Abstract
Although campylobacters have been isolated from a wide range of animal hosts, the association between campylobacters isolated from humans and animals in the farm environment is unclear. We used flagellin gene typing and pulsed-field gel electrophoresis (PFGE) to investigate the genetic diversity among isolates from animals (cattle, sheep, and turkey) in farm environments and sporadic cases of campylobacteriosis in the same geographical area. Forty-eight combined fla types were seen among the 315 Campylobacter isolates studied. Six were found in isolates from all four hosts and represented 50% of the total number of isolates. Seventy-one different SmaI PFGE macrorestriction profiles (mrps) were observed, with 86% of isolates assigned to one of 29 different mrps. Fifty-seven isolates from diverse hosts, times, and sources had an identical SmaI mrp and combined fla type. Conversely, a number of genotypes were unique to a particular host. We provide molecular evidence which suggests a link between campylobacters in the farm environment with those causing disease in the community.
Campylobacters are the most frequent cause of acute bacterial diarrhea in the United Kingdom (18) and other industrialized countries (20). Since the early 1980s, the number of cases of Campylobacter-related diarrhea reported to the Public Health Laboratory Service Communicable Disease Surveillance Centre by laboratories in England and Wales has increased nearly fivefold and has exceeded levels of Salmonella diarrhea. By 1998, the yearly total had reached 58,211 (7). The true incidence of infection is thought to be significantly higher and in the United States an estimated 2.4 million people are affected by Campylobacter spp. each year. Campylobacters are a common component in the intestines of a wide range of warm-blooded mammals and birds. They have been isolated from the feces of domestic farm animals, including beef cattle (8, 10), dairy cows, and sheep (21). In order to elucidate possible sources of sporadic Campylobacter infection, phenotypic typing methods such as biotyping and serotyping have been applied to Campylobacter isolates from animal and environmental sources, as well as clinical strains (2, 10, 23). Epidemiological traceback conclusions based on these typing methods are tentative because of the limited discriminatory potential of the techniques.
In an attempt to find an environmental reservoir of Campylobacter infection in Lancaster, England, Jones et al. (16) suggested the involvement of animals. Further investigations showed that campylobacters were readily isolated in large numbers from dairy cattle (32); sheep grazing on salt marsh, fell, and farm pasture (17, 31); and turkeys (37). All of these isolates were obtained within the Lancaster area. Although these studies were useful in quantifying the role of farm animals as reservoirs of Campylobacter, it was not possible to ascertain whether they were important in either the dissemination of campylobacters from the farm environment or the causing of disease in the community. To address this, two high-resolution genotypic methods, PCR-restriction fragment length polymorphism (PCR-RFLP) analysis of the flaA gene and macrorestriction analysis by pulsed-field gel electrophoresis (PFGE) in combination with Preston biotyping, were used to investigate the diversity of genotypes occurring in the farm environment in a defined geographical area (Lancaster) and to compare these to clinical isolates collected from the same area in the same time frame.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The Campylobacter isolates examined in this study comprised four sets: 88 isolates from dairy cattle, 79 isolates from turkeys, 71 isolates from sheep, and 77 clinical isolates. The cattle came from four different farms in the Lancaster area (32); the sheep from salt marsh, fell, and farmland grazing areas (17, 31); the turkeys from a mixed farm (37); and the clinical isolates from Lancaster Royal Infirmary. All isolates were selected at random from isolates that had been collected during the period 1993 to 1996. All isolates were cultivated at 37°C for 48 h on 5% (vol/vol) defibrinated horse blood agar under microaerobic conditions (10% CO2, 10% H2, and 80% N2) in stainless steel anaerobic gas canisters (Don Whitley Scientific, Leeds, England) using the gas replacement method (5).
Identification and biotyping.
Presumptive identification of isolates was based on Gram stain and on catalase and cytochrome oxidase production. The modified Preston biotyping scheme (5) was used to speciate and biotype the isolates.
DNA preparation, PCR, and PFGE.
Bacterial suspensions were made in sterile distilled water (800 μl), and 5 μl was added to 50 μl of the standard PCR mixture described below. Oligonucleotide PCR primers were based on those previously described (1, 22). The sequence of the forward primer was 5′-GGATTTCGTATTAACAC(AC)AATGTTGC-3′, and the reverse primer was 5′-GCACC(CT)TTAAG(AT)GT(AG)GTTACACCTGC-3′. The template DNA for each strain was added to 50 μl of reaction mixture containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl, 0.4 μM concentrations of each primer, 200 μM concentrations of each deoxynucleotide, and 2.5 U of Thermus aquaticus (Taq) polymerase. PCR was performed on a Perkin-Elmer System 2400 thermal cycler. Samples were first incubated for 2 min at 94°C and then cycled 30 times at 92°C for 30 s, at 40°C for 1 min, and at 72°C for 5 min and were maintained at 4°C until processed. Then, 7 μl from the PCR mixture was checked for the presence of the expected 1.45-kbp amplicon representing the Campylobacter flagellin gene (flaA) by electrophoresis on a 0.7% minigel. If a sufficient amount of DNA was observed, 8 μl of the PCR product was digested with the restriction enzyme DdeI (New England Biolabs) or doubly digested with EcoRI/PstI (New England Biolabs) according to the manufacturer's instructions. The digested products were analyzed by agarose gel electrophoresis using 3% (wt/vol) Nusieve 3:1 agarose (FMC Bioproducts) in 1× TBE buffer (89 mM Tris-HCl, 89 mM boric acid, 2 mM EDTA; pH 8.3). The DNA fragments were stained with ethidium bromide and visualized using a UV transilluminator. Macrorestriction using the restriction enzyme SmaI and PFGE was performed as previously described for Campylobacter jejuni (27). Electrophoresis was carried out for 22 h at 200 V and 14°C constant temperature in a CHEF-DRII system (Bio-Rad) with pulsed times ramped from 10 to 35 s.
RESULTS
Distribution of strains by biotype.
