Abstract
Most cancer cases occur in low- and middle-income countries (LMICs). The sophisticated technical and human infrastructure needed for optimal diagnosis, treatment, and monitoring of cancers is difficult enough in affluent countries; it is especially challenging in LMICs. In Western, educated, industrial, rich, democratic countries, there is a growing emphasis on and success with precision medicine, whereby targeted therapy is directed at cancers based on the specific genetic lesions in the cancer. Can such precision approaches be delivered in LMICs? We offer some examples of novel partnerships and creative solutions that suggest that precision medicine may be possible in LMICs given heavy doses of will, creativity, and persistence and a little luck.
Keywords: precision medicine, LMICs, diagnostics
INTRODUCTION
The biggest determinant of cancer outcome is one’s place of birth. While cancer causes 20% of all deaths worldwide, approximately 70% of cancer deaths occur in low-income countries (1). The top five most common cancers worldwide are lung, breast, colorectal, prostate, and stomach. All require standard pathohistological diagnoses, and management decisions require additional immunohistochemistry (IHC) [e.g., estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2)] as well as molecular testing (e.g., epidermal growth factor receptor mutation testing), according to the World Health Organization (WHO) list of essential diagnostic tests (1, 2). Low- and middle-income countries (LMICs) suffer the heaviest burden of cancer deaths, and much of this is attributed to the lack of pathology services, which results in delayed and sometimes incomplete or inappropriate diagnoses. This can lead to progression of disease with worsening prognosis and incorrect treatment decisions, ultimately contributing to higher morbidity and mortality rates (3). A systematic review showed that earlier diagnosis is associated with earlier-stage disease and improved survival benefits for breast, colorectal, head and neck, and prostate cancers and melanoma (4). In 2017, only one-quarter of low-income countries reported having readily available access to pathology services (2, 5). In response, WHO published a guide to early cancer diagnosis emphasizing its importance not only in ensuring access to care but also for evaluating the disease and providing subsequent treatment (6). Gross disparities in cancer deaths in LMICs are partially attributed to these diagnostic barriers but can also be attributed to environmental factors, infectious agents, occupational exposures, and lifestyle behaviors. As a result, molecular variation based on environmental exposures, genetic factors, and regional habits has been hypothesized to contribute to carcinogenesis, and this also warrants further study (7).
The study of cancer genomes has identified scores of potentially targetable proteins and pathways activated by gene mutations, methylation changes, and gene expression deregulation. The concept of precision medicine is to rapidly study each patient’s cancer for their private genetic landscape and find agents active against that particular cancer. This relies on sophisticated and expensive diagnostics and agents. Is there a way to bring modern, genetics-based cancer care to the entire world? In this review, we discuss some of the solutions we have adopted that focus on new approaches and technologies that may close rather than widen the cancer care gap. We especially concentrate on chronic myeloid leukemia (CML), since it was the first cancer to have a clear genetic target with a therapeutic solution and is an example of bringing both diagnostics and targeted agents to LMICs.
CHRONIC MYELOID LEUKEMIA AS A MODEL FOR DELIVERING PRECISION MEDICINE TO LOW-INCOME AREAS
CML is the poster child of the bench-to-bedside paradigm (sadly found much more often in grants and papers than in real life). The t(9;22) reciprocal translocation is the basis of pathology, diagnosis, and monitoring in CML. Diagnosis and monitoring can be performed by metaphase cytogenetics (the shortened chromosome 22 is the Philadelphia chromosome) or, much more sensitively, by reverse transcription polymerase chain reaction (RT-PCR) of the BCR-ABL1 transcript. All patients with CML have the BCR-ABL1 fusion gene, and thus every CML patient has a recognizable molecular target for diagnosis and monitoring, as well as a protein target for inhibition. The use of tyrosine kinase inhibitors (TKIs) in CML has made a revolutionary change in the natural history of the disease, turning the natural history from an average life span of approximately 7 years to an expectation of a near-normal life span (8).
Quantitative RT-PCR of BCR-ABL1 mRNA is the most sensitive method for monitoring CML (8) and is a fast, accurate, and clinically useful measure of disease burden. BCR-ABL1 levels during therapy offer robust signposts of outcomes. Achievement of a major molecular response, which is a three-log reduction on the standardized international scale (IS) (BCR-ABL1 ≤ 0.1% IS), reflects a safe harbor of disease reduction associated with low rates of future resistance or progression (9, 10). Deeper molecular responses identify patients who may successfully discontinue TKI therapy (11, 12). Resistance can occur and may be either primary (poor initial response to therapy) or secondary (acquired resistance after initial response to TKI therapy). In 25–50% of cases, secondary resistance occurs from point mutations in the ABL1 tyrosine kinase domain that affect TKI binding and activity (13). Upon resistance, changing therapy to another TKI can produce a cytogenetic remission about 50% of the time (14). However, once resistance occurs, further outgrowths of resistance often occur, often with a new mutation. Remarkably, approximately half of CML patients with a sustained deep molecular remission can successfully discontinue TKI therapy without subsequent relapse (15).
In Western, educated, industrial, rich, democratic (WEIRD) countries, the most pressing clinical questions in CML management are when to change therapy and when to end therapy. The major obstacles in LMICs are simply diagnosing CML and finding available and affordable treatment. The obstacles are myriad and formidable: low income, poor access to medical care, questionable travel infrastructure, and lack of availability of diagnostic testing and TKIs.
This dilemma is shared across all cancers in LMICs, but the experience in CML demonstrates a model of delivering precision medicine in a very challenging setting. There is a concrete example of how a team of divergent partners—a nonprofit nongovernmental organization, academia, pharma, and biotech—can cooperate and make the seemingly impossible possible.
The core of this effort began with the partnership of Novartis, the maker of imatinib, and The Max Foundation, a nonprofit originally serving as an information and support center for international CML patients. The arrangement was for The Max Foundation to find physicians and patients who needed imatinib provided by Novartis through the Glivec International Patient Assistance Program. The Max Foundation had to find and diagnose CML cases, whereby Novartis would provide free drug for life or until the drug became available through local authorities (16). It quickly became clear that making the diagnosis was not going to be easy. The usual tools available to WEIRD country–located doctors to diagnose CML—marrow aspirate, cytogenetics, fluorescent in situ hybridization (FISH), and RT-PCR—require biopsy needles, microscopes, culture equipment, and thermocyclers, not to mention the trained expertise to perform these tasks. How could diagnosis and monitoring get done?
The potential strategies for testing in low-income countries have clear advantages and disadvantages (Table 1). Home-brew assays often use different sets of primers, probes, and standards, often optimized for an individual lab’s needs for test characteristics (e.g., sensitivity, speed, cost). Adapting a home-brew assay in an area of low resources has far more disadvantages than advantages. The main advantage of the home-brew test is that it is often relatively cheap—if you already have all the equipment you need (e.g., thermocycler, refrigerator), not to mention trained technicians (and assuming a reliable electrical grid).
Table 1.
Different approaches to molecular testing in low- and middle-income countries
| Strategy | Pros | Cons |
|---|---|---|
| Home-brew assay | Optimized in expert labs Sensitive and reproducible in the inventor’s hands |
Technical expertise needed Expensive machines and reagents Often performs worse at new location |
| Automated assays | Less technical training needed Self-contained reagents Can be run without batching Fast |
Machines, cartridges, and reagents are expensive to buy and maintain |
| Shipping to specialized centers | Robust and reliable test performance | Shipping can affect sample quality Batching is difficult Expensive |
A huge leap in the potential to diagnose CML (and other cancers and infectious diseases) came with the development of cartridge PCR assays run on the GeneXpert, developed by Cepheid (Sunnyvale, CA) (17). Prepackaged cartridges have the great advantages of less operator handling (less sophisticated training needed) and increased speed and efficiency (so-called plug and play, avoiding the need to batch). Other advantages include avoiding amplimer cross contamination and potential same-day turnaround times (an important feature when patients must travel long distances for their test). The quantitative RT-PCR assay for BCR-ABL was Cepheid’s first oncology test (18) and greatly increased The Max Foundation’s ability to diagnose and monitor CML cases, since WHO had partnered with Cepheid to put over 25,000 machines across the globe for infectious disease testing (e.g., HIV, tuberculosis). However, while the advantages of using the Cepheid platform are numerous, the system is relatively costly (compared to a home-brew assay) and needs electricity (see the sections titled Breast Cancer and Point-of-Care Diagnostics).
