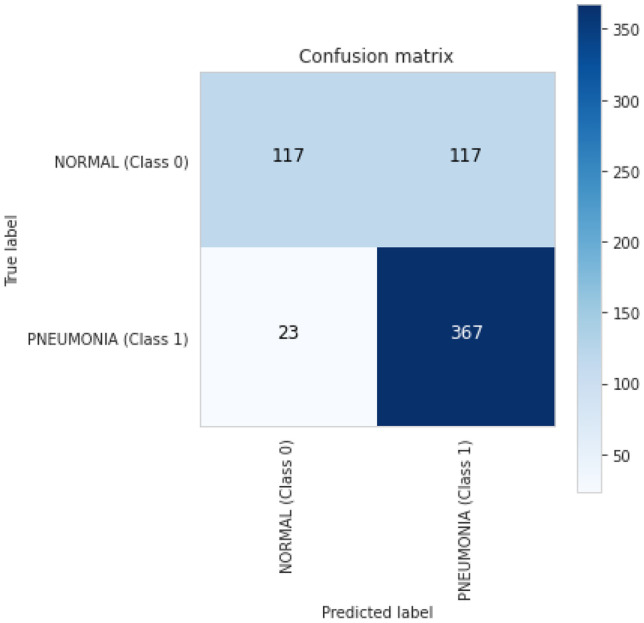
ResNet-50 confusion matrix
Inception-V3
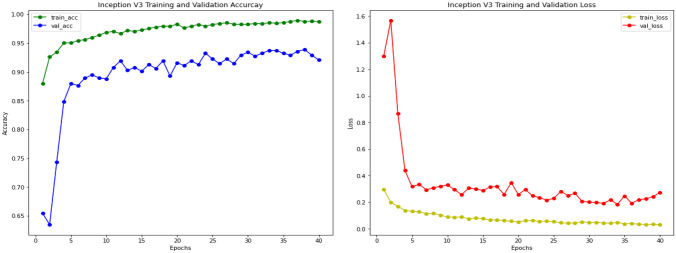
Inception-V3 experimental results on pediatric pneumonia are also based on a pre-trained model on ImageNet dataset. We have used Inception-V3 as base model with an extra classifier implemented on top of that base model to classify images of chest X-rays into normal and pneumonia classes. Figure 11 shows the training and validation accuracy curves for the Inception-V3 model. Accuracy and loss are computed during 30 training epochs.
Fig. 11.
Inception-V3 training and validation accuracy and loss
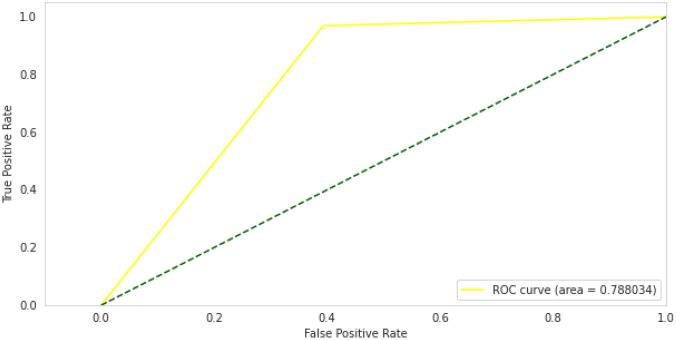
We also computed a receiver operating characteristic curve (ROC) for Inception-V3 model after training and validation, and it demonstrates performance above 0.5 and an area under the ROC curve (AUC) of 0.78, as shown in Fig. 12.
Fig. 12.
Inception-V3 ROC
For a better understanding of the results, Inception-V3’s confusion matrix is produced, and it indicates the number of TP, FP, TN, and FN for classifying X-ray pictures into normal and pneumonia classes, as shown in Fig. 13.
Fig. 13.