Fifty-four different Preston biotypes were identified among the 315 isolates comprising the cattle, turkey, sheep, and clinical sets. The number of Preston biotypes belonging to each host is shown in Table 1. The predominant biotype in the clinical, cattle, and sheep sets was biotype 6000, and this was also found in 4% of the turkey set. Other biotypes, i.e., biotypes 6010, 6004, and 6014, were also found in all of the hosts. Biotype 6012, the second most common type in the sheep set, was found in 1% of the cattle isolates and 1% of the turkey isolates but was not found in the clinical set. The predominant biotype in the turkey set, biotype 6112, was found in 3% of the human isolates and 1% of the cattle isolates. It was not found in the sheep set. Thirteen biotypes were unique to the human set, although each biotype comprised three isolates or less. Similarly, there were 14, 6, and 4 unique biotypes in the cattle, turkey, and sheep sets, respectively.
TABLE 1.
Number and predominant Preston biotypes belonging to isolates of C. jejuni and C. coli from each host
| Set | No. of isolates | No. of Preston biotypes | Predominant Preston biotypes | % Biotype within set |
|---|---|---|---|---|
| Clinical | 77 | 29 | 6000 | 21 |
| 6010 | 16 | |||
| Cattle | 88 | 26 | 6000 | 27 |
| 6010 | 14 | |||
| Sheep | 71 | 11 | 6000 | 34 |
| 6012 | 20 | |||
| Turkey | 79 | 16 | 6112 | 32 |
| 6010 | 15 |
Flagellin gene polymorphism.
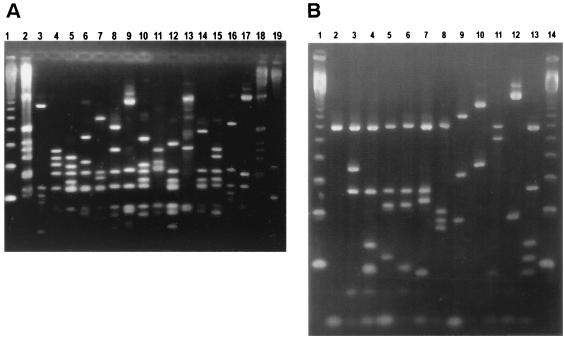
Flagellin gene (flaA) PCR amplicons of 1.45-kbp were generated from all isolates, i.e., the 302 C. jejuni and 13 C. coli isolates. Amplicons were digested with either DdeI or doubly digested with EcoRI and PstI restriction endonucleases. Designated fla types were assigned according to the different fragment sizes. Restriction analysis of the flaA amplicons by DdeI gave 35 different digest profiles (assigned fla types 1 to 35), and restriction by EcoRI and PstI gave 26 different digest profiles (assigned fla types A to Z). Examples of these PCR-RFLP profiles are shown in Fig. 1. The DdeI and EcoRI/PstI polymorphisms were then used to designate 48 combined fla types (Table 2).
FIG. 1.
(A) DdeI flagellin gene PCR-RFLP patterns. Lanes 1 and 19, 123-bp ladder (New England Biolabs); lanes 2 and 18, 1-kbp ladder (New England Biolabs); lane 3, 940801/01 (D2); lane 4, 950823/05 (D21); lane 5, T941017/110d (D33); lane 6, T941017/14d (D24); lane 7, T941017/05 (D4); lane 8, 950717/10d (D3); lane 9, T941017/19e (D28); lane 10, T941017/25d (D5); lane 11, 950717/2d (D8); lane 12, 950605/12e (D18); lane 13, 941024/05 (D14); lane 14, 940902/01 (D34); lane 15, T941017/04 (D25); lane 16, 941024/01 (D10); lane 17, 941024/03 (D27). Letters in parentheses show a particular designated type for DdeI flagellin gene PCR-RFLP patterns. (B) EcoRI/PstI flagellin gene PCR-RFLP patterns. Lanes 1 and 14, 123-bp ladder (New England Biolabs); lane 2, 950619/5e (L); lane 3, T941017/03 (Z); lane 4, T941017/110d (W); lane 5, 941024/03 (V); lane 6, T941017/14d (E); lane 7, 950823/diad (G); lane 8, 950717/2d (H); lane 9, 940801/01 (I); lane 10, 941024/06 (J); lane 11, T941017/210d (N); lane 12, T941017/19e (F); lane 13; T941017/25d (P). Letters in parentheses show a particular designated type for EcoRI/PstI flagellin gene PCR-RFLP patterns.
TABLE 2.
Summary of the number of isolates from each host which belong to a particular combined fla type
| Combined fla typea | No. of isolates
|
|||
|---|---|---|---|---|
| Human | Sheep | Cattle | Turkey | |
| 3G∗ | 13 | 3 | 20 | 8 |
| 13A | 6 | |||
| 12B | 1 | |||
| 11C | 2 | |||
| 21N | 1 | 12 | ||
| 22R | 1 | |||
| 7K | 1 | |||
| 6J | 2 | |||
| 10D | 2 | 17 | ||
| 14J∗ | 4 | 4 | 1 | 3 |
| 18L | 2 | 33 | ||
| 29Q | 2 | 1 | ||
| 15A | 1 | |||
| 16Q | 1 | |||
| 7A | 2 | 3 | ||
| 8H∗ | 4 | 1 | 2 | 5 |
| 28F | 1 | 1 | ||
| 2I∗ | 3 | 26 | 32 | 2 |
| 1V | 1 | |||
| 5G | 1 | |||
| 4E | 2 | 1 | ||
| 9W | 1 | |||
| 23F | 1 | |||
| 24E∗ | 1 | 4 | 4 | 2 |
| 25N∗ | 10 | 2 | 2 | 2 |
| 29O | 1 | |||
| 1M | 1 | |||
| 21U | 2 | |||
| 3T | 1 | |||
| 27V | 1 | 1 | ||
| 17O | 1 | |||
| 26P | 1 | |||
| 29S | 1 | 2 | ||
| 30S | 2 | |||
| 20S | 2 | 4 | ||
| 33W | 3 | 2 | ||
| 5P | 1 | 11 | 1 | |
| 17A | 1 | |||
| 35A | 3 | |||
| 20Z | 3 | |||
| 19X | 1 | |||
| 11X | 1 | |||
| 31N | 3 | |||
| 10I | 2 | 2 | ||
| 14Y | 1 | |||
| 32H | 1 | |||
| 34L | 1 | |||
| 4V | 1 | |||
Combined fla types found in all four hosts are indicated by an asterisk.