What is the alternative to setting up a home-brew assay in an LMIC’s hospital or clinic? One option is the mail. Shipping a blood sample to a centralized center has the advantage of doing the assay at a dedicated location where an optimized test can routinely be performed. However, again, disadvantages prevail. Even if sent by express air courier with proper packaging, samples degrade (especially a problem for RNA-based tests). Even more problematic is the cost. For example, sending one tube of blood by air from Africa to Seattle, WA, costs about $500! However, we recently developed a new option: dried blood spots (DBS). While DNA is stable on commercially available paper (e.g., Guthrie spots), RNA is not. However, despite degradation, enough RNA persists after weeks of DBS storage that BCR-ABL measurements are accurate (17) (Figure 1). For example, samples can be obtained from patients from over a week of clinic visits and can then be batched and mailed, allowing for a huge savings compared to rushing fresh blood across the seas. Since RNA and DNA can be obtained from the same paper, BCR-ABL1 quantification and ABL mutation detection can be done from the same DBS paper. Recently, we have successfully used DBS to extract DNA for performing next-generation sequencing (NGS) of a panel of genes often mutated in myeloid malignancies, further expanding the potential of DBS in clinical and research work. Our lab has partnered with The Max Foundation and the International CML Foundation to diagnose and monitor more than 800 samples in the Spot On CML project. All told, these DBS have come from 22 countries and have traveled nearly a half a million miles (unfortunately, these cannot be claimed on my mileage program) (https://spotoncml.shinyapps.io/SPOTonCML/).
Figure 1.
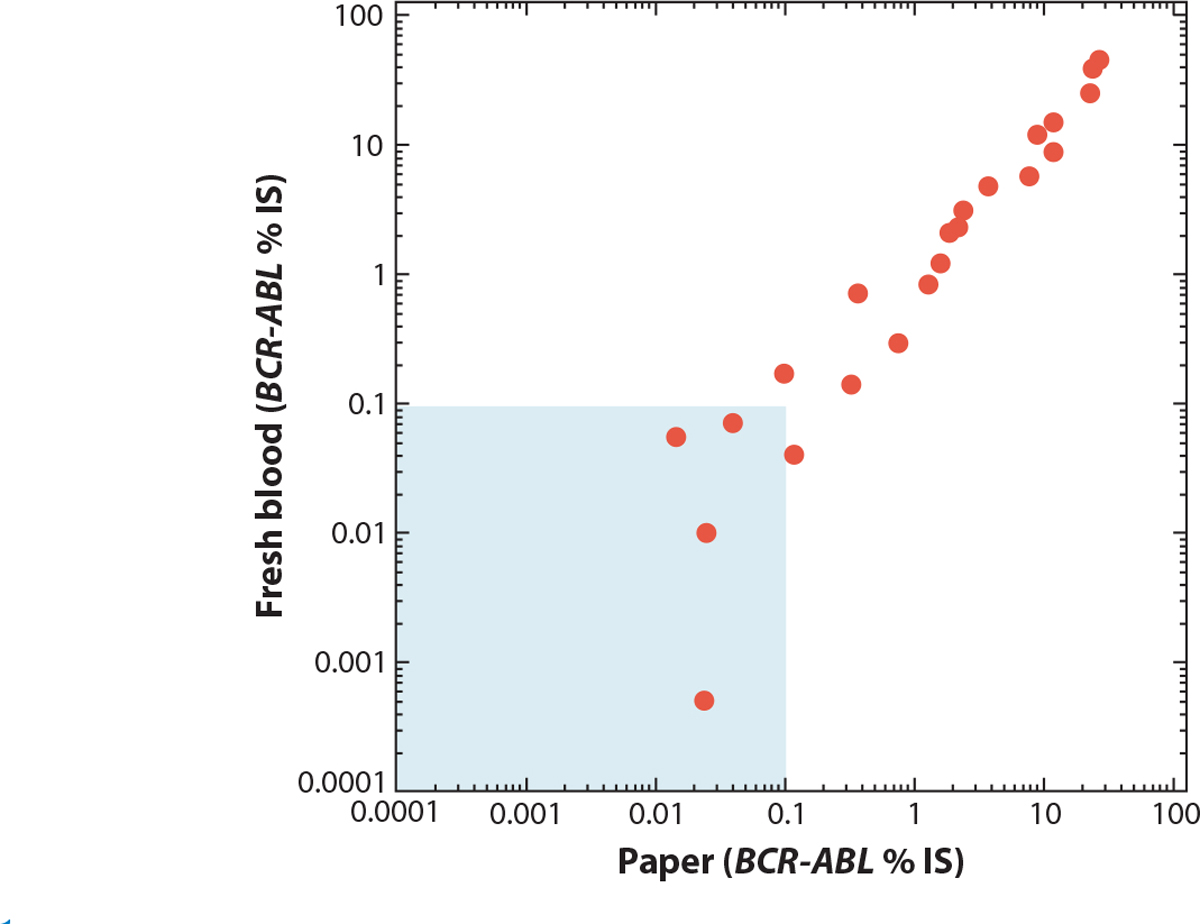
Comparison of BCR-ABL RNA from fresh peripheral blood versus dried blood spots. Samples were taken from patients in Adelaide, South Australia. One aliquot was tested immediately, and the other was spotted onto paper, dried, and shipped by mail to Seattle, WA. The average transit time was over a month. The values are BCR-ABL/ABL percentage on the international scale (% IS). The shaded area is the level at or below a major molecular response. Figure adapted with permission from Reference 17.
As noted above, DNA obtained from DBS can be used for NGS assays. Testing for ABL1 mutations in DBS CML specimens came as a natural extension of our Spot On CML project. Requests for ABL1 mutation detection originated as patients treated with TKIs started to present suboptimal responses or relapsed. While the quality of specimens collected as DBS is good for amplification of the BCR-ABL1 RNA fusion, ABL1 mutation analysis in RNA requires the study of a much bigger gene segment, and the RNA obtained from DBS is too fragmented. DNA is much more stable than RNA and can be extracted from filter paper using commercially available DNA extraction kits. In our lab, we use the QIAamp DNA Blood Mini Kit (Qiagen) and follow its “DNA Purification from Dried Blood Spots” protocol (19, p. 42). Initially, we designed specific primers to amplify ABL1 exons in which clinically relevant mutations had been described, but new mutations have been discovered, and six PCR and Sanger reactions per specimen are now utilized to study exons 4–9. Currently, we screen specimens by NGS with the ArcherDX VariantPlex Myeloid panel, a targeted sequencing assay that includes ABL1 among 75 targeted genes. Archer DX chemistry includes an anchored multiplex PCR (AMP™) that incorporates molecular barcodes and is sequenced on an Illumina sequencer. Although the yield of DNA obtained from DBS is variable, we usually obtain enough to input 100–250 ng into the Archer DX reaction, extracting 2–4 spots (equivalent to 100–200 μL of blood). Another advantage is the detection of additional mutations that can be relevant to the disease.