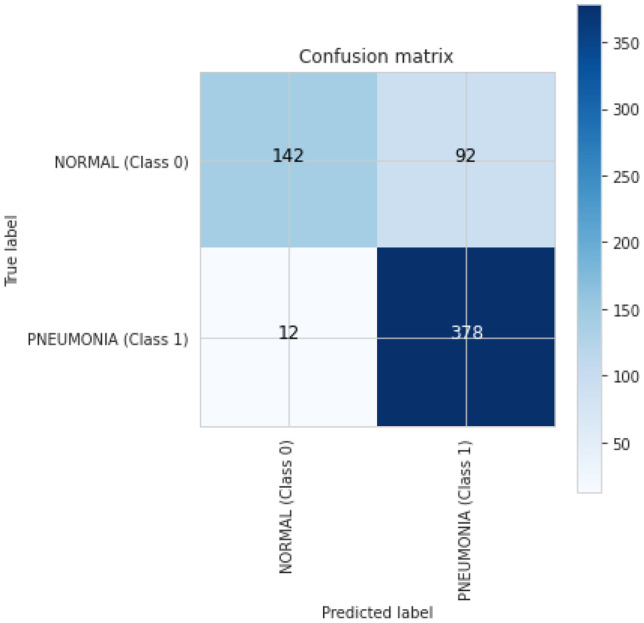
Inception-V3 confusion matrix
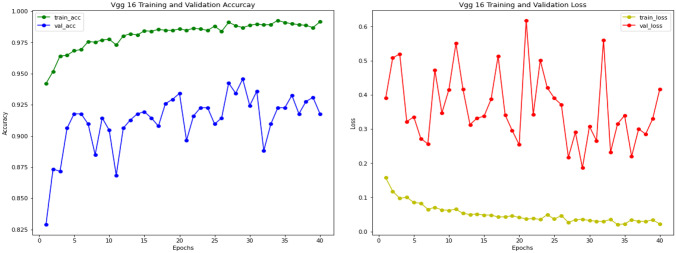
VGG-16
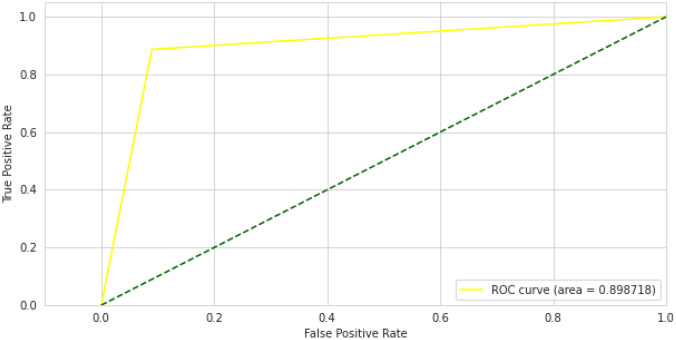
The results of the VGG-16 on pediatric pneumonia experiment are also based on a pre-trained model on the ImageNet dataset. To classify images of chest X-rays into normal and pneumonia classes, we utilized VGG-16 as the base model and added an additional classifier on top of it as a transfer learning technique. The training and validation accuracy curves for the VGG-16 model are shown in Fig. 14. During 30 training epochs, accuracy and loss are calculated. After training and validation, we also computed a receiver operating characteristic curve (ROC) for the VGG-16 model, which shows performance above 0.5 and an area under the ROC curve (AUC) of 0.8, as shown in Fig. 15. The confusion matrix of VGG-16 is produced and shows the number of TP, FP, TN or FN classified into normal and pneumonia classes for the classification of X-ray images as shown in Fig. 16.
Fig. 14.
VGG-16 training and validation accuracy and loss
Fig. 15.
VGG-16 ROC
Fig. 16.
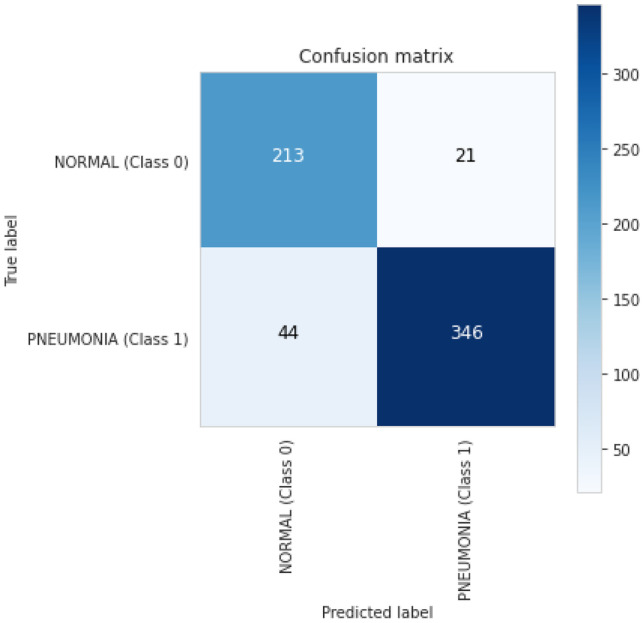
VGG-16 confusion martix
Vision Transformer
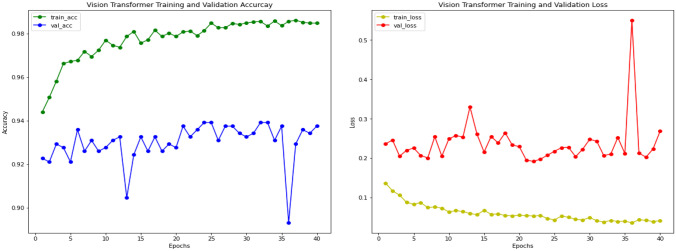
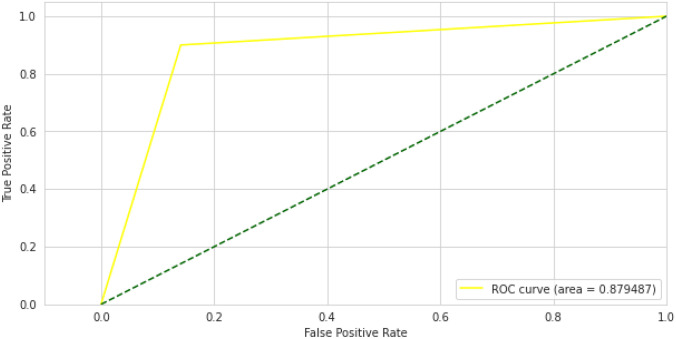
The results of the Vision transformer experiment with pediatric pneumonia dataset are also based on the pre-trained ImageNet model. We used Vision transformer as the basic model to classify images of chest X-rays into normal and pneumonia classes for that we added a further classifier head on top of vision transformer as a transfer