Abstract
Pseudomonads from environmental sources vary widely in their sensitivity to cadmium, but the basis for this resistance is largely uncharactarized. A chromosomal fragment encoding cadmium resistance was cloned from Pseudomonas putida 06909, a rhizosphere bacterium, and sequence analysis revealed two divergently transcribed genes, cadA and cadR. CadA was similar to cadmium-transporting ATPases known mostly from gram-positive bacteria, and to ZntA, a lead-, zinc-, and cadmium-transporting ATPase from Escherichia coli. CadR was related to the MerR family of response regulators that normally control mercury detoxification in other bacterial systems. A related gene, zntR, regulates zntA in E. coli, but it is not contiguous with zntA in the E. coli genome as cadA and cadR were in P. putida. In addition, unlike ZntA and other CadA homologs, but similar to the predicted product of gene PA3690 in the P. aeruginosa genome, the P. putida CadA sequence had a histidine-rich N-terminal extension. CadR and the product of PA3689 of P. aeruginosa also had histidine-rich C-terminal extensions not found in other MerR family response regulators. Mutational analysis indicated that cadA and cadR are fully responsible for cadmium resistance and partially for zinc resistance. However, unlike zntA, they did not confer significant levels of lead resistance. The cadA promoter was responsive to Cd(II), Pb(II), and Zn(II), while the cadR promoter was only induced by Cd(II). CadR apparently represses its own expression at the transcriptional level. However, CadR apparently does not repress cadA. Homologs of the cadmium-transporting ATPase were detected in many other Pseudomonas species.
The cadmium cation is toxic to most microorganisms, probably by binding to essential respiratory proteins (54) and through oxidative damage by production of reactive oxygen species (50). Cadmium enters bacterial cells by the transport systems for essential divalent cations such as Mn2+ (53) or Zn2+ (22). Microbial resistance to cadmium is usually based on energy-dependent efflux mechanisms (46).
One of the best-characterized bacterial cadmium resistance mechanisms is determined by the cadmium-transporting ATPase found initially in gram-positive bacteria (47). The cadmium-transporting ATPase is a P-type ATPase, a member of the cation-transporting ATPases found in both Bacteria and Eucarya (48). It is widespread in Staphylococcus aureus (36) and Listeria monocytogenes (23). The ATPase is encoded by cadA, which is usually plasmid-borne and associated with transposons in L. monocytogenes (23, 24). The cadmium efflux genes in S. aureus are both plasmid-borne and chromosomal. The chromosomal locus of S. aureus is similar to cadAC of the plasmid-borne genes but confers resistance to low concentrations (MIC of 128 μg/ml) of cadmium nitrate (56). CadC, encoded immediately downstream of cadA, is a regulatory protein, which is also required for cadmium resistance in gram-positive bacteria. CadC binds to the promoter-operator area of the cadA gene and works as a transcriptional repressor in vitro (12).
Another class of cadmium resistance genes in S. aureus includes cadB or the cadB-like cadD, which confers a different mechanism of resistance (11, 39). The function of CadB is not well defined, but it may protect bacterial cells by binding cadmium in the membrane (39). A positive response regulator gene, cadX, was found in the cadB-like operon on plasmid pLUG10 in Staphylococcus lugdunensis. CadX is similar to CadC of the cadA operon but acts as a positive regulator (7). CadD of S. aureus is similar to CadB of S. lugdunensis. Hydropathy analysis of the CadD from plasmid pRW001 revealed transmembrane domains with potential cadmium cation-binding motifs in the cytosolic domain (11).
A well-characterized cadmium resistance system in gram-negative bacteria is the cadmium, zinc, and cobalt (czc) resistance determinant of Alcaligenes eutrophus. The CzcC, CzcB, and CzcA proteins comprise an active efflux mechanism driven by a cation-proton antiporter, rather than a cation-transporting ATPase (35). Homologs of the czc genes, called czr, which conferred cadmium and zinc resistance, were recently identified in the chromosome of Pseudomonas aeruginosa and appear to be highly conserved in environmental isolates of that species (14). In addition, a homolog of the cadAC operon, found previously only in gram-positive bacteria, was identified in the gram-negative bacterium Stenotrophomonas maltophilia (1). The flanking insertion sequences and unusual G+C content of the locus was suggestive of its transfer from gram-positive bacteria (10).