LYMPHOMA
Lymphoma presents a unique challenge to diagnostics in LMICs. Over 90 subtypes of lymphoma are defined by WHO, and treatment strategies span from simple observation to aggressive chemotherapy. Despite the increasing use of core needle biopsies, excisional or incisional biopsies are still the standard of care, especially in settings where molecular techniques are limited. The tissue is then interrogated with multiple diagnostic techniques, including hematoxylin and eosin stain, IHC, and FISH. All of these are limited or absent in many LMICs. In addition to costly and labor-intensive diagnostic techniques, the subtle differences among lymphoma subtypes often require the expertise of a dedicated hematopathologist for interpretation. Even with ideal resources, discordance among this highly trained group reaches 10% (20, 21). We recently partnered with colleagues at the Instituto de Cancerología y Hospital Dr. Bernardo del Valle S. (INCAN) in Guatemala for a secondary review of all biopsies obtained for a suspicion of lymphoma over a 12-year period (22). Cases were reevaluated with additional stains and assays as required for WHO gold-standard diagnosis. Indeed, following this review, <40% of cases were concordant, and the vast majority of this discordance was due to an incomplete rather than misclassified diagnosis. Of note, INCAN has more recently benefited from an on-site hematopathologist, which is extremely rare.
In many LMICs, an improvement in diagnostics alone would have an immediate impact on patients with lymphoma. Broadly applicable systemic regimens such as cyclophosphamide, vincristine, and prednisone with or without doxorubicin (for diffuse large B cell, follicular, marginal zone, mantle cell, and peripheral T cell lymphoma) and adriamycin, bleomycin, vinblastine, and dacarbazine (for Hodgkin’s lymphoma) are available today in many LMICs and may be adequate treatments for some patients. A definitive diagnosis of early-stage indolent disease may prevent many patients from being overtreated. However, maximizing the benefit of improved diagnostics will inevitably depend on reliable access to appropriate anticancer agents.
With regard to lymphoma, the history of access to the anti-CD20 agent rituximab is an informative example. Rituximab is the first-line therapy in several CD20+ B cell lymphomas as a monotherapy or in combination with radiation for early-stage indolent disease. Additionally, it improves outcomes when added to chemotherapy backbones in several more aggressive or late-stage CD20+ B cell lymphomas (23). Despite the fact that rituximab was first approved in 1997, its incredible efficacy across a range of lymphomas, and its addition to the WHO essential medicine list in 2015, use in LMICs has been extremely limited. Recent patent expiration and the subsequent infusion of several biosimilars into the marketplace raise hope that the identification of CD20+ malignancies, rather than cost, will be the primary barrier to broader rituximab use. However, without innovative solutions to both the difficulty of identifying new targets at diagnosis and the lack of access to affordable drugs, newer agents risk this same decades-long delay before they can deliver appropriate lifesaving treatment to the majority of patients. For example, the CD30-targeting antibody brentuximab has a clear benefit in patients with peripheral T cell lymphoma expressing CD30 in the first-line setting (24). Given the observed overall-survival benefit of brentuximab, only 11 patients would need to be treated to improve overall survival. Yet borders, rather than biology, currently preclude saving the eleventh patient in most places. Similar challenges should be anticipated for other promising therapies including, but not limited to, agents targeting Bruton tyrosine kinase, phosphoinositide 3-kinase, and immune checkpoints. Access to locations able to safely deliver traditional chemotherapy and supportive interventions for treatment complications are also limitations in many LMICs. Therefore, any new agent that generates similar efficacy with reduced toxicity or that can be taken at home will likely have an even greater impact in LMICs.
BREAST CANCER
Breast cancer, the most common cancer among women globally, is characterized by poor survival, with less than half of women diagnosed with the disease alive at 5 years in many LMICs (25–27). While the incidence of breast cancer in LMICs is lower than the incidence in most resource-abundant countries, the mortality rate from breast cancer in LMICs is considerably higher (28). Indeed, the estimated 5-year overall survival from breast cancer in certain countries is less than half of what it is in resource-abundant countries. For example, the 5-year survival for breast cancer in Uganda stands at 46%, in contrast to resource-rich countries, where survival is upwards of 90% (27). Similar disparities are observed when comparing breast cancer survival by a country’s human development index (29). A variety of factors are responsible for this survival disparity among women with breast cancer in LMICs compared with resource-rich regions and include a poor health care infrastructure that leads to late stage at presentation. However, inadequate methods to diagnose and effectively treat the disease remain burdensome (30). The ability to more accurately diagnose and implement breast cancer treatment strategies, via precision medicine, can help bridge this gap.
The Breast Health Global Initiative and the National Comprehensive Care Network have previously published guidelines for the diagnosis of breast cancer in LMICs (31–34). In both resource-abundant and resource-limited regions, the histologic examination of tumor tissue and the description of hormone receptor status (i.e., ER, PR, and potentially the expression of HER2 via IHC) are recommended. An accurate determination of hormone receptor status is essential to guide therapeutic decision making and provides both prognostic data with regard to survival and predictive data with regard to response to therapy. Notably, the selective ER modulator tamoxifen, the aromatase inhibitor anastrozole, and the HER2-targeted agent trastuzumab are listed on the WHO Model List of Essential Medicines (35). Indeed, given the widespread availability of tamoxifen, it is often given empirically to patients with breast cancer. However, the optimal use of these medications requires the determination of receptor status—data often currently unavailable in many LMICs. Even when available, IHC results have often been limited by poor specimen quality attributable to tissue degradation and improper storage (36, 37). Novel diagnostic options are necessary.
Although WHO has determined that the pathologic evaluation of tumor tissue is an essential laboratory service globally, obstacles to testing (and the ability to conduct precision medicine) include the relatively high cost, lack of essential equipment and necessary reagents, and limited number of trained personnel (37, 38). Therefore, diagnostic modalities that can overcome these limitations are necessary. Indeed, WHO specifies that diagnostic tests, in resource-limited regions, be characterized by the following criteria: affordable, sensitive, specific, user-friendly, rapid, equipment-free (no large, electricity-dependent machinery), and delivered to the target population (forming the acronym ASSURED) (39). Fortunately, diagnostic options are currently available that meet some of these criteria (the most difficult to achieve is obviously the equipment-free criterion; see the section titled Point-of-Care Diagnostics).
The use of diagnostic platforms that currently exist and are available in LMICs, including RT-PCR, which has been favorably compared to IHC and is less prone to error, has yielded promising results in the detection of ER, PR, and HER2 (40–42). Importantly, the testing infrastructure for RT-PCR, which is used to provide quantitative measurements of HIV, currently exists and is widely available in many LMICs. Similarly, the use of a rapid and automated molecular diagnostic test for the detection of Mycobacterium tuberculosis via the Cepheid GeneXpert, with its high sensitivity and specificity when compared to standard testing, has revolutionized the care and treatment of patients with tuberculosis in highly endemic countries (43, 44). This cartridge-based testing combines sample processing and testing (PCR) and can yield results within two hours. Cepheid has recently developed a real-time quantitative PCR (RT-qPCR) assay, STRAT4, which measures ER, PR, HER2, and Ki67 (a marker of cell proliferation) and has yielded promising results in both LMICs and resource-rich countries (45, 46). While much of the initial assessments of the STRAT4 assay utilized formalin-fixed paraffin-embedded (FFPE) tissue from a core-needle biopsy, the use of the assay on fine-needle aspirate specimens is under investigation.
The application of NGS to breast cancer specimens in the United States has led to the identification of many molecular mechanisms for breast cancer and has enabled the delivery of precision medicine. The Cancer Genome Atlas Network analyzed breast cancer specimens via six separate platforms, including whole-exome sequencing and gene expression analyses, and identified known mutations associated with breast cancer as well as novel mutations (47, 48). Such molecular testing has rarely been applied to tumors from LMICs; however, this remains a promising opportunity for future applications. Similarly, the ability to detect cancer-associated mutations in the peripheral blood, via the liquid biopsy, is a promising and minimally invasive diagnostic method utilizing cell-free DNA and circulating tumor DNA (ctDNA). Dawson and colleagues (49) have demonstrated the utility of ctDNA in monitoring the tumor burden of women receiving therapy for metastatic breast cancer. These researchers found that ctDNA was highly sensitive and specific and enabled an early assessment of treatment response (49). These alternative diagnostic methods and methods to monitor treatment response are particularly attractive in LMICs given the limited availability of clinical pathology and radiography in these countries.