learning strategy. The training and validation curves are shown in Fig. 17 for the vision transformer model. Accuracy and loss are calculated during 30 training periods. After training and validation, we also computed a receiver operating characteristic curve (ROC) for the Vision Transformer model, which shows performance above 0.5 and an area under the ROC curve (AUC) of 0.87, as shown in Fig. 18. The confusion matrix of Vision Transformer is produced and shows the number of TP, FP, TN or FN classified into normal and pneumonia classes for the classification of X-ray images as shown in Fig. 19.
Fig. 17.
Vision transformer training and validation accuracy and loss
Fig. 18.
Vision transformer ROC
Fig. 19.
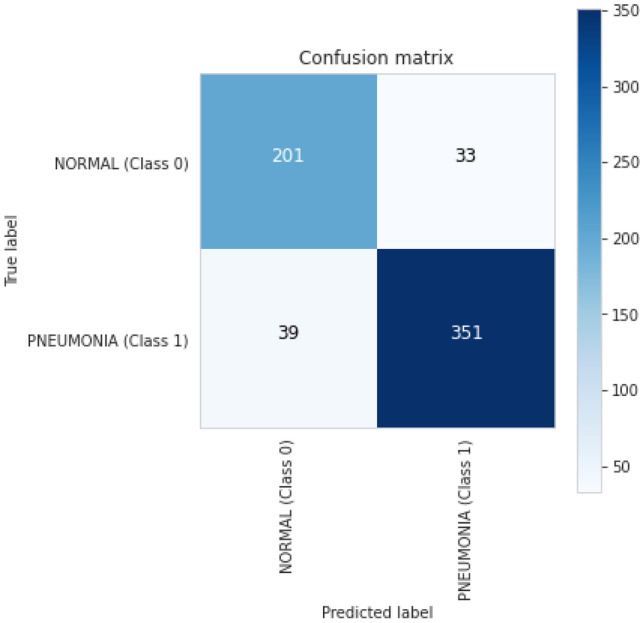
Vision transformer confusion matrix
Pre-trained Vision Transformer vs Training from Scratch
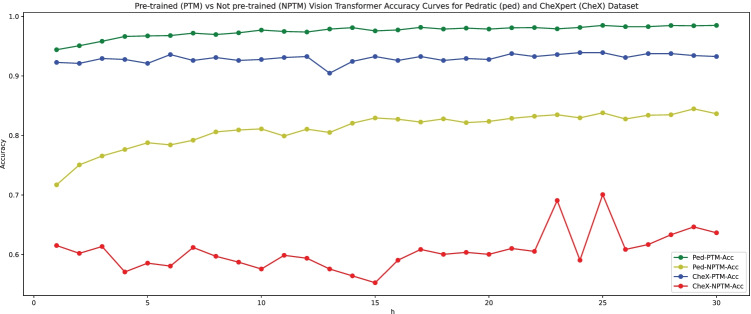
The results of the pre-trained vs training from scratch of Vision transformer on pediatric pneumonia dataset are shown in Fig. 7. We used Vision transformer as the basic model to classify images of chest X-rays into normal and pneumonia classes for that we added a further classifier head on top of vision transformer as a transfer learning strategy. The training and validation accuracy curves are shown in Fig. 7 for the vision transformer pre-trained model (ViT-PTM and vision transformer not pre-trained model (ViT-NPTM). Accuracy is calculated during 30 training periods model stops after convergence of 10 epochs. Results shows that pre-trained vision transformer performs better as compared to training a model from scratch.