Recently, the genome sequences of several gram-negative bacteria have revealed homologs of cadA. Functional analysis of their role in metal resistance has been conducted in Helicobacter pylori (16) and with the Escherichia coli cadA homolog, zntA (42). ZntA was originally described as a zinc-transporting ATPase, but it also confers resistance to cadmium and lead. Recent studies proposed that CadA of S. aureus and ZntA of E. coli are Pb(II)-transporting ATPases (40, 41, 45). In contrast to cadA of gram-positive bacteria, zntA expression is regulated by zntR, encoding a MerR homolog, but located in another region of the E. coli chromosome from zntA (6, 37).
In this study, and in previous reports (14, 18, 28, 43), Pseudomonas spp. from the soil and other environments have been shown to vary widely in sensitivity to cadmium. In addition to the finding of czc homologs in P. aeruginosa, one report identified cadA-encoded cadmium resistance in a Pseudomonas sp. from river sediment (61). For most pseudomonads, the basis of cadmium resistance has not been characterized, but the recent finding of cadA homologs in several bacterial genome sequences suggests that CadA may play a broader role in cadmium resistance in gram-negative bacteria. We report here homologs of CadA-ZntA and ZntR that are adjacent in the chromosome of a rhizosphere strain of Pseudomonas putida 06909 (59) and confer higher levels of cadmium resistance than ZntA of E. coli but not lead resistance. In addition, we investigated the specificity of metal ion induction of cadA and cadR transcription and the presence of cadA homologs in various Pseudomonas species.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids are listed in Table 1. E. coli strains were cultured at 37°C on Luria-Bertani (LB) agar or in LB broth supplemented with the appropriate antibiotics (29). Antibiotic concentrations used for E. coli strains were as follows: tetracycline, 15 μg/ml; kanamycin, 50 μg/ml; ampicillin, 100 μg/ml; or gentamicin, 20 μg/ml. E. coli strain GM2163 (dam mutant) was a host strain used for manipulation of the plasmid carrying a ClaI site that was resistant to the restriction enzyme due to an overlapping Dam methylation site. P. putida strains were grown at 28°C on mannitol-glutamate medium (MG) (20) supplemented with yeast extract (0.25 g/liter) (MGY) or in MGY broth. Antibiotic concentrations used in MGY were as follows: tetracycline, 20 μg/ml; kanamycin, 30 μg/ml; or gentamicin, 20 μg/ml. To conduct maker exchange mutagenesis, P. putida strains were cultured in LB broth under the same conditions without antibiotics.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | F−recA1 Δ(lacZYA-argF) U169 hsdR17 thi-1 gyra66 supE44 endA1 relA1 Δ(lacZ)M15 | 44 |
| GM2163 | F−ara-14 leuB6 thi-1 fhuA31 lacY1 tsx-78 galK2 galT22 supE44 hisG4 rpsL 136 (Strr) xyl-5 mtl-1 dam13::Tn9 (Camr) dcm-6 mcrB1 hsdR2 mcrA | New England Biolabs |
| HB101 | F−hsdS20 (r−m−) recA13 ara-14 proA2 lacY1 galK2 rpsL20 (Strr) xyl-5 mtl-1 supE44 λ− | 5 |
| P. putida | ||
| 06909 | Ampr, wild type | 59 |
| 06909s21x | Kanr, kanamycin cassette inserted into the LysR family response regulator in wild-type strain 06909 chromosomal locus | This study |
| 06909s22x | Kanr, kanamycin cassette inserted into cadmium-transporting ATPase in wild-type strain 06909 chromosomal locus | This study |
| 06909s23 | Genr, gentamicin cassette inserted into cadR in wild-type strain 06909 chromosomal locus | This study |
| Plasmids | ||
| pUC119 | Apr, cloning vector | 60 |
| pRK415 | Tcr, RK2 derived broad-host-range cloning vector | 21 |
| pRK2013 | Kmr, mobilization helper | 13 |
| pHRP311 | Gmr Smr, an RSF1010 derivative, broad-host-range transcriptional fusion vector | 38 |
| pRKL1 | Tcr, a 3.5-kb HindIII fragment carrying a promoterless lacZ gene from pHRP311 cloned into pRK415 | This study |
| pMGm | Apr Gmr, plasmid carrying a gentamicin resistance gene cassette | 33 |