TELEPATHOLOGY AND TUMOR BOARDS
Telepathology and digital pathology have revolutionized the field of pathology by enabling whole-slide scanning and diagnoses to occur anywhere in the world. The advent of coronavirus disease 2019 (COVID-19) and resultant relaxing of regulations have demonstrated that accurate diagnoses are feasible in the majority of cases across the United States and the world (50). A study performed at the Butaro Cancer Center of Excellence in Rwanda showed greater than 95% agreement in classification and diagnosis between glass slides and a telepathology system, demonstrating feasibility for such a system to be implemented in a low-resource country (51). Milner & Holladay (52) describe how, in response to a dire shortage of skilled personnel, the American Society for Clinical Pathology (ASCP) deployed automated histopathology, whole-slide imaging, and cloud-based diagnostic systems with the enrollment of 600 pathologists (ASCP members) to assist in reviewing cases covering the spectrum of surgical pathology. Fischer et al. (53) describe establishing a robotic telepathology platform in Botswana and highlight the feasibility of such a system for overcoming particular challenges with internet connectivity, stable power sources, and governmental support personnel with technical expertise.
At the Uganda Cancer Institute weekly leukemia/lymphoma tumor boards, hematology-oncology fellows from the Uganda Cancer Institute present clinically challenging cases for discussion to a multidisciplinary team including oncologists from Uganda, Seattle, and other sites around the world. During these tumor boards, pathologists from Fred Hutchinson Cancer Research Center review slides that are digitally scanned and uploaded through a cloud service for viewing by the international audience during the conference. Additionally, molecular testing results performed at the Radich lab in Seattle are presented and discussed by a molecular pathologist in relation to the clinical case. This multidisciplinary approach is necessary not only for patient care but also for improving education and establishing best standards for the resource-limited region.
Common language is a frequent barrier to such programs in non-English-speaking countries. Colleagues from non-English-speaking countries specifically noted this barrier when discussing past tumor board partnerships at their institution. They were often asked to prepare all cases in English, which resulted in a significantly greater time burden for primary participants and the exclusion of critical local discussants. Multiple potential solutions exist, including language-specific tumor boards, investment in translation services, and support of regional consortiums. An excellent example of the latter approach is the Grupo de Estudio Latinoamericano de Linfoproliferativos, which was formed in 2018 by physicians in South and Central America. While not specifically focused only on clinical case review, this consortium of lymphoma-focused physicians has created a regional network of lymphoma-focused expertise and academic research. Such solutions not only overcome language barriers but also, given the variable geographic distribution of lymphoma subtypes, provide a higher level of expert opinion for regional case management.
NEW METHODOLOGICAL SOLUTIONS
Point-of-Care Diagnostics
Some ongoing areas of work could further expand diagnostic testing for cancers and infectious diseases. As noted above, WHO has introduced the ASSURED standards for developing assays for low-resource areas, and the obvious problem is that much of the world has no or unreliable electricity (i.e., tests must meet the equipment-free criterion in the ASSURED acronym). Fortunately, some bacterial polymerases can amplify nucleic acid targets at a single temperature (an isothermal reaction) rather than needing to cycle through various temperatures, as in a PCR. This isothermal PCR affords considerable creative license to the design of simple assays that require little in the way of training and equipment, perfect for point-of-care (POC) devices (54, 55). Current POC diagnostics include infectious disease tests, pregnancy tests, hematology assays, and glucose monitoring urinalysis. The demand for increased laboratory testing at the bedside or for deployment in the field is increasing, as many patient-care decisions are reliant on laboratory results. POC devices cost less than conventional ones, require little to no operator training, can be built to withstand rough handling, are highly portable, require no additional electricity or refrigeration, and typically do not need costly accessory equipment—all advantages for easier implementation in resource-constrained regions. However, a major point of caution with the design of POC devices is the need for many of these tests to have follow-up confirmatory studies, as the POC assays are not truly infallible.
Studies using loop-mediated isothermal amplification (LAMP) to detect the fusion gene promyelocytic leukemia–retinoic acid receptor alpha (PML-RARA) transcript in acute promyelocytic leukemia reliably demonstrate isothermal nucleic acid analysis feasibility for rapid and simple diagnostics (55). Detection of the BCR-ABL fusion transcript has been performed by LAMP (56) and by a slightly different isothermal method: nucleic acid sequence-based amplification (NASBA) (26). NASBA works by directly amplifying RNA molecules by first utilizing reverse transcriptase in a series of initial steps to create a double-stranded DNA molecule, then utilizing T7 RNA polymerase to continuously amplify complementary RNA transcripts. In contrast, LAMP utilizes specialized primers and the Bst polymerase enzyme, a combination that allows for increased strand displacement and high replication activity, thereby increasing the amount of DNA produced, which can be detected by DNA stains or UV light with recombinase polymerase amplification (RPA), which uses a mix of three enzymes: recombinase, a single-stranded DNA-binding protein, and strand-displacing polymerase. An example of an RPA reaction run on a passive lateral flow device is shown in Figure 2 for an assay identifying the PML-RARA fusion transcript.
Figure 2.
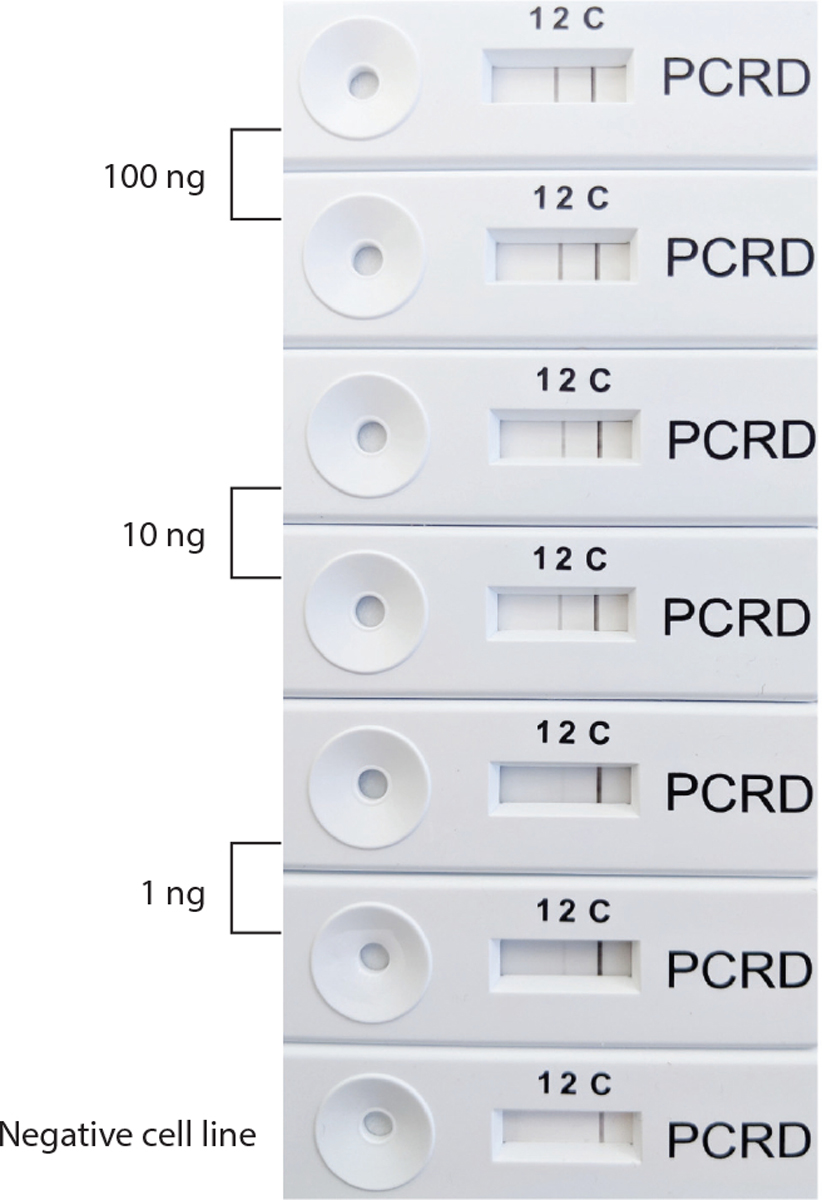
A recombinase polymerase amplification (RPA) reaction for the PML-RARA fusion mRNA found in acute promyelocytic leukemia. The RPA enzyme mix uses three enzymes: recombinase, a single-stranded DNA-binding protein, and strand-displacing polymerase. For the reverse transcription step, Superscript IV was used at 37°C. The reaction is run on a PCRD nucleic acid detection lateral flow device. On the PCRD device, the band at C is the negative control product, while the PML-RARA amplification band is seen under the number 2 label. For the reaction, the NB4 cell line, which contains PML-RARA, was diluted into a negative control cell line mRNA.