Fig. 7.
Pre-trained vision transformer vs training from scratch
Experiments on CheXpert Dataset
CheXpert is a multi-label classification task in which X-rays images are classified in 14 observations. Through experimentation on a CheXpert dataset it is revealed that pre-trained transformer transfer learning performs better as compared to other state-of-the-art CNN-based vision models. Performance comparison of vision transformer and other CNN-based deep learning models are shown in Table 4
Table 4.
Performance comparison on CheXpert dataset
| Model | Precision | Recall | F1 Score |
|---|---|---|---|
| VGG-16 | 0.64 | 0.51 | 0.57 |
| ResNet-50 | 0.63 | 0.49 | 0.55 |
| Vision Transformer | 0.67 | 0.53 | 0.59 |
ResNet-50
Experimental results of ResNet-50 on CheXpert are done using pre-trained ResNet-50 base model. ResNet model uses residual connections or skip connection for learning representation. On top of that base model an additional classifier is added to classify images of chest X-rays into 14 classes. Classification report of ResNet model can be seen in Table 5; accuracy and loss are computed on 30 epochs of training.
Table 5.
ResNet-50 classification report
| Pathology | Precision | Recall | F1 Score | Support |
|---|---|---|---|---|
| No Finding | 0.49 | 0.30 | 0.37 | 808 |
| Enlarged Cardiom | 0.00 | 0.00 | 0.00 | 821 |
| Cardiomengaly | 0.62 | 0.33 | 0.43 | 1322 |
| Lung Opacity | 0.61 | 0.68 | 0.64 | 3895 |
| Lung Lesion | 0.00 | 0.00 | 0.00 | 357 |
| Edema | 0.61 | 0.47 | 0.53 | 2298 |
| Consolidation | 0.29 | 0.04 | 0.07 | 1498 |
| Pneumonia | 0.29 | 0.07 | 0.11 | 848 |
| Atelectasis | 0.44 | 0.24 | 0.31 | 2377 |
| Pneumothorax | 0.36 | 0.51 | 0.42 | 859 |
| Pleural Effusioin | 0.74 | 0.70 | 0.72 | 3515 |
| Pleural Other | 0.00 | 0.00 | 0.00 | 230 |
| Fracture | 0.00 | 0.00 | 0.00 | 341 |
| Support Devices | 0.75 | 0.83 | 0.79 | 4222 |
VGG-16
The results of the VGG-16 on CheXpert are also based on a pre-trained model on the ImageNet dataset. To classify images of chest X-rays into 14 classes, we utilized Vgg-16 as the base model and added an additional classifier on top of it as a transfer learning technique. The classification report for the VGG-16 model is shown in Table 6. During 30 training epochs, accuracy and loss are calculated.
Table 6.
VGG-16 classification report
| Pathology | Precision | Recall | F1 Score | Support |
|---|---|---|---|---|
| No Finding | 0.50 | 0.28 | 0.36 | 828 |
| Enlarged Cardiom | 0.00 | 0.00 | 0.00 | 834 |
| Cardiomengaly | 0.54 | 0.44 | 0.48 | 1253 |
| Lung Opacity | 0.60 | 0.77 | 0.68 | 3875 |
| Lung Lesion | 0.00 | 0.00 | 0.00 | 362 |
| Edema | 0.60 | 0.63 | 0.62 | 2339 |
| Consolidation | 0.34 | 0.03 | 0.06 | 1561 |
| Pneumonia | 0.32 | 0.11 | 0.16 | 911 |
| Atelectasis | 0.47 | 0.30 | 0.37 | 2387 |
| Pneumothorax | 0.58 | 0.27 | 0.37 | 919 |
| Pleural Effusioin | 0.75 | 0.71 | 0.73 | 3455 |
| Pleural Other | 0.00 | 0.00 | 0.00 | 245 |
| Fracture | 0.00 | 0.00 | 0.00 | 374 |
| Support Devices | 0.77 | 0.79 | 0.78 | 4079 |
Vision Transformer
The results of the Vision transformer experiment with CheXpert dataset are also based on the pre-trained ImageNet model. We used Vision transformer as the basic model to classify images of chest X-rays into 14 classes for that we added a further classifier head on top of vision transformer as a transfer learning strategy. The classification report can be seen in Table 7 for the vision transformer model. Accuracy and loss are calculated during 30 training periods.