| pMKm | Apr Kmr, plasmid carrying a kanamycin resistance gene cassette | 33 |
| pRIVS2 | Tcr, pRIV16 carrying a clone expressed on cultured Phytophthora parasitica | 25 |
| pUIVS2 | Apr, pUC119 carrying a 5.5-kb EcoRI fragment from pRIVS2 | 25 |
| pUIVS21 | Apr, pUC119 carrying a 2-kb SphI fragment from pUIVS2 | 25 |
| pUIVS22 | Apr, pUC119 carrying a 1-kb SphI fragment from pUIVS2 with a partial cadmium-transporting ATPase gene | This study |
| pUIVS21K | Apr Kmr, pUIVS21 carrying a 1.8-kb kanamycin cassette from pMKm cloned in a blunt-ended XhoI site of lysR | This study |
| pUIVS22K | Apr Kmr, pUIVS22 carrying a 1.8-kb kanamycin cassette from pMKm cloned in a blunt-ended XhoI site of cadA | This study |
| pRS21k | Tcr Kmr, pRK415 carrying a 3.8-kb SphI fragment from pUIVS21K | This study |
| pRS22k | Tcr Kmr, pRK415 carrying a 2.8-kb SphI fragment from pUIVS22K | This study |
| pRS23s | Tcr Gmr, pRCD12 carrying a 1.9-kb PstI fragment blunt-ended gentamicin cassette from pMGm cloned in a blunt-ended ClaI site of cadR | This study |
| pRCD12 | Tcr, a 4.2-kb PstI fragment carrying cadmium resistance genes in pRK415 | This study |
| pRCD22 | Tcr, the 4.2-kb PstI fragment in pRCD12 is reverse oriented against lac promoter in pRK415 | This study |
| pRCD13 | Tcr, a 3.0-kb PstI-HindIII fragment of pRCD12 cloned in pRK415 | This study |
| pRCD14 | Tcr, a 1.0-kb PstI-XhoI fragment was removed from pRCD13 | This study |
| pUCD12 | Apr, a 3.0-kb PstI-HindIII fragment of pRCD12 in pUC119 | This study |
| pUCD30 | Apr, pUC129 carrying a 2-kb ClaI-XhoI fragment of pRCD12 | This study |
| pRCD31 | Tcr, pRKL1 carrying cadA promoter region, an 800-bp EcoRI-KpnI fragment of pUCD30 | This study |
| pRCD32 | Tcr, pRKL1 carrying cadR promoter region, an 800-bp PstI-KpnI fragment of pUCD30 | This study |
Ampr, chromosomal ampicillin resistance; Kanr, chromosomal kanamycin resistance; Genr, chromosomal gentamicin resistance; Apr, ampicillin resistance; Tcr, tetracycline resistance; Kmr, kanamycin resistance; Gmr, gentamicin resistance; Smr, streptomycin resistance.
General DNA manipulations and DNA sequencing.
Standard recombinant DNA techniques were carried out for restriction endonuclease digestion, ligation, transformation of plasmid DNA, and isolation of total DNA (44). A DNA sequencing facility at the University of California, Berkeley, was used for DNA sequencing of cadmium resistance genes. The DNA sequences were analyzed with a software package from the Genetics Computer Group of the University of Wisconsin and the BLAST programs provided by the National Center for Biotechnology Information. The primers used for DNA sequencing were synthesized commercially (Genosys Biotechnologies, Inc., Woodlands, Tex). Preliminary sequence data was obtained from The Institute for Genomic Research website at http://www.tigr.org.
Plasmid construction for marker exchange mutagenesis.
Insertional mutations in cadA and the gene downstream from cadA encoding a putative LysR family response regulator were constructed by cloning a kanamycin resistance gene cassette into XhoI sites in each of these genes, with the cassette in the opposite orientation relative to the transcription of the target genes (Table 1). The cadR gene was similarly mutated by insertion of a gentamicin resistance gene cassette. After subcloning of these constructs into the broad-host-range plasmid pRK415, the plasmids were introduced into the wild-type strain P. putida 06909 by triparental mating with pRK2013 as a helper plasmid. Marker exchange mutagenesis was carried out as described previously (57). Mutants sensitive to tetracycline but resistant to kanamycin or gentamicin were selected, and the correct gene replacement was confirmed by Southern blot hybridization. P. putida 06909s21x, 06909s22x, and 06909s23 were selected as LysR, CadA, and CadR mutants, respectively.