Distribution of fla types by host.
The most common EcoRI/PstI fla type I (found in 21% of the total number of isolates) was found in all four hosts. It was the predominant type in the sheep (37%) and cattle (39%) sets. The second most common EcoRI/PstI fla type was G (14%), and it again was seen in all four hosts. It was the predominant fla type in the human clinical set (18%) and the second largest in the cattle set (23%). The predominant fla type in the turkey set was L (43%). This did not occur in either the cattle or sheep sets but was found in 3% of the human set.
The most common DdeI fla type, type 2 (20%), was found in all four hosts and was the predominant fla type in both the cattle (36%) and the sheep (37%) sets. The second most common DdeI type was type 3 and again was seen in all four hosts. This was the predominant fla type in the human set (18%) and the second largest type in the cattle set (23%). The most common DdeI type in the turkey set was 18 (42%) and, although this type did not occur in either the cattle or the sheep sets, it was seen in 3% of the human set. Strains belonging to the human set were more evenly distributed among the restriction fla types, while the animal hosts each had two or three predominant fla types.
All hosts exhibited a diversity of combined fla types. Six were found in all four hosts and represented 50% (158 of 315) of the total number of isolates. The human set was the most heterogeneous group and contained 34 combined fla types. This compared with 14 combined fla types in the sheep, 16 fla types in the cattle, and 16 fla types in the turkey sets. The number of combined fla types unique to the human set was 19 (although each type contained six isolates or less). There were five unique combined fla types in the cattle set, three in the turkey set, and two in the sheep set.
PFGE analysis.
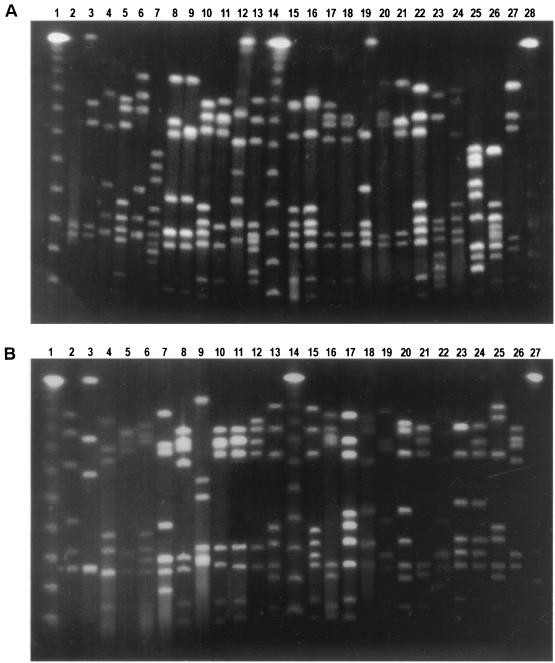
PFGE analysis of SmaI-digested DNA from all the isolates yielded between 6 and 12 fragments that ranged in size from ∼40 to ∼485 kbp. Eighty-six percent of the isolates were assigned to 29 different macrorestriction profile (mrp) types, which were observed among multiple strains (Fig. 2A). Forty-two mrps were found in only one strain and were assigned a type number from U1 to U42 (Fig. 2B).
FIG. 2.
(A) PFGE restriction profiles of SmaI-digested DNA found in more than one isolate. Lanes 1, 14, and 28, 48.5-kb ladder (New England Biolabs); lane 2, 930621/03 (PFGE type 1); lane 3, T941017/14d (PFGE type 2); lane 4, 940815/03 (PFGE type 3); lane 5, 940902/14 (PFGE type 4); lane 6, 52/96 (PFGE type 5); lane 7, 51/96 (PFGE type 6); lane 8, 941024/05 (PFGE type 7); lane 9, 24/10 5e (PFGE type 8); lane 10, 25/95 (PFGE type 9); lane 11, 27/95 (PFGE type 10); lane 12, 436/94 (PFGE type 11); lane 13, 950807/1d (PFGE type 12); lane 15, 930505/07 (PFGE type 13); lane 16, 7/95 (PFGE type 14); lane 17, 931214/11 (PFGE type 15); lane 18, 941118/02 (PFGE type 17); lane 19, 950607/29d (PFGE type 18); lane 20, 317/93 (PFGE type 19); lane 21 950403/2d (PFGE type 20); lane 22, 931025/03 (PFGE type 21); lane 23, 32/95 (PFGE type 22); lane 24, 1175/93 (PFGE type 23); lane 25, T941017/29d (PFGE type 24); lane 26, 950622/40e (PFGE type 25); lane 27, 810/93 (PFGE type 26). (B) Lanes 2 to 3, 5 to 7, and 9 to 25, unique PFGE restriction profiles of SmaI-digested DNA; lanes 1, 14, and 27, 48.5-kb ladder (New England Biolabs); lane 2, 306/94; lane 3, T941017/04; lane 4, 940721/04 (PFGE type 29); lane 5, 13/96; lane 6, 14/96; lane 7, 39/95; lane 8, 950410/2e (PFGE type 15); lane 9, 3/94; lane 10, 10/95; lane 11, 428/94; lane 12, 969/94; lane 13, 37/92; lane 15, 31/10 19d; lane 16, 940726/05; lane 17, T941017/25d; lane 18, 14/93; lane 19, 950717/4e; lane 20, 941024/03; lane 21, 332/93; lane 22, 960610/30; lane 23, 731/93; lane 24, 930505/05; lane 25, 940118/05; lane 26, 940318/04 (PFGE type 15).
Distribution of mrp types by host.
The association between mrp type and host is summarized in Table 3. All hosts exhibited a diversity of mrp's. Three were found in all four hosts. The most common mrp, type 15 (Fig. 2A, lane 17), contained 62 isolates and was diverse with respect to host and source. The human set was the most heterogeneous and contained 20 different mrp types observed among multiple strains and 28 unique mrps seen for only one strain. In the cattle, sheep, and turkey sets the numbers of mrp types were 14, 12, and 9, respectively.