Formalin-Fixed Paraffin-Embedded Tissue Use in Diagnostics
Improvements in transcriptional profiling of FFPE tissue have made this a viable possibility for broader diagnostics. Gene expression assays had previously been used for binary distinctions between subtypes of lymphoma; however, these still required prior pathology review, thereby limiting their use in many LMICs (57–61). We recently developed an inexpensive assay in Guatemala using a selective cohort of genes, multiplex PCR, capillary electrophoresis, and a machine learning algorithm to classify biopsies obtained for clinical suspicion of lymphoma. Depending on the stringency of accepted probability, the assay was able to classify cases into aggressive B cell lymphoma, diffuse large B cell lymphoma, Hodgkin’s lymphoma, T cell lymphoma, follicular lymphoma, marginal zone lymphoma, extranodal natural killer/T cell lymphoma, mantle cell lymphoma, or nonmalignant bins with an accuracy of 94% (22). The majority of cases that did not fall into any of these diagnostic bins generated a low-probability call that would give an output of indeterminant. In practice, these relatively infrequent cases could be reflexed for standard pathology review. This proof-of-principle investigation supports inexpensive gene expression assays as viable initial rather than ancillary tests for cancer diagnostics.
Noninvasive Blood Collection
To do diagnostic testing, one needs blood. In many LMICs, the basics of blood drawing (needles and syringes) are in very short supply. A new attractive alternative are minimally invasive blood collection systems. Typically, these have an adherent surface that allows the device to secure to skin (for example, the arm), and with activation, a spring-loaded lancet pierces the skin. Blood is drawn into a chamber, which can contain either a buffer or paper. We are currently studying the feasibility of a self-applied Tasso-M20 blood sampling device (Tasso, Inc., Seattle, WA) for the diagnosis and monitoring of CML. In a feasibility study conducted in our lab, we tested the performance of a previous version of the current Tasso-M20 device, the HemoLink. The HemoLink was a self-appliable blood collection device with precut filter paper in the blood reservoir that, with simple drying instructions, collected DBS. The reservoir contained four filter paper fragments that, if fully soaked, would amount to an estimated 100 μL of blood. Applied on the upper arm, it took approximately 5 min to complete a collection. We tested 20 volunteers and used the Cepheid GeneXpert system to detect only the ABL1 housekeeping gene from an average of 3.2 filter paper DBS per person. ABL1 was detected in 18 out of 20 patients, with one of the detection failures amplifying slightly over the arbitrary cutoff of 16 amplification cycles. The average cycle threshold was 15.5 (range: 13.5–18.1). Encouraged by these results, we are moving to compare BCR-ABL1 test results for CML patients between fresh blood and blood collected with a readily available Tasso-M20 device that collects blood in a volumetrically controlled substrate instead of on filter paper. Pairing the self-applied device with a solid matrix for blood collection can obviate the need for a phlebotomist and extraction rooms and eliminates the monetary and time expenses of patients traveling long distances to the clinic for a blood draw, a frequent hurdle in many countries. Additionally, this system eliminates the cost of rushed sample transportation, as BCR-ABL1 RNA in dried blood is stable over long periods of time and can even be mailed. If successful, applications are myriad, especially in the age of COVID-19, in which potential patients even in WEIRD countries may do best to avoid the clinic.
PCR
Digital PCR (dPCR) is an emerging technology that is driving advances in nucleic acid quantitation and, unlike qPCR, is adaptable for deployment to low-resource settings (62). It works by partitioning nucleic acids in a sample into thousands or even millions of nanoliter compartments containing one or more nucleic acid molecules, a process referred to as sample digitization. After the amplification reaction, any compartment that initially contained at least one target molecule produces a positive signal. The number of compartments with a positive signal over the total number of compartments is used to calculate the absolute concentration of the target nucleic acid. Unlike qPCR, dPCR achieves absolute quantitation without the need for a standard curve. Adaptations of current dPCR systems are required for POC device use in low-resource settings to make sample preparation, partitioning, and analysis simple and automated with devices that are low-cost and portable. We have recently developed a simple, inexpensive self-digitization device that allows sample digitization without the need for precise and complex instrumentation (e.g., valves and flow control) (63–65). Sample digitization is achieved by a combination of centrifugal force, fluidic forces, interfacial tension, channel geometry, and the final stabilization of the partitioned sample in isolated nanoliter chambers containing PCR reagents for target amplification and detection (Figure 3). The self-digitization discs can be made with material similar to conventional CDs and using similar manufacturing processes. The optical disc–style dPCR reader is portable and affordable. In addition to the cost and portability advantages of the self-digitization device and dPCR reader and the many advantages of absolute quantitation (such as better site-to-site comparison of results), dPCR is more tolerant of sample contamination and inhibitors of amplification than qPCR, which relaxes the constraints on the sample preparation step. Importantly, dPCR is much more sensitive than qPCR, allowing it to detect nucleic acid sequences with very low copy numbers. Therefore, dPCR with a self-digitization chip is inherently robust, highly sensitive, and adaptable for nucleic acid detection and quantitation in low-resource settings.
Figure 3.