Plasmid construction to measure promoter activity.
A low-copy-number broad-host-range transcriptional fusion vector, pRKL1, was constructed and used to analyze promoter activity from cadA and cadR. A promoterless lacZ gene was cloned into pRK415 in the opposite orientation to the lac promoter, and the promoter regions of divergently transcribed cadA and cadR were cloned in front of lacZ in this vector (Table 1). The resulting pRCD31 carries the cadA promoter, and pRCD32 carries the cadR promoter, as transcriptional fusions with lacZ.
MIC determination.
The MICs of several metals, along with cadmium chloride, were determined on mannitol-glutamate agar (20) supplemented with yeast extract at 0.25 g/liter (MGY agar) and one of the following chemicals, at various concentrations, as described previously (9): CdCl2 · 2.5H2O, CuSO4 · 5H2O, ZnSO4 · 7H2O, HgCl2, AgNO3, CoCl2 · 6H2O, NiSO4, KCl, NaCl, FeCl3 · 6H2O, CaCl2 · 2H2O, Pb(C2H3O2) · 3H2O, and MnCl2 · 4H2O.
β-Galactosidase assays.
P. putida 06909 or 06909s23 transconjugants carrying various plasmids were grown at 28°C overnight in 5 ml of MGY broth supplemented with appropriate antibiotics. A 0.3-ml sample of each culture was transferred into a fresh 5-ml portion of MGY broth supplemented with appropriate antibiotics and one of the metal salts at a subinhibitory concentration. The subinhibitory concentration chosen for each metal was the highest level that allowed the same growth rate of the strain as obtained without metals added, based on the MIC studies described above. The supplemented concentration of each metal for the induction studies was as follows: CdCl2 · 2.5H2O, 12.5 μM; CuSO4 · 5H2O, 25 μM; ZnSO4 · 7H2O, 50 μM; HgCl2, 0.5 μM; CoCl2 · 6H2O, 5 μM; NiSO4, 10 μM; Pb(C2H3O2) · 3H2O, 10 μM; and MnCl2 · 4H2O, 25 μM. The bacterial culture was further shaken under the same conditions for 6 h. Finally, the grown cells were resuspended into sterile water and lysed to measure the β-galactosidase activity as described by Miller (29) with o-nitrophenyl-β-d-galactopyranoside (ONPG) as the substrate.
Detection of cadA homologs from Pseudomonas species.
Total genomic DNA from various bacteria was isolated as described previously (25). The SphI fragment of pUIVS22 was gel purified and labeled by random primed labeling with a digoxigenin-dUTP DNA-labeling kit (Boehringer GmbH, Mannheim, Germany). Southern blot analysis of BamHI-digested genomic DNA from various bacteria was performed on nylon membranes (MSI, Westboro, Mass.). Posthybridization washes were carried out at relatively low stringency (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]–0.1% sodium dodecyl sulfate at 62°C). The hybridization was detected with a chemiluminescent substrate, disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)-tricyclo(3.3.1.13,7)decan}-4-yl)phenyl phosphate (CSPD; Boehringer), as described in the manufacturer's instructions.
Nucleotide sequence accession number.
The DNA sequence containing the P. putida 06909 cadA and cadR genes has been assigned GenBank accession no. AF333961.
RESULTS AND DISCUSSION
Identification and mutagenesis of a cadmium-transporting ATPase.