TABLE 3.
Distribution of mrp types by host
| mrp typea | No. of isolates
|
|||
|---|---|---|---|---|
| Cattle | Sheep | Turkey | Human | |
| 1 | 4 | 4 | 3 | |
| 2 | 2 | 1 | ||
| 3 | 2 | 6 | ||
| 4 | 12 | 10 | 5 | |
| 5 | 2 | |||
| 6 | 1 | 1 | 1 | |
| 7∗ | 2 | 2 | 3 | 2 |
| 8 | 2 | 1 | ||
| 9∗ | 3 | 3 | 1 | 4 |
| 10 | 3 | 2 | 4 | |
| 11 | 2 | |||
| 12 | 11 | |||
| 13 | 2 | |||
| 14 | 2 | 1 | ||
| 15∗ | 33 | 25 | 1 | 3 |
| 16 | 2 | |||
| 17 | 4 | |||
| 18 | 31 | 2 | ||
| 19 | 2 | |||
| 20 | 17 | 1 | ||
| 21 | 7 | |||
| 22 | 2 | |||
| 23 | 5 | |||
| 24 | 7 | |||
| 25 | 3 | |||
| 26 | 3 | 2 | ||
| 27 | 4 | 2 | 3 | |
| 28 | 4 | |||
| 29 | 4 | |||
| Unique | 5 | 3 | 6 | 28 |
| Unknown | 1 | 1 | ||
mrp types seen in all four hosts are indicated by an asterisk.
Association between fla type and mrp type.
The associations among the isolates of C. jejuni and C. coli with respect to fla type and mrp type are shown in Table 4. Fourteen mrp's were observed among strains that had different combined fla types. The other 15 mrp's corresponded to a single fla type. The largest single group was formed by 57 isolates, which each exhibited mrp type 15 and combined fla type 2I. However, fla type 2I was also seen in strains belonging to four other mrp types. Most other mrp types comprised just one or two fla types. In contrast, combined fla type 3G was more heterogeneous with respect to mrp type and contained strains from 13 different mrp types. Most other fla types comprised one or two mrp's. Three C. coli isolates which had an mrp type 6 had a combined fla type 8H, which was also seen in eight C. jejuni isolates. This observation deserves further study and is currently being investigated.
TABLE 4.
Distribution of isolates by fla type and mrp type for C. jejuni and C. coli
| Combined fla type | mrp typea
|
|||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | U | |
| 24E | 9 | 2 | ||||||||||||||||||||||||||||
| 8H | 4 | 2 | 2 | 3∗ | 1 | |||||||||||||||||||||||||
| 25N | 8 | 3 | 1 | 4 | ||||||||||||||||||||||||||
| 14J | 8 | 3 | 1 | |||||||||||||||||||||||||||
| 3G | 1 | 22 | 4 | 2 | 3 | 4 | 2 | 6 | ||||||||||||||||||||||
| 2I | 1 | 2 | 57 | 2 | 1 | |||||||||||||||||||||||||
| 21N | 1 | 11 | 1 | |||||||||||||||||||||||||||
| 18L | 1 | 1 | 32 | 1 | ||||||||||||||||||||||||||
| 28F | 2 | |||||||||||||||||||||||||||||
| 27V | 1 | 1 | ||||||||||||||||||||||||||||
| 20S | 5 | 1 | ||||||||||||||||||||||||||||
| 5P | 3 | 2 | 7 | 1 | ||||||||||||||||||||||||||
| 10D | 18 | 1 | ||||||||||||||||||||||||||||
| 7A | 5 | |||||||||||||||||||||||||||||
| 4E | 2 | 1 | ||||||||||||||||||||||||||||
| 29Q | 3 | |||||||||||||||||||||||||||||
| 29S | 3 | |||||||||||||||||||||||||||||
| 33W | 2∗ | 3∗ | ||||||||||||||||||||||||||||
| 10I | 2 | 2 | ||||||||||||||||||||||||||||
| 35A | 3 | |||||||||||||||||||||||||||||
| 31N | 3 | |||||||||||||||||||||||||||||
| 21U | 2 | |||||||||||||||||||||||||||||
| 11C | 1 | 1 | ||||||||||||||||||||||||||||
| 13A | 5 | 1 | ||||||||||||||||||||||||||||
| 6J | 2 | |||||||||||||||||||||||||||||
| 30S | 2 | |||||||||||||||||||||||||||||
| 20Z | 3∗ | |||||||||||||||||||||||||||||
| U | 1 | 1 | 1 | 1 | 1 | 1 | 13† | |||||||||||||||||||||||
∗, C. coli isolates; †, 2 of 13 isolates that had both a unique combined fla type and a PFGE type were C. coli isolates.
By using a combination of mrp type and fla type, 12 distinct clusters of five or more isolates could be seen. Their composition and distribution among the different hosts is shown in Table 5. Isolates within clusters 7, 9, and 10 were found in all of the hosts. The isolates in cluster 3 came from the clinical set only and included strains isolates from 1992, 1993, and 1995. The isolates in cluster 4 came from the cattle set only and contained isolates from three of the four farms during 1993 and 1994. The isolates in cluster 8 came from the turkey set only and included isolates all from the same farm. Strains isolated from this farm formed several other clusters. These included cluster 6, which contained 31 turkey isolates, and cluster 11, which contained 6 turkey isolates; 8 of the 12 cattle isolates; and 4 clinical isolates from 1994, 1995, and 1996.
TABLE 5.