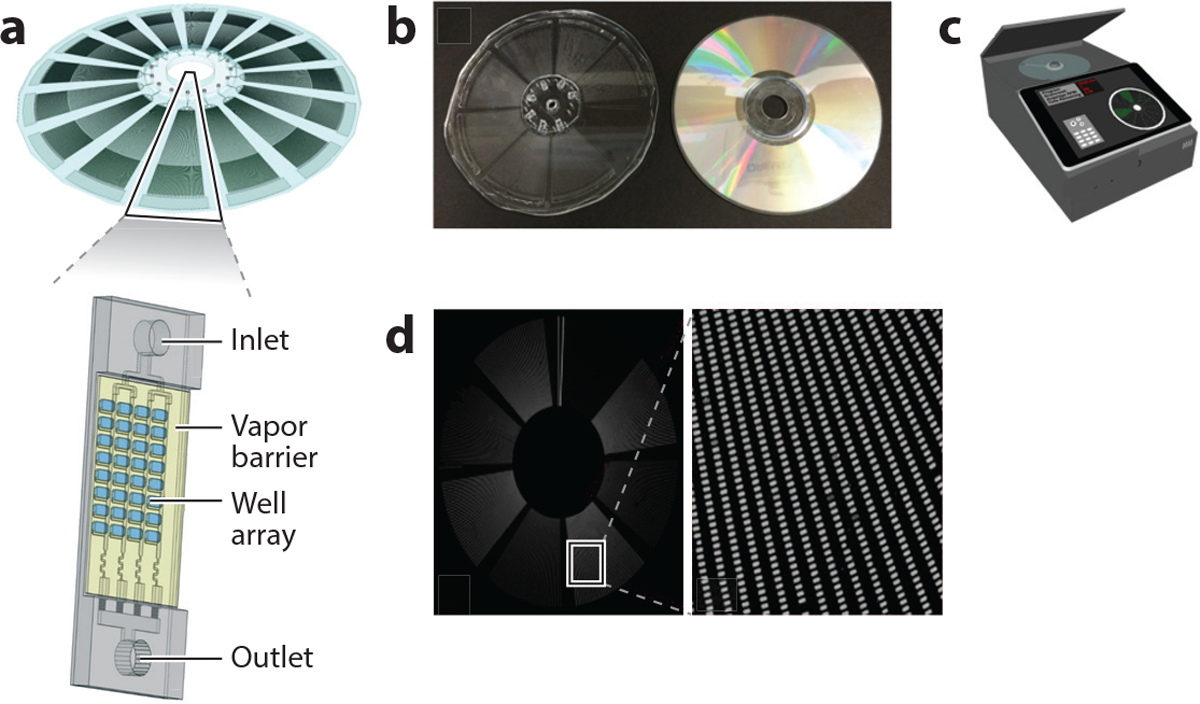
Self-digitization disc and digital polymerase chain reaction (dPCR) reader for nucleic acid detection and quantitation in low-resource settings. (a) Self-digitization disc. (b) The self-digitization disc (left) can be made with material similar to a conventional CD (right) and using a similar process. (c) The optical disc–style dPCR reader is portable and affordable. (d) dPCR works by partitioning nucleic acids in a sample into thousands or even millions of nanoliter compartments containing one or more nucleic acid molecules. After amplification, any compartment that contains at least one of the target molecules will produce a positive signal.
CONCLUSION
We have outlined various strategies for bringing precision medicine to LMICs, with a few examples of applications and progress. The next question is, If and when we can bring precision medicine to LMICs, will it work? Will we see the same types of advances that this treatment strategy brings to WEIRD countries?
The experience from CML says yes. If necessity is the mother of invention, the unique partnership between patients, doctors, The Max Foundation, pharma, biotech, and academia might be the mother of all inventions. The sum of this activity is that this novel alliance has been remarkably successful, with TKIs going to >80,000 cases worldwide. As shown in Figure 4, these patients miraculously have similar survival to CML patients treated in economically advanced nations! As more oral targeted therapies are developed, we will need as a community to quickly invent technical diagnostic solutions so that these drugs can get to the appropriate patients. The experience in CML shows that with imagination and determination, solutions to daunting problems can be overcome. We expect similar successes will be seen in other malignancies as we democratize precision medicine.
Figure 4.
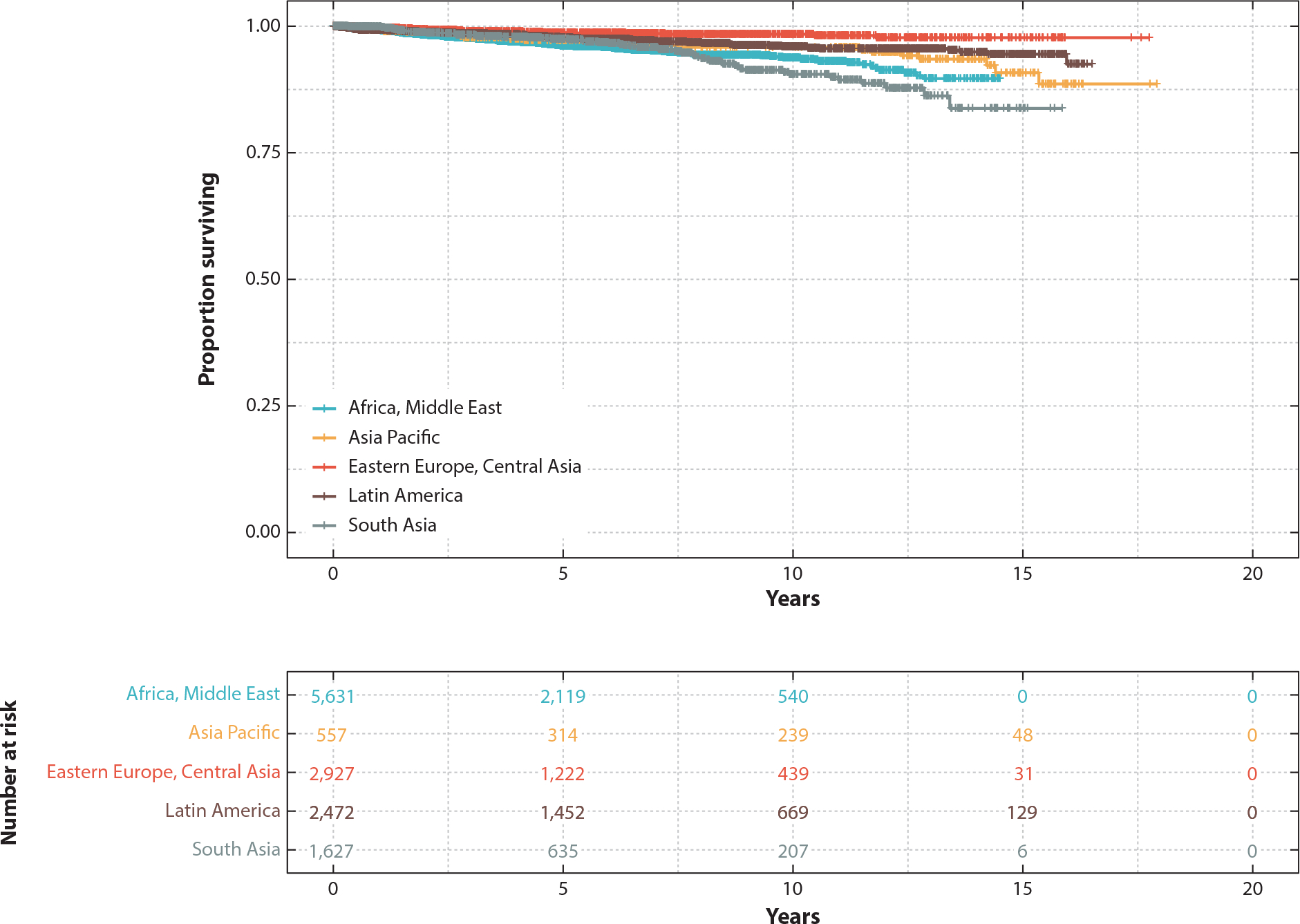
Survival in chronic myeloid leukemia (CML) patients currently treated by the Max Access Solutions program, using imatinib as first-line therapy. At-risk patients from different areas are shown. Individual data points indicate deaths. Figure adapted with permission from Reference 16.
ACKNOWLEDGMENTS
J.P.R. and D.T.C. gratefully acknowledge support from the National Institutes of Health (UH3CA211139). E.H.W. acknowledges National Cancer Institute award U54CA190146.
Footnotes
DISCLOSURE STATEMENT
M.P.M. received research support from Cepheid and from GlaxoSmithKline’s Africa Non-Communicable Disease Open Lab. D.T.C. is a founder of and has financial interest in MiCareo and Lamprogen, both of which are companies that develop new diagnostic platforms. C.C.S.Y. has consulted for TwinStrand Biosciences.