Table 7.
Vision transformer classification
| Pathology | Precision | Recall | F1 Score | Support |
|---|---|---|---|---|
| No Finding | 0.50 | 0.44 | 0.42 | 728 |
| Enlarged Cardiom | 0.00 | 0.00 | 0.00 | 794 |
| Cardiomengaly | 0.54 | 0.49 | 0.51 | 1278 |
| Lung Opacity | 0.63 | 0.78 | 0.68 | 3950 |
| Lung Lesion | 1.00 | 0.00 | 0.01 | 354 |
| Edema | 0.60 | 0.61 | 0.62 | 2358 |
| Consolidation | 0.39 | 0.09 | 0.14 | 1485 |
| Pneumonia | 0.32 | 0.11 | 0.12 | 852 |
| Atelectasis | 0.47 | 0.35 | 0.40 | 2341 |
| Pneumothorax | 0.58 | 0.33 | 0.39 | 935 |
| Pleural Effusioin | 0.75 | 0.70 | 0.73 | 3463 |
| Pleural Other | 0.00 | 0.00 | 0.00 | 247 |
| Fracture | 0.32 | 0.02 | 0.04 | 355 |
| Support Devices | 0.80 | 0.79 | 0.72 | 4251 |
Conclusions
The transfer learning of transformers for medical imaging is evaluated in this paper. For this purpose, a transformer-based strategy is used to classify chest X-ray images. To assess performance, CheXpert and the Pediatric Pneumonia dataset are used. Transfer learning in the proposed vision transformer outperforms existing CNN-based models in identifying medical images. Our method is based on the original architecture of the transformer as well as transfer learning techniques. For the image classification model, the transformer’s Encoder block is used. In the future different new models, as well as a combination of CNN and transformer architectures, may be used to evaluate model efficacy in medical imaging.
Supplementary Information
Below is the link to the electronic supplementary material.
Biographies
Mohammad Usman
received the B.S. degree in computer sciences from IUB, Pakistan. He is currently pursuing the MS degree in computer science with COMSATS University, Islamabad. His areas of interests include computer vision, deep learning.
Tehseen Zia
is Assistant Professor Computer Science at COMSATS University Islamabad, Pakistan. He is also co-Principle Investigator at Medical Imaging and Diagnostic Lab, established under National Center of Artificial Intelligence, a major project of Higher Education Commission of Pakistan. His research interests involve machine-learning (A.I.), natural language processing and computer vision. He holds a Ph.D. in Computer Science (2010) from Vienna University of Technology, Austria, and Master in Computer Science from University of Engineering and Technology, Texila (2003).
Syed Ali Tariq
received the B.S. degree in computer and information sciences from PIEAS, Pakistan, and the M.S. degree in computer science from Abasyn University, Islamabad. He is currently pursuing the Ph.D. degree with COMSATS University, Islamabad. He is also working with the Medical Imaging and Diagnostics Lab, COMSATS. His areas of interests include image processing, deep learning, biometric systems, and GPU-based parallel computing.
Appendix Extended results on CheXpert and pediatric pneumonia datasets
Further results on pediatric pneumonia dataset
Funding
This work was supported by the Higher Education Commission under National Center of Artificial Intelligence, Grant 2(1064).