We have previously cloned genes from P. putida 06909 induced during colonization of a plant pathogenic fungus, Phytophthora parasitica (25). One of the clones, pUIVS2, carried at its 5′ end a partial sequence of a heavy-metal-transporting ATPase gene. The nucleotide sequence was determined for 1,043 bp of the pUIVS22 clone. The deduced amino acid sequence of the 1,030-bp open reading frame (ORF) was highly similar to the C-terminal half of cadmium-transporting ATPases and other heavy-metal-transporting ATPases from many bacterial species. Comparison of the deduced amino acid sequences showed that the ORF in clone pUIVS22 is not complete, missing the N-terminal sequences and its promoter. Based on the 342-amino-acid peptide sequence, our clone was the most similar to the cadmium-transporting ATPases of gram-positive bactaria. Since experiments to detect plasmids from wild-type strain P. putida 06909 were unsuccessful (data not shown), the ORF is likely to reside in the bacterial chromosome. An insertional mutation of the partial clone was made by inserting a kanamycin resistance gene cassette in the bacterial chromosome through marker exchange mutagenesis as described in Materials and Methods. Mutational analysis of the partial clone showed that the ATPase mutant, 06909s22x, was highly sensitive to cadmium and had moderately decreased zinc resistance (Table 2). The analysis of the 06909s22x strain, along with other mutants, is further described in the next section. Therefore, we designated the gene cadA, encoding a cadmium-transporting ATPase. The ATPase was not important for fungal hyphae colonization (data not shown).
TABLE 2.
MICs of different metals for P. putia 06909, P. putida 06909s22x, P. putida 06909s21x, and P. putida 06909s23
| Metal | MIC (mM) for P. putida strain:
|
|||
|---|---|---|---|---|
| 06909 | 06909s22x | 06909s21x | 06909s23 | |
| CdCl2 · 2.5H2O | 1.7 | 0.05 | 1.7 | 1.3 |
| CuSO4 · 5H2O | 2.0 | 2.0 | 2.0 | 2.0 |
| ZnSO4 · 7H2O | 11.5 | 7.0 | 11.5 | 10.5 |
| HgCl2 | 0.03 | 0.03 | 0.03 | 0.03 |
| AgNO3 | <0.01 | <0.01 | <0.01 | <0.01 |
| CoCl2 · 6H2O | 0.3 | 0.3 | 0.3 | 0.3 |
| NiSO4 | 1.0 | 1.0 | 1.0 | 1.0 |
| KCl | >100 | >100 | >100 | >100 |
| NaCl | >100 | >100 | >100 | >100 |
| FeCl3 · 6H2O | >2.0 | >2.0 | >2.0 | >2.0 |
| CaCl2 · 2H2O | >25 | >25 | >25 | >25 |
| Pb(C2H3O2) · 3H2O | 2.4 | 2.3 | 2.4 | 2.3 |
| MnCl2 · 4H2O | >2.5 | >2.5 | >2.5 | >2.5 |
Organization and nucleotide sequences of genes required for cadmium resistance.
Since we had only a partial fragment of the cadmium-transporting ATPase gene, the clone carrying the full-length gene was obtained by complementing the cadmium-sensitive mutant 06909s22x with a subgenomic library of wild-type strain P. putida 06909 DNA constructed in a broad-host-range plasmid pRK415. The subgenomic library was constructed with PstI inserts of ca. 4 to 6 kb, since Southern hybridizations showed that a 4.2-kb PstI fragment of the wild-type strain 06909 hybridized with the cadA probe from pUIVS22 (data not shown). The plasmid pRCD12 complemented the cadmium sensitivity of mutant 06909s22x. When the orientation of the 4.2-kb insert of pRCD12 was reversed in pRCD22, the clone still complemented the mutation in 06909s22x. A subcloned 3-kb PstI-HindIII fragment in pRCD13 also complemented the cadmium sensitivity of 06909s22x, but a 2-kb subclone in pRCD14 did not (Fig. 1B). This indicated that the 3-kb DNA fragment was sufficient for cadmium resistance in P. putida 06909. DNA sequence analysis of the insert in pUCD12 revealed two ORFs (Fig. 1A). One was cadA, encoding the cadmium-transporting ATPase, and the other was cadR, named after sequence comparison and mutational analysis. cadR was divergently transcribed from cadA, indicating the presence of separate promoters for the two genes. Another ORF found downstream of cadA was a partial fragment of a LysR family response regulator, which may be involved in bacterium-fungus interactions (25).
FIG. 1.