Main genotypic subgroups and their distribution among the different hosts
| Cluster | No. of isolates | mrp type | Combined fla type | No. of isolates
|
|||
|---|---|---|---|---|---|---|---|
| Clinical | Cattle | Turkey | Sheep | ||||
| 1 | 5 | 27 | 20S | 3 | 2 | ||
| 2 | 5 | 26 | 7A | 2 | 3 | ||
| 3 | 5 | 23 | 13A | 5 | |||
| 4 | 7 | 21 | 5P | 7 | |||
| 5 | 18 | 20 | 10D | 1 | 17 | ||
| 6 | 32 | 18 | 18L | 1 | 31 | ||
| 7 | 57 | 15 | 2I | 3 | 28 | 1 | 25 |
| 8 | 11 | 12 | 21N | 11 | |||
| 9 | 8 | 9 | 25N | 4 | 1 | 1 | 2 |
| 10 | 8 | 7 | 14J | 2 | 1 | 3 | 2 |
| 11 | 22 | 4 | 3G | 4 | 12 | 6 | |
| 12 | 9 | 1 | 24E | 1 | 4 | 4 | |
DISCUSSION
A seemingly simple question to ask regarding the Campylobacter isolates examined in this study was “Are these animal campylobacters the same as those causing disease in the community?” Unfortunately, unlike the question, the answer is somewhat more complex, since the choice of typing method, the analysis and interpretation of the results, and a consideration of the biology of the organism are all factors that have to be taken into account. In the absence of a definitive typing scheme for Campylobacter spp., two genotypic typing methods, PFGE and PCR-RFLP analysis of flaA gene sequences, were chosen to help us answer this question; both methods have proved useful in other epidemiological investigations of Campylobacter (22, 27, 30).
Several studies have examined polymorphisms within the flagellar genes of both C. jejuni and C. coli (29, 30). The discriminatory power of flagellin gene typing is influenced by the restriction endonuclease used. Other workers have used a variety of enzymes including HinfI (26), AluI (4, 6), DdeI (22, 29), and EcoRI/PstI (1, 30). In the present study DdeI was used since it appeared to be more discriminatory than HinfI (29), and EcoRI/PstI was used rather than AluI because the latter can produce complex restriction profiles (6). The results showed that DdeI digestion was more discriminatory than EcoRI/PstI with 35 different digest profiles produced by DdeI compared to 26 produced by EcoRI/PstI. The number of fla types increased to 48 when the profiles for both sets of restriction endonucleases were combined.
Ayling et al. (3) developed a PCR-RFLP technique based on the flaA and flaB genes. It is unclear whether the inclusion of the flaB sequence, as well as flaA, would significantly increase the overall discriminatory power of this method (40). Since the flaA and flaB genes have been shown to have a high degree of DNA sequence homology (1), DdeI and EcoRI/PstI restriction analysis of flaB is unlikely to produce significantly more polymorphisms. Analysis of RFLPs within flaA gene sequences alone appears to give sufficient discrimination for its use as a subtyping method for C. jejuni and C. coli.
An overlap of flagellin gene polymorphisms between human and animal isolates of C. jejuni and C. coli has been shown (6, 29, 30), but these studies included only a few strains isolated from animal hosts. In the present study 238 Campylobacter isolates from animals in the farm environment were examined. Of the 34 combined fla types seen in the human set, 15 were found in at least one animal host and 6 were found in each of the hosts.
Of the two methods for typing C. jejuni and C. coli used here, PFGE was more discriminatory than fla typing. Seventy-three different SmaI restriction profiles were found compared to 46 combined fla types. Similar findings have been reported elsewhere for C. jejuni (29) and C. coli (30).
A number of recent studies report the identification of genetically indistinguishable sporadic isolates from human and animal hosts (13, 25, 27). In our study, when the results of the two genotyping methods were combined several clusters of isolates of C. jejuni and C. coli could be identified (Table 5). The largest of these, cluster 7, contained 57 isolates of C. jejuni and was found on all the farms and in each of the hosts throughout the 3-year period of sampling, suggesting that this C. jejuni strain is capable of colonizing a broad host range. In contrast, a number of genotypes were unique to each of the farm animals, suggesting a commensal host-adapted type of C. jejuni strains. The large number of unique human genotypes seen is indicative of the sporadic nature of Campylobacter infections and probably reflects the variety of different sources of Campylobacter.
Two isolates, one from cluster 7 and another from cluster 9, were isolated from patients who became ill after returning from Goa and Egypt, respectively, suggesting that these genotypes, which are found in farm animals and humans in the Lancaster area, have a global distribution.
An analysis of the genotypic data provides some clues to the epidemiology of Campylobacter on the farm. For example, clusters 6 and 8 contain isolates from an investigation of the colonization of turkeys with Campylobacter spp. (37). Newly hatched chicks were free of Campylobacter, but after day 6 the bacteria appeared in the flock and spread rapidly. For the next 5 weeks, only one Campylobacter genotype (mrp type 18 and fla type 18L) was found, accounting for 29 of the 31 isolates seen in cluster 6. Interestingly, this genotype was not detected in any other Campylobacter strains on the farm. After day 49, turkeys from another brooder shed were mixed in with the original flock. By day 54, the dominant genotype of mrp type 18 and combined fla type 18L (cluster 6) had nearly disappeared, being found in only one isolate, and had been replaced with a variety of new genotypes, although with no one genotype particularly dominant. By day 75 a new genotype, with mrp type 12 and combined fla type 21N (cluster 8) was predominant and remained so until the turkeys were sold after day 90. Two litter samples from its floor and three drinking water samples obtained from the waterers also had this genotype, but it was seen in the water only after the turkeys had become infected, so it is probable that the contamination came from the turkeys. The change in Campylobacter genotype was paralleled at the phenotypic level with a change in Preston biotype from 6112 to 6010. These results provide molecular evidence for the horizontal transmission of Campylobacter in turkeys.
Cluster 5 contained 17 isolates from sheep feces. These all came from the same farm in early spring 1995 and included isolates from ewes and their lambs, suggesting that the campylobacters are passed from ewe to lamb (17).
Five turkey isolates from 1994 and 1995 and 9 of the 12 cattle isolates from 1994 in cluster 11 and four turkey isolates and one cattle isolate in cluster 12 all came from the same farm. This indicates either that there is cross-contamination between the turkeys and cattle or that the campylobacters come from a common source. The turkey litter was often put in the calf sheds as bedding, and this is a likely vehicle for the transfer of campylobacters from the turkeys to the calves. The presence of farm animals, including cattle and sheep, other than broiler chickens, was found to be independently associated with an increased risk of Campylobacter infection in broiler flocks in an epidemiological investigation of risk factors in Dutch broiler flocks (35).