References
- 1.Irvin J, Rajpurkar P, Ko M, Yu Y, Ciurea-Ilcus S, Chute C, Marklund H, Haghgoo B, Ball R, Shpanskaya K et al (2019) Chexpert: A large chest radiograph dataset with uncertainty labels and expert comparison. In: Proceedings of the AAAI Conference on Artificial Intelligence, pp 590–597
- 2.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM (2017a) Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2097–2106
- 3.Abideen ZU, Ghafoor M, Munir K, Saqib M, Ullah A, Zia T, Tariq SA, Ahmed G, Zahra A (2020a) Uncertainty assisted robust tuberculosis identification with bayesian convolutional neural networks. Ieee Access 8:22812–22825 [DOI] [PMC free article] [PubMed]
- 4.Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLoS medicine. 2018;15(11):e1002683. doi: 10.1371/journal.pmed.1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gordienko Y, Gang P, Hui J, Zeng W, Kochura Y, Alienin O, Rokovyi O, Stirenko S. International Conference on Computer Science. Engineering and Education Applications: Springer; 2018. Deep learning with lung segmentation and bone shadow exclusion techniques for chest x-ray analysis of lung cancer; pp. 638–647. [Google Scholar]
- 6.Que Q, Tang Z, Wang R, Zeng Z, Wang J, Chua M, Gee TS, Yang X, Veeravalli B (2018) Cardioxnet: automated detection for cardiomegaly based on deep learning. In: 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), IEEE, pp 612–615 [DOI] [PubMed]
- 7.Organization WH (2016) Chest radiography in tuberculosis detection: summary of current WHO recommendations and guidance on programmatic approaches. World Health Organization
- 8.Brady A, Laoide RÓ, McCarthy P, McDermott R. Discrepancy and error in radiology: concepts, causes and consequences. The Ulster medical journal. 2012;81(1):3. [PMC free article] [PubMed] [Google Scholar]
- 9.He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
- 10.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Advances in neural information processing systems. 2012;25:1097–1105. [Google Scholar]
- 11.Tan M, Le Q (2019) Efficientnet: Rethinking model scaling for convolutional neural networks. In: International Conference on Machine Learning, PMLR, pp 6105–6114
- 12.Gu Y, Lu X, Yang L, Zhang B, Yu D, Zhao Y, Gao L, Wu L, Zhou T. Automatic lung nodule detection using a 3d deep convolutional neural network combined with a multi-scale prediction strategy in chest cts. Computers in biology and medicine. 2018;103:220–231. doi: 10.1016/j.compbiomed.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 13.Isensee F, Kickingereder P, Wick W, Bendszus M, Maier-Hein KH (2017) Brain tumor segmentation and radiomics survival prediction: Contribution to the brats 2017 challenge. In: International MICCAI Brainlesion Workshop, Springer, pp 287–297
- 14.Wang J, Li F, Li Q. Automated segmentation of lungs with severe interstitial lung disease in ct. Medical physics. 2009;36(10):4592–4599. doi: 10.1118/1.3222872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, Dehghani M, Minderer M, Heigold G, Gelly S, et al. (2020) An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:201011929
- 16.Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser L, Polosukhin I (2017) Attention is all you need. arXiv preprint arXiv:170603762
- 17.Carion N, Massa F, Synnaeve G, Usunier N, Kirillov A, Zagoruyko S (2020) End-to-end object detection with transformers. In: European Conference on Computer Vision, Springer, pp 213–229
- 18.Liu Z, Lin Y, Cao Y, Hu H, Wei Y, Zhang Z, Lin S, Guo B (2021) Swin transformer: Hierarchical vision transformer using shifted windows. arXiv:210314030
- 19.Zheng S, Lu J, Zhao H, Zhu X, Luo Z, Wang Y, Fu Y, Feng J, Xiang T, Torr PH et al. (2021) Rethinking semantic segmentation from a sequence-to-sequence perspective with transformers. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp 6881–6890
- 20.Dai Y, Gao Y (2021) Transmed: Transformers advance multi-modal medical image classification. arXiv preprint arXiv:210305940 [DOI] [PMC free article] [PubMed]
- 21.Park J, Kim Y (2021) Styleformer: Transformer based generative adversarial networks with style vector. arXiv preprint arXiv:210607023
- 22.Chen J, Lu Y, Yu Q, Luo X, Adeli E, Wang Y, Lu L, Yuille AL, Zhou Y (2021) Transunet: Transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:210204306
- 23.Jun E, Jeong S, Heo DW, Suk HI (2021) Medical transformer: Universal brain encoder for 3d mri analysis. arXiv preprint arXiv:210413633 [DOI] [PubMed]