LITERATURE CITED
- 1.Lancet. 2018. GLOBOCAN 2018: counting the toll of cancer. Lancet 392(10152):985. [DOI] [PubMed] [Google Scholar]
- 2.WHO (World Health Organ.). 2019. Second WHO model list of essential in vitro diagnostics. Publ. WHO/MVP/EMP/2019.05, WHO, Geneva [Google Scholar]
- 3.Nelson AM, Milner DA, Rebbeck TR, Iliyasu Y. 2016. Oncologic care and pathology resources in Africa: survey and recommendations. J. Clin. Oncol 34(1):20–26 [DOI] [PubMed] [Google Scholar]
- 4.Neal RD, Tharmanathan P, France B, Din NU, Cotton S, et al. 2015. Is increased time to diagnosis and treatment in symptomatic cancer associated with poorer outcomes? Systematic review. Br. J. Cancer 112(Suppl. 1):S92–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO (World Health Organ.). Cancer Fact Sheet 2020. Geneva: WHO. https://www.who.int/news-room/fact-sheets/detail/cancer [Google Scholar]
- 6.WHO (World Health Organ.). 2017. Guide To Cancer Early Diagnosis. Geneva: WHO [Google Scholar]
- 7.Drake TM, Knight SR, Harrison EM, Soreide K. 2018. Global inequities in precision medicine and molecular cancer research. Front. Oncol 8:346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luu MH, Press RD. 2013. BCR–ABL PCR testing in chronic myelogenous leukemia: molecular diagnosis for targeted cancer therapy and monitoring. Expert Rev. Mol. Diagn 13(7):749–62 [DOI] [PubMed] [Google Scholar]
- 9.Hughes TP, Kaeda J, Branford S, Rudzki Z, Hochhaus A, et al. 2003. Frequency of major molecular responses to imatinib or interferon alfa plus cytarabine in newly diagnosed chronic myeloid leukemia. N. Engl. J. Med 349(15):1423–32 [DOI] [PubMed] [Google Scholar]
- 10.Hughes T, Deininger M, Hochhaus A, Branford S, Radich J, et al. 2006. Monitoring CML patients responding to treatment with tyrosine kinase inhibitors: review and recommendations for harmonizing current methodology for detecting BCR-ABL transcripts and kinase domain mutations and for expressing results. Blood 108(1):28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Branford S, Yeung DT, Ross DM, Prime JA, Field CR, et al. 2013. Early molecular response and female sex strongly predict stable undetectable BCR-ABL1, the criteria for imatinib discontinuation in patients with CML. Blood 121(19):3818–24 [DOI] [PubMed] [Google Scholar]
- 12.Ross DM, Branford S, Seymour JF, Schwarer AP, Arthur C, et al. 2013. Safety and efficacy of imatinib cessation for CML patients with stable undetectable minimal residual disease: results from the TWISTER study. Blood 122(4):515–22 [DOI] [PubMed] [Google Scholar]
- 13.Jabbour E, Kantarjian H, Jones D, Talpaz M, Bekele N, et al. 2006. Frequency and clinical significance of BCR-ABL mutations in patients with chronic myeloid leukemia treated with imatinib mesylate. Leukemia 20(10):1767–73 [DOI] [PubMed] [Google Scholar]
- 14.Milojkovic D, Apperley JF, Gerrard G, Ibrahim AR, Szydlo R, et al. 2012. Responses to second-line tyrosine kinase inhibitors are durable: an intention-to-treat analysis in chronic myeloid leukemia patients. Blood 119(8):1838–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahon FX, Réa D, Guilhot J, Guilhot F, Huguet F, et al. 2010. Discontinuation of imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: the prospective, multicentre Stop Imatinib (STIM) trial. Lancet Oncol. 11(11):1029–35 [DOI] [PubMed] [Google Scholar]
- 16.Malhotra H, Radich JP, Garcia-Gonzalez P. 2019. Meeting the needs of CML patients in resource-poor countries. In Hematolology 2019: American Society of Hematology Education Program, pp. 433–42. Washington, DC: Am. Soc. Hematol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sala Torra O, Beppu L, Smith JL, Welden L, Georgievski J, et al. 2016. Paper or plastic? BCR-ABL11 quantitation and mutation detection from dried blood spots. Blood 127(22):2773–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winn-Deen ES, Helton B, Van Atta R, Wong W, Peralta J, et al. 2007. Development of an integrated assay for detection of BCR-ABL RNA. Clin. Chem 53(9):1593–600 [DOI] [PubMed] [Google Scholar]
- 19.Qiagen. 2016. QIAamp® DNA mini and blood mini handbook. Handb., Qiagen, Germantown, MD [Google Scholar]
- 20.Laurent C, Baron M, Amara N, Haioun C, Dandoit M, et al. 2017. Impact of expert pathologic review of lymphoma diagnosis: study of patients from the French Lymphopath Network. J. Clin. Oncol 35(18):2008–17 [DOI] [PubMed] [Google Scholar]
- 21.Strobbe L, van der Schans SAM, Heijker S, Meijer JWR, Mattijssen EJMV, et al. 2014. Evaluation of a panel of expert pathologists: review of the diagnosis and histological classification of Hodgkin and non-Hodgkin lymphomas in a population-based cancer registry. Leuk. Lymphoma 55(5):1018–22 [DOI] [PubMed] [Google Scholar]
- 22.Valvert F, Silva O, Solórzano-Ortiz E, Puligandla M, Siliézar Tala MM. et al. 2021. Low-cost transcriptional diagnostic to accurately categorize lymphomas in low- and middle-income countries. Blood Adv. 5(10):2447–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salles G, Barrett M, Foa R, Maurer J, O’Brien S, et al. 2017. Rituximab in B-cell hematologic malignancies: a review of 20 years of clinical experience. Adv. Ther 34(10):2232–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horwitz S, O’Connor OA, Pro B, Illidge T, Fanale M, et al. 2019. Brentuximab vedotin with chemotherapy for CD30-positive peripheral T-cell lymphoma (ECHELON-2): a global, double-blind, randomised, phase 3 trial. Lancet 393(10168):229–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, et al. 2015. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136(5):E359–86 [DOI] [PubMed] [Google Scholar]
- 26.Gakwaya A, Kigula-Mugambe JB, Kavuma A, Luwaga A, Fualal J, et al. 2008. Cancer of the breast: 5-year survival in a tertiary hospital in Uganda. Br. J. Cancer 99(1):63–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sankaranarayanan R, Swaminathan R, Brenner H, Chen K, Chia KS, et al. 2010. Cancer survival in Africa, Asia, and Central America: a population-based study. Lancet Oncol. 11(2):165–73 [DOI] [PubMed] [Google Scholar]
- 28.DeSantis CE, Bray F, Ferlay J, Lortet-Tieulent J, Anderson BO, Jemal A. 2015. International variation in female breast cancer incidence and mortality rates. Cancer Epidemiol. Biomarkers Prev 24(10):1495–506 [DOI] [PubMed] [Google Scholar]
- 29.Joko-Fru WY, Miranda-Filho A, Soerjomataram I, Egue M, Akele-Akpo MT, et al. 2020. Breast cancer survival in sub-Saharan Africa by age, stage at diagnosis and human development index: a population-based registry study. Int. J. Cancer 146(5):1208–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foerster M, Anderson BO, McKenzie F, Galukande M, Anele A, et al. 2019. Inequities in breast cancer treatment in sub-Saharan Africa: findings from a prospective multi-country observational study. Breast Cancer Res. 21(1):93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson BO, Carlson RW. 2007. Guidelines for improving breast health care in limited resource countries: the Breast Health Global Initiative. J. Natl. Compr. Cancer Netw 5(3):349–56 [DOI] [PubMed] [Google Scholar]
- 32.Carlson RW, Larsen JK, McClure J, Fitzgerald CL, Venook AP, et al. 2014. International adaptations of NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw 12(5):643–48 [DOI] [PubMed] [Google Scholar]
- 33.Eniu A, Carlson RW, El Saghir NS, Bines J, Bese NS, et al. 2008. Guideline implementation for breast healthcare in low- and middle-income countries: treatment resource allocation. Cancer 113(8 Suppl):2269–81 [DOI] [PubMed] [Google Scholar]