Maslow et al. (19) stated that “the utility of a particular characteristic for typing is related to its stability within a strain and its diversity within the species.” The question of instability in the genome of C. jejuni has been raised recently, both at the single loci level, i.e., the flagellin genes (15), and in analysis of the whole of the Campylobacter genome by PFGE (24, 39).
Using cassette-insertion mutant experiments, several investigators report evidence of exchange between the flaA and flaB genes of both C. coli (1) and C. jejuni (38) which have involved either intragenomic recombinational events or natural-transformation-mediated intergenomic recombinational events. Similarly, both mechanisms of recombination were observed by Harrington et al. (15) when they examined C. jejuni fla sequences for mosaic structure. Since reproducibility is one of several important criteria in evaluating the usefulness of a typing system (19), the effect of these findings on the use of RFLP analysis of the flagellin gene as an epidemiological tool deserves careful consideration (15), especially when inferences are formed about clonal ancestry. However, our findings and those of other workers (6, 34) show that some fla types are seen over long periods of time and are stable. This suggests that the susceptibility of campylobacters to genomic rearrangements is strain specific. Although it has been demonstrated that not all C. coli (24) and C. jejuni (14, 39) strains have the same genetic stability, our data in addition to a number of other studies, have identified stable clones of C. jejuni (14, 25, 27) and C. coli (30). Thus, the factors responsible for genetic change and the rate at which it occurs are unclear. Even so, it is important to be aware of the capacity of Campylobacter to undergo such changes, especially when one is interpreting data based on a single locus. A number of recent publications address the need to use different enzymes or additional methods to check the validity of PFGE data based on the use of a single restriction enzyme (12, 25). It is therefore advisable to use more than one method when investigating strain relatedness and to be aware of the possibility of genomic rearrangements when PFGE data are analyzed. Consequently, any link between isolates with indistinguishable SmaI profiles in this study is at this stage only suggestive and will require further investigation to confirm strain relatedness.
The mrps and flaA types from strains in this study can be compared with those published elsewhere. Stanley et al. (30) identified clonal groups of C. coli. Three mrp types and their corresponding fla types were indistinguishable from the PFGE and fla types in our study. For example, the mrp type V, fla type fIV, of Stanley et al., which was found in two sheep isolates from Bristol and in two human isolates from Lancaster University, corresponds to the mrp type Y and the fla type W that were uniquely associated with three C. coli sheep isolates in the present study. mrp type I, fla type fI, which was found in four sheep isolates and one cattle isolate from Lancaster University, two cattle isolates from the Central Veterinary Laboratory in Weybridge (England), 5 sheep isolates from Bristol University, and a human isolate from the Ashford Public Health Laboratory was the same as our mrp type F and EcoRI/PstI fla type H found in three C. coli isolates from the human, sheep, and cattle sets. mrp type II, fla type fIII, which was found in seven sheep isolates from Lancaster University was also seen in one human C. coli isolate in the present study.
The SmaI PFGE restriction types observed in our study are indistinguishable from several PFGE profiles reported by Gibson et al. (11). mrp type 27, which comprises three isolates from sheep, five isolates from cattle, and three isolates from humans, corresponds to the main outbreak type A that was isolated from children who had been on a school visit to a farm in the Cardiff area. The consumption of raw milk was suspected as the source of infection (9).
It is of interest that the most common mrp type, i.e., type 15, in our study, which was seen in 62 isolates, was also the most common PFGE profile isolated from HS4 strains in a study investigating subtypes of C. jejuni from sporadic cases of diarrheal disease from three locations in the United Kingdom, one of which was Lancaster (28). Ten of the isolates with this mrp type in our study were serotyped, and all were determined to be Penner serotype O4 complex. It would seem therefore that this represents a stable clone of C. jejuni which has been isolated from a diverse host range and diverse sources. More recently, Wareing et al. (36) identified a C. jejuni strain (Penner serotype HS4 complex, Preston phage group 55) that has been frequently associated with human gastroenteritis in the United Kingdom over a 7-year period. The organism had also been isolated in Canada, Australia, and New Zealand and has been shown to be the cause of three milkborne outbreaks. The SmaI PFGE profile of this strain is indistinguisable from mrp type 15 in our study in Lancaster. It would certainly be of interest to see whether our Lancaster strain was Preston phage group 55 and to investigate further the pathogenic capability of this strain.
The amount of molecular information regarding PCR-RFLP analysis of flagellin genes and PFGE profiles for strains of C. jejuni and C. coli seems to be increasing rapidly, as reflected by the present study. In order to make sense of the increasing amount of data produced, there is a need for an internationally recognized standardization of methods and analysis of data so that results can be compared between different laboratories. In Europe, a consortium called CAMPYNET has recently been established (40) in order to address such issues. Public health laboratories in the United States at The Centers for Disease Control and Prevention, the Food and Drug Administration, the U.S. Department of Agriculture, and state health departments share a database called PulseNet that contains information on genetic fingerprints of bacteria involved in foodborne outbreaks (33). The establishment of a similar internationally available computer network for Campylobacter and other important pathogenic bacteria would greatly assist our understanding of their epidemiology and, more specifically, enable us to arrive at meaningful and relevant epidemiological conclusions.
The combined use of polymorphisms within flaA gene sequences, which is a simple, quick, and reproducible method, and the highly discriminatory PFGE provides us with information that allows us to monitor the fate of C. jejuni and C. coli strains through the environment. The stability of the SmaI restriction sites and the flaA gene sequences was evident from the results, since many genotypes were seen continuously over the 3-year period. None of the four farms or the three sheep sampling sites was uniquely associated with a particular Campylobacter genotype. Many were seen at a variety of the sampling sites over the 3-year period, indicating that Campylobacter strains are in the farm environment consistently, and the genotypes of C. jejuni and C. coli in it are very diverse. The contribution of farm animals and the role of the environment in the transmission of Campylobacter may be more significant than is currently thought. The isolation of some of these genotypes from sporadic human cases in the community provides molecular evidence linking disease in humans with campylobacters in the farm environment.