- 24.Karimi D, Vasylechko S, Gholipour A (2021) Convolution-free medical image segmentation using transformers. arXiv preprint arXiv:210213645
- 25.Zhang Y, Liu H, Hu Q (2021a) Transfuse: Fusing transformers and cnns for medical image segmentation. arXiv preprint arXiv:210208005
- 26.Gao X, Qian Y, Gao A (2021) Covid-vit: Classification of covid-19 from ct chest images based on vision transformer models. arXiv preprint arXiv:210701682
- 27.Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
- 28.Ghassemi N, Shoeibi A, Khodatars M, Heras J, Rahimi A, Zare A, Pachori RB, Gorriz JM (2021) Automatic diagnosis of covid-19 from ct images using cyclegan and transfer learning. arXiv preprint arXiv:210411949 [DOI] [PMC free article] [PubMed]
- 29.Kalkreuth R, Kaufmann P (2020) Covid-19: a survey on public medical imaging data resources. arXiv preprint arXiv:200404569
- 30.Li J, Zhu G, Hua C, Feng M, Li P, Lu X, Song J, Shen P, Xu X, Mei L, et al. (2021a) A systematic collection of medical image datasets for deep learning. arXiv preprint arXiv:210612864
- 31.Deng J, Dong W, Socher R, Li LJ, Li K, Fei-Fei L (2009) Imagenet: A large-scale hierarchical image database. In: 2009 IEEE conference on computer vision and pattern recognition, Ieee, pp 248–255
- 32.Raghu M, Zhang C, Kleinberg J, Bengio S (2019b) Transfusion: Understanding transfer learning for medical imaging. arXiv preprint arXiv:190207208
- 33.Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:14091556
- 34.Abràmoff MD, Lou Y, Erginay A, Clarida W, Amelon R, Folk JC, Niemeijer M. Improved automated detection of diabetic retinopathy on a publicly available dataset through integration of deep learning. Investigative ophthalmology & visual science. 2016;57(13):5200–5206. doi: 10.1167/iovs.16-19964. [DOI] [PubMed] [Google Scholar]
- 35.De Fauw J, Ledsam JR, Romera-Paredes B, Nikolov S, Tomasev N, Blackwell S, Askham H, Glorot X, O, Donoghue B, Visentin D, et al. (2018) Clinically applicable deep learning for diagnosis and referral in retinal disease. Nature medicine 24(9):1342–1350 [DOI] [PubMed]
- 36.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, Venugopalan S, Widner K, Madams T, Cuadros J, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. Jama. 2016;316(22):2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 37.Raghu M, Blumer K, Sayres R, Obermeyer Z, Kleinberg B, Mullainathan S, Kleinberg J (2019a) Direct uncertainty prediction for medical second opinions. In: International Conference on Machine Learning, PMLR, pp 5281–5290
- 38.Rajpurkar P, Irvin J, Zhu K, Yang B, Mehta H, Duan T, Ding D, Bagul A, Langlotz C, Shpanskaya K, et al. (2017) Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv preprint arXiv:171105225
- 39.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM (2017b) Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2097–2106
- 40.Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nature medicine. 2019;25(1):44–56. doi: 10.1038/s41591-018-0300-7. [DOI] [PubMed] [Google Scholar]
- 41.Van Der Heijden AA, Abramoff MD, Verbraak F, van Hecke MV, Liem A, Nijpels G. Validation of automated screening for referable diabetic retinopathy with the idx-dr device in the hoorn diabetes care system. Acta ophthalmologica. 2018;96(1):63–68. doi: 10.1111/aos.13613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chouhan V, Singh SK, Khamparia A, Gupta D, Tiwari P, Moreira C, Damaševičius R, De Albuquerque VHC. A novel transfer learning based approach for pneumonia detection in chest x-ray images. Applied Sciences. 2020;10(2):559. doi: 10.3390/app10020559. [DOI] [Google Scholar]
- 43.Ding Y, Sohn JH, Kawczynski MG, Trivedi H, Harnish R, Jenkins NW, Lituiev D, Copeland TP, Aboian MS, Mari Aparici C, et al. A deep learning model to predict a diagnosis of alzheimer disease by using 18f-fdg pet of the brain. Radiology. 2019;290(2):456–464. doi: 10.1148/radiol.2018180958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen S, Ma K, Zheng Y (2019) Med3d: Transfer learning for 3d medical image analysis. arXiv preprint arXiv:190400625
- 45.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S (2017) Dermatologist-level classification of skin cancer with deep neural networks. nature 542(7639):115–118 [DOI] [PMC free article] [PubMed]