- 34.Shyyan R, Sener SF, Anderson BO, Garrote LM, Hortobagyi GN, et al. 2008. Guideline implementation for breast healthcare in low- and middle-income countries: diagnosis resource allocation. Cancer 113(Suppl. 8):2257–68 [DOI] [PubMed] [Google Scholar]
- 35.WHO Expert Comm. Sel. Use Essent. Med. 2014. The selection and use of essential medicines: report of the WHO Expert Committee, 2013 (including the 18th WHO Model List of Essential Medicines and the 4th WHO Model List of Essential Medicines for Children). WHO Tech. Rep. Ser. No. 985, WHO, Geneva [Google Scholar]
- 36.Bird PA, Hill AG, Houssami N. 2008. Poor hormone receptor expression in East African breast cancer: evidence of a biologically different disease? Ann. Surg. Oncol 15(7):1983–88 [DOI] [PubMed] [Google Scholar]
- 37.Ziegenhorn HV, Frie KG, Ekanem IO, Ebughe G, Kamate B, et al. 2020. Breast cancer pathology services in sub-Saharan Africa: a survey within population-based cancer registries. BMC Health Serv. Res 20(1):912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martei YM, Pace LE, Brock JE, Shulman LN. 2018. Breast cancer in low- and middle-income countries: why we need pathology capability to solve this challenge. Clin. Lab. Med 38(1):161–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kosack CS, Page AL, Klatser PR. 2017. A guide to aid the selection of diagnostic tests. Bull. World Health Organ 95(9):639–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barberis M, Pellegrini C, Cannone M, Arizzi C, Coggi G, Bosari S. 2008. Quantitative PCR and HER2 testing in breast cancer: a technical and cost-effectiveness analysis. Am. J. Clin. Pathol 129(4):563–70 [DOI] [PubMed] [Google Scholar]
- 41.Menon MP, Niyonzima N, Gralow J, Orem J. 2021. Breast cancer clinical trials: the landscape at the Uganda Cancer Institute and lessons learned. JCO Glob. Oncol 7:127–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muller BM, Kronenwett R, Hennig G, Euting H, Weber K, et al. 2011. Quantitative determination of estrogen receptor, progesterone receptor, and HER2 mRNA in formalin-fixed paraffin-embedded tissue—a new option for predictive biomarker assessment in breast cancer. Diagn. Mol. Pathol 20(1):1–10 [DOI] [PubMed] [Google Scholar]
- 43.Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, et al. 2010. Rapid molecular detection of tuberculosis and rifampin resistance. N. Engl. J. Med 363(11):1005–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jobbagy Z, van Atta R, Murphy KM, Eshleman JR, Gocke CD. 2007. Evaluation of the Cepheid GeneXpert BCR-ABL assay. J. Mol. Diagnost 9(2):220–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta S, Mani NR, Carvajal-Hausdorf DE, Bossuyt V, Ho K, et al. 2018. Macrodissection prior to closed system RT-qPCR is not necessary for estrogen receptor and HER2 concordance with IHC/FISH in breast cancer. Lab. Investig 98(8):1076–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu NC, Wong W, Ho KE, Chu VC, Rizo A, et al. 2018. Comparison of central laboratory assessments of ER, PR, HER2, and Ki67 by IHC/FISH and the corresponding mRNAs (ESR1, PGR, ERBB2, and MKi67) by RT-qPCR on an automated, broadly deployed diagnostic platform. Breast Cancer Res. Treat 172(2):327–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cancer Genome Atlas Netw. 2012. Comprehensive molecular portraits of human breast tumours. Nature 490(7418):61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma CX, Ellis MJ. 2013. The Cancer Genome Atlas: clinical applications for breast cancer. Oncology 27(12):1263–69, 1274–79 [PubMed] [Google Scholar]
- 49.Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, et al. 2013. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med 368(13):1199–209 [DOI] [PubMed] [Google Scholar]
- 50.Hanna MG, Reuter VE, Ardon O, Kim D, Sirintrapun SJ, et al. 2020. Validation of a digital pathology system including remote review during the COVID-19 pandemic. Mod. Pathol 33(11):2115–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mpunga T, Hedt-Gauthier BL, Tapela N, Nshimiyimana I, Muvugabigwi G, et al. 2016. Implementation and validation of telepathology triage at cancer referral center in rural Rwanda. J. Glob. Oncol 2(2):76–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Milner DA Jr., Holladay EB. 2018. Laboratories as the core for health systems building. Clin. Lab. Med 38(1):1–9 [DOI] [PubMed] [Google Scholar]
- 53.Fischer MK, Kayembe MK, Scheer AJ, Introcaso CE, Binder SW, Kovarik CL. 2011. Establishing telepathology in Africa: lessons from Botswana. J. Am. Acad. Dermatol 64(5):986–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hashimoto Y, Hatayama Y, Kojima N, Morishita S, Matsumoto S, et al. 2016. Development of reverse transcription loop-mediated isothermal amplification for simple and rapid detection of Promyelocytic Leukemia–Retinoic Acid Receptor α mRNA. Yonago Acta Med. 59(4):262–69 [PMC free article] [PubMed] [Google Scholar]
- 55.Stevens WS, Scott LE, Crowe SM. 2010. Quantifying HIV for monitoring antiretroviral therapy in resource-poor settings. J. Infect. Dis 201(Suppl. 1):S16–26 [DOI] [PubMed] [Google Scholar]
- 56.Sooknanan R, Malek L, Wang XH, Siebert T, Keating A. 1993. Detection and direct sequence identification of BCR-ABL1 mRNA in Ph+ chronic myeloid leukemia. Exp. Hematol 21(13):1719–24 [PubMed] [Google Scholar]
- 57.Bobée V, Ruminy P, Marchand V, Viailly PJ, Abdel Sater A, et al. 2017. Determination of molecular subtypes of diffuse large B-cell lymphoma using a reverse transcriptase multiplex ligation-dependent probe amplification classifier: a CALYM study. J. Mol. Diagn 19(6):892–904 [DOI] [PubMed] [Google Scholar]
- 58.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, et al. 2008. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol 26(3):317–25 [DOI] [PubMed] [Google Scholar]
- 59.Mareschal S, Ruminy P, Bagacean C, Marchand V, Cornic M, et al. 2015. Accurate classification of germinal center B-cell–like/activated B-cell–like diffuse large B-cell lymphoma using a simple and rapid reverse transcriptase–multiplex ligation-dependent probe amplification assay: a CALYM study. J. Mol. Diagn 17(3):273–83 [DOI] [PubMed] [Google Scholar]
- 60.Morgan EA, Sweeney MP, Tomoka T, Kopp N, Gusenleitner D, et al. 2016. Targetable subsets of non-Hodgkin lymphoma in Malawi define therapeutic opportunities. Blood Adv. 1(1):84–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mottok A, Wright G, Rosenwald A, Ott G, Ramsower C, et al. 2018. Molecular classification of primary mediastinal large B-cell lymphoma using routinely available tissue specimens. Blood 132(22):2401–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tzonev S 2018. Fundamentals of counting statistics in digital PCR: I just measured two target copies—what does it mean? In Digital PCR: Methods and Protocols, ed. Karlin-Neumann G, Bizouarn F, pp. 25–43. New York: Humana; [DOI] [PubMed] [Google Scholar]
- 63.Kreutz JE, Wang J, Sheen AM, Thompson AM, Staheli JP, et al. 2019. Self-digitization chip for quantitative detection of human papillomavirus gene using digital LAMP. Lab Chip 19(6):1035–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J, Kreutz JE, Thompson AM, Qin Y, Sheen AM, et al. 2018. SD-chip enabled quantitative detection of HIV RNA using digital nucleic acid sequence-based amplification (dNASBA). Lab Chip 18(22):3501–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gansen A, Herrick AM, Dimov IK, Lee LP, Chiu DT. 2012. Digital LAMP in a sample self-digitization (SD) chip. Lab. Chip 12(12):2247–54 [DOI] [PMC free article] [PubMed] [Google Scholar]