ACKNOWLEDGMENTS
We are grateful to the Department of Health Project 221 for funding this work.
We are greatly indebted to Joanne Wallace and several undergraduate project students for providing Campylobacter strains from a variety of farm environments and to David Telford, Consultant Microbiologist at the Lancaster Royal Infirmary, for the provision of clinical isolates.
REFERENCES
- 1.Alm R A, Guerry P, Trust T J. Distribution and polymorphism of the flagellin genes from isolates of Campylobacter jejuni. J Bacteriol. 1993;175:3051–3057. doi: 10.1128/jb.175.10.3051-3057.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arimi S M, Fricker C R, Park R W A. Occurrence of thermophilic campylobacters in sewage and their removal by treatment processes. Epidemiol Infect. 1988;101:279–286. doi: 10.1017/s0950268800054194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayling R D, Woodward M J, Evans S, Newell D G. Restriction fragment length polymorphism of polymerase chain reaction products applied to the differentiation of poultry campylobacters for epidemiological investigations. Res Vet Sci. 1996;60:168–172. doi: 10.1016/s0034-5288(96)90013-2. [DOI] [PubMed] [Google Scholar]
- 4.Birkenhead D, Hawkey P M, Heritage J, Gascoyne-Binzi D M, Kite P. PCR for the detection and typing of campylobacters. Lett Appl Microbiol. 1993;17:235–237. doi: 10.1111/j.1472-765x.1993.tb01455.x. [DOI] [PubMed] [Google Scholar]
- 5.Bolton F J, Wareing D R A, Skirrow M B, Hutchinson D N. Identification and biotyping of campylobacters. In: Board R G, Jones D, Skinner F A, editors. Identification methods in applied and environmental microbiology. SAB Technical Series 29. London, England: Academic Press, Ltd.; 1992. pp. 151–161. [Google Scholar]
- 6.Burnens A P, Wagner J, Lior H, Nicolet J, Frey J. Restriction fragment length polymorphisms among the flagellar genes of the Lior heat-labile serogroup reference strains and field strains of Campylobacter jejuni and C. coli. Epidemiol Infect. 1995;114:423–431. doi: 10.1017/s0950268800052134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Communicable Disease Surveillance Centre. Surveillance of Campylobacter infection in England and Wales. CDSC Communicable Dis Rep. 1999;9:194. [Google Scholar]
- 8.Dilworth C R, Lior H, Belliveau M A. Campylobacter enteritis acquired from cattle. Can J Public Health. 1988;79:60–62. [PubMed] [Google Scholar]
- 9.Evans M R, Roberts R J, Ribeiro C D, Gardner D, Kembrey D. A milk-borne Campylobacter outbreak following an educational farm visit. Epidemiol Infect. 1996;117:457–462. doi: 10.1017/s0950268800059112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia M M, Lior H, Stewart R B, Ruckerbauer G M, Trudel J R R, Skljarevski A. Isolation, characterisation and serotyping of Campylobacter jejuni and Campylobacter coli from slaughter cattle. Appl Environ Microbiol. 1985;49:667–672. doi: 10.1128/aem.49.3.667-672.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibson J, Fitzgerald C, Owen R J. Genomic fingerprinting of Campylobacter jejuni serogroup HS2 by pulsed field gel electrophoresis and comparison with phagetyping and rRNA gene profiling. Epidemiol Infect. 1995;115:215–225. doi: 10.1017/s0950268800058349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibson J, Lorenz E, Owen R J. Lineages within Campylobacter jejuni defined by numerical analysis of pulsed field gel electrophoresis DNA profiles. J Med Microbiol. 1997;46:157–163. doi: 10.1099/00222615-46-2-157. [DOI] [PubMed] [Google Scholar]
- 13.Hanninen M L, Pajarre S, Klossner M J, Rautelin H. Typing of human Campylobacter jejuni isolates in Finland by pulsed-field gel electrophoresis. J Clin Microbiol. 1998;36:1787–1789. doi: 10.1128/jcm.36.6.1787-1789.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanninen M L, Hakkinen M, Rautelin H. Stability of related human and chicken Campylobacter jejuni genotypes after passage through chick intestine studied by pulsed-field gel electrophoresis. Appl Environ Microbiol. 1999;65:2272–2275. doi: 10.1128/aem.65.5.2272-2275.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrington C S, Thomson-Carter F M, Carter P E. Evidence for recombination in the flagellin locus of Campylobacter jejuni: implications for the flagellin gene typing scheme. J Clin Microbiol. 1997;35:2386–2392. doi: 10.1128/jcm.35.9.2386-2392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones K, Betaieb M, Telford D R. Correlation between Environmental monitoring of thermophilic campylobacters in sewage effluent and the incidence of Campylobacter infection in the community. J Appl Bacteriol. 1990;69:235–240. doi: 10.1111/j.1365-2672.1990.tb01514.x. [DOI] [PubMed] [Google Scholar]
- 17.Jones K, Howard S, Wallace J S. Intermittent shedding of thermophilic campylobacters by sheep at pasture. J Appl Microbiol. 1999;86:531–536. doi: 10.1046/j.1365-2672.1999.00702.x. [DOI] [PubMed] [Google Scholar]
- 18.Ketley J M. Pathogenesis of enteric infection by Campylobacter. Microbiology. 1997;143:5–21. doi: 10.1099/00221287-143-1-5. [DOI] [PubMed] [Google Scholar]
- 19.Maslow J N, Mulligan M E, Arbeit R D. Molecular epidemiology: application of contemporary techniques to the typing of microorganisms. J Clin Infect Dis. 1993;17:153–164. doi: 10.1093/clinids/17.2.153. [DOI] [PubMed] [Google Scholar]