- 46.Khosravi P, Kazemi E, Zhan Q, Toschi M, Malmsten JE, Hickman C, Meseguer M, Rosenwaks Z, Elemento O, Zaninovic N, et al. (2018) Robust automated assessment of human blastocyst quality using deep learning. bioRxiv p 394882
- 47.Kim D, MacKinnon T. Artificial intelligence in fracture detection: transfer learning from deep convolutional neural networks. Clinical radiology. 2018;73(5):439–445. doi: 10.1016/j.crad.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 48.Abid MMN, Zia T, Ghafoor M, Windridge D (2021) Multi-view convolutional recurrent neural networks for lung cancer nodule identification. Neurocomputing
- 49.Abideen ZU, Ghafoor M, Munir K, Saqib M, Ullah A, Zia T, Tariq SA, Ahmed G, Zahra A (2020b) Uncertainty assisted robust tuberculosis identification with bayesian convolutional neural networks. Ieee Access 8:22812–22825 [DOI] [PMC free article] [PubMed]
- 50.Mamalakis M, Swift AJ, Vorselaars B, Ray S, Weeks S, Ding W, Clayton RH, Mackenzie LS, Banerjee A (2021) Denrescov-19: A deep transfer learning network for robust automatic classification of covid-19, pneumonia, and tuberculosis from x-rays. arXiv preprint arXiv:210404006 [DOI] [PMC free article] [PubMed]
- 51.Palaskar R, Vyas R, Khedekar V, Palaskar S, Sahu P (2020) Transfer learning for oral cancer detection using microscopic images. arXiv preprint arXiv:201111610
- 52.Geirhos R, Rubisch P, Michaelis C, Bethge M, Wichmann FA, Brendel W (2018) Imagenet-trained cnns are biased towards texture; increasing shape bias improves accuracy and robustness. arXiv preprint arXiv:181112231
- 53.He K, Girshick R, Dollár P (2019) Rethinking imagenet pre-training. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp 4918–4927
- 54.Huh M, Agrawal P, Efros AA (2016) What makes imagenet good for transfer learning? arXiv preprint arXiv:160808614
- 55.Kornblith S, Shlens J, Le QV (2019) Do better imagenet models transfer better? In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp 2661–2671
- 56.Ngiam J, Peng D, Vasudevan V, Kornblith S, Le QV, Pang R (2018) Domain adaptive transfer learning with specialist models. arXiv preprint arXiv:181107056
- 57.Ophthalmoscopy D, Levels E (2002) International clinical diabetic retinopathy disease severity scale detailed table. Ophthalmology
- 58.Parmar N, Vaswani A, Uszkoreit J, Kaiser L, Shazeer N, Ku A, Tran D (2018) Image transformer. In: International Conference on Machine Learning, PMLR, pp 4055–4064
- 59.Wu B, Xu C, Dai X, Wan A, Zhang P, Tomizuka M, Keutzer K, Vajda P (2020) Visual transformers: Token-based image representation and processing for computer vision. arXiv preprint arXiv:200603677
- 60.Devlin J, Chang MW, Lee K, Toutanova K (2018) Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:181004805
- 61.Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, Kang J. Biobert: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2020;36(4):1234–1240. doi: 10.1093/bioinformatics/btz682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beltagy I, Lo K, Cohan A (2019) Scibert: A pretrained language model for scientific text. arXiv preprint arXiv:190310676
- 63.Huang K, Altosaar J, Ranganath R (2019) Clinicalbert: Modeling clinical notes and predicting hospital readmission. arXiv preprint arXiv:190405342
- 64.Brown TB, Mann B, Ryder N, Subbiah M, Kaplan J, Dhariwal P, Neelakantan A, Shyam P, Sastry G, Askell A, et al. (2020) Language models are few-shot learners. arXiv preprint arXiv:200514165
- 65.Chen M, Radford A, Child R, Wu J, Jun H, Luan D, Sutskever I (2020) Generative pretraining from pixels. In: International Conference on Machine Learning, PMLR, pp 1691–1703
- 66.Touvron H, Cord M, Douze M, Massa F, Sablayrolles A, Jégou H (2020) Training data-efficient image transformers & distillation through attention. arXiv preprint arXiv:201212877
- 67.Li S, Sui X, Luo X, Xu X, Liu Y, Goh RSM (2021b) Medical image segmentation using squeeze-and-expansion transformers. arXiv preprint arXiv:210509511
- 68.Zhang Z, Yu L, Liang X, Zhao W, Xing L (2021b) Transct: Dual-path transformer for low dose computed tomography. arXiv preprint arXiv:210300634
- 69.Kermany D, Zhang K, Goldbaum M, et al. (2018) Labeled optical coherence tomography (oct) and chest x-ray images for classification. Mendeley data 2(2)
- 70.(April, 2021 (accessed on May , 2021)) Pneumonia in children statistics. https://data.unicef.org/topic/child-health/pneumonia/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.