- 20.Mead P S, Slutsker L, Dietz V, McCaig L F, Bresee J S, Shapiro C, Griffin P M, Tauxe R V. Food-related illness and death in the United states. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meanger J D, Marshall R B. Seasonal prevalence of thermophilic Campylobacter infections in dairy cattle and a study of infection of sheep. N Z Vet J. 1988;37:18–20. doi: 10.1080/00480169.1989.35540. [DOI] [PubMed] [Google Scholar]
- 22.Nachamkin I, Ung H, Patton C M. Analysis of HL and O serotypes of Campylobacter strains by the flagellin gene typing system. J Clin Microbiol. 1996;34:277–281. doi: 10.1128/jcm.34.2.277-281.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nielsen E M, Engberg J, Madsen M. Distribution of serotypes of Campylobacter jejuni and C. coli from Danish patients, poultry, cattle and swine. FEMS Immunol Med Microbiol. 1997;19:47–56. doi: 10.1111/j.1574-695X.1997.tb01071.x. [DOI] [PubMed] [Google Scholar]
- 24.On S W L. In vitro genotypic variation of Campylobacter coli documented by pulsed field gel electrophoresis DNA profiling: implications for epidemiological studies. FEMS Microbiol Lett. 1998;165:341–346. doi: 10.1111/j.1574-6968.1998.tb13167.x. [DOI] [PubMed] [Google Scholar]
- 25.On S L W, Nielsen E M, Engberg J, Madsen M. Validity of SmaI-defined genotypes of Campylobacter jejuni examined by SalI, KpnI, and BamHI polymorphisms: evidence of identical clones infecting humans, poultry and cattle. 1998. Epidemiol Infect. 1998;120:232–237. doi: 10.1017/s0950268898008668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Owen R J, Fitzgerald C, Sutherland K, Borman P. Flagellin gene polymorphism analysis of Campylobacter jejuni infecting man and other hosts and comparison with biotyping and somatic antigen serotyping. Epidemiol Infect. 1994;113:221–234. doi: 10.1017/s0950268800051657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Owen R J, Sutherland K, Fitzgerald C, Gibson J, Borman P, Stanley J. Molecular subtyping scheme for serotypes HS1 and HS4 of Campylobacter jejuni. J Clin Microbiol. 1995;33:872–877. doi: 10.1128/jcm.33.4.872-877.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Owen R J, Slater E, Telford D, Donovan T, Barnham M. Subtypes of Campylobacter jejuni from sporadic cases of diarrhoeal disease at different locations in England are highly diverse. Eur J Epidemiol. 1997;13:837–840. doi: 10.1023/a:1007497005152. [DOI] [PubMed] [Google Scholar]
- 29.Santesteban E, Gibson J, Owen R J. Flagellin gene profiling of Campylobacter jejuni heat-stable serotype 1 and 4 complex. Res Microbiol. 1996;147:641–649. doi: 10.1016/0923-2508(96)84021-6. [DOI] [PubMed] [Google Scholar]
- 30.Stanley J, Linton D, Sutherland K, Jones C, Owen R J. High-resolution genotyping of Campylobacter coli identifies clones of epidemiologic and evolutionary significance. J Infect Dis. 1995;172:1130–1134. doi: 10.1093/infdis/172.4.1130. [DOI] [PubMed] [Google Scholar]
- 31.Stanley K N, Wallace J S, Currie J E, Diggle P J, Jones K. Seasonal variation of thermophilic campylobacters in lambs at slaughter. J Appl Microbiol. 1998;84:1111–1116. doi: 10.1046/j.1365-2672.1998.00450.x. [DOI] [PubMed] [Google Scholar]
- 32.Stanley K N, Wallace J S, Currie J, Diggle P J, Jones K. The seasonal variation of thermophilic campylobacters in beef cattle, dairy cows and calves. J Appl Microbiol. 1998;85:472–480. doi: 10.1046/j.1365-2672.1998.853511.x. [DOI] [PubMed] [Google Scholar]
- 33.Swaminathan B, Barrett T J . the CDC Pulsenet Task Force. A national molecular subtyping network for food-borne bacterial disease surveillance in the United States. In: Nachamkin I, Blaser M J, editors. Campylobacter. 2nd ed. Washington, D.C.: American Society for Microbiology; 2000. pp. 529–535. [Google Scholar]
- 34.Thomas L M, Long K A, Good R T, Panaccio M, Widders P R. Genotypic diversity among Campylobacter jejuni isolates in a commercial broiler flock. Appl Environ Microbiol. 1997;63:1874–1877. doi: 10.1128/aem.63.5.1874-1877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van de Giessen A W, Bloemberg B P M, Ritmeester W S, Tilburg J J H C. Epidemiological study on risk factors and risk reducing measures for Campylobacter infections in Dutch broiler flocks. Epidemiol Infect. 1996;117:245–250. doi: 10.1017/s0950268800001412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wareing D, Bolton F J, Fox A J, Wright P A. Abstracts of the 10th International workshop on Campylobacter, Helicobacter and related organisms. 1999. The annual incidence and seasonal distribution of a Campylobacter jejuni strain associated with human infections. A proposal for a bovine reservoir and potential sources of transmission. [Google Scholar]
- 37.Wallace J S, Stanley K N, Jones K. The colonisation of turkeys by thermophilic campylobacters. J Appl Microbiol. 1998;85:224–230. doi: 10.1046/j.1365-2672.1998.00470.x. [DOI] [PubMed] [Google Scholar]
- 38.Wassenaar T M, Fry B N, van der Zeijst B A M. Variation of the flagellin gene locus of Campylobacter jejuni by recombination and horizontal gene transfer. Microbiology. 1995;141:95–101. doi: 10.1099/00221287-141-1-95. [DOI] [PubMed] [Google Scholar]
- 39.Wassenaar T M, Geilhausen B, Newell D G. Evidence of genomic instability in Campylobacter jejuni isolated from poultry. Appl Environ Microbiol. 1998;64:1816–1821. doi: 10.1128/aem.64.5.1816-1821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wassenaar T M, Newell D G. Genotyping of Campylobacter spp. Appl Environ Microbiol. 1999;66:1–9. doi: 10.1128/aem.66.1.1-9.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]