Complementation of cadium-sensitive mutant 06909s22x (cadA::km). (A) Map of a 4.2-kb PstI fragment carrying cadA and cadR. (B) Complementation of cadmium sensitivity by different subclones. Plac indicates lac promoter in pRK415. Restriction endonuclease sites of the subclones are not indicated. A “+” indicates complementation of cadmium sensitivity to cadmium resistance, but a “−” indicates no complementation. Abbreviations: C, ClaI; E, EcoRI; H, HindIII; K, KpnI; P, PstI; Sp, SphI; Xh, XhoI.
The space between the ATG codons for cadA and cadR was 84 bp, which should contain the promoter regions for the two transcriptional units. The organization and nucleotide sequence of this area was similar to a region in contig 10704 of the unfinished microbial genome of bioremediation strain P. putida KT2440 (32). A similar region was also found in the recently published sequence of P. aeruginosa (51). However, to our knowledge, the role of these homologs in cadmium resistance has not been determined. No significant similarities were found in the unfinished genome sequences of P. putida RPS1. The organization of cadA and cadR in P. putida 06909 is different from cadmium resistance determinants reported from other bacterial systems. The most similar system may be zntA and zntR of E. coli, but these two genes are separated from each other in the bacterial chromosome (6, 37). The divergent transcriptional orientation of cadR with respect to the adjacent cadA in P. putida is reminiscent of the organization of merR and the genes encoding mercury detoxification in other bacteria (47).
Similarity to genes for heavy metal resistance.
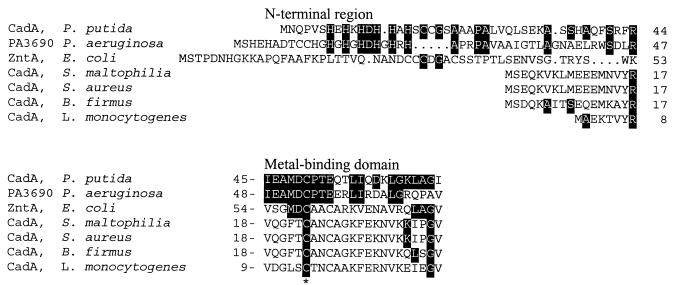
cadA encodes 737 amino acids, and the deduced amino acid sequence shared strong similarity with other known heavy-metal-transporting ATPases, especially cadmium-transporting ATPases and zinc-transporting ATPases (Fig. 2). Alignment of CadA of P. putida 06909 showed the conserved motifs and residues for P-type ATPase function, including metal binding (Fig. 2), ATP binding, and aspartyl phosphorylation sites (data not shown) (49). Initially, ZntA was identified as a zinc-transporting ATPase in E. coli. However, it was also shown that ZntA transports Cd(II) and Pb(II) (40, 41). CadA of pI258 in S. aureus, a homolog of ZntA, also transports Cd(II), Zn(II), and Pb(II) (40). In contrast, CadA of P. putida 06909 provided more specific resistance to cadmium. It was partially responsible for zinc resistance, but its contribution to lead resistance was negligible. The level of cadmium resistance in P. putida 06909 was also 17-fold higher than the cadmium resistance of E. coli K-12 carrying zntA.
FIG. 2.
Alignment of the N-terminal region and metal-binding domains of CadA or ZntA from P. putida, P. aeruginosa (51), E. coli (48), S. maltophilia (1), S. aureus (42), B. firmus (19), and L. monocytogenes (27). Identical bases are shown as white letters on a dark background. Asterisks indicate identical residues among all seven proteins.
An unusual feature of CadA of P. putida 06909 and of P. aeruginosa, compared with other known CadA sequences, was a histidine-rich N-terminal extension (Fig. 2). It will be interesting to test whether this histidine-rich motif (HEHKHDHHAH) contributes to the higher levels of cadmium resistance conferred by CadA in P. putida or to the differences in the specificity of metal resistance.
cadR is predicated to encode a 147-amino-acid protein that is similar to MerR family response regulators (Fig. 3), which control the expression of mercury-detoxifying genes both positively and negatively (27, 47, 52). It was also similar to ZntR of E. coli (6) and to the predicted product of gene PA3689 of P. aeruginosa (51). The conserved helix-turn-helix motif for DNA binding, probably binding to the promoter-operator region of cadA and cadR, was found in the CadR sequence. There were also three conserved cysteine residues (Cys77, Cys112, and Cys119 of CadR), which are putative cation-binding sites (62). The five heptad repeats between Cys77 and Cys 112 form a potential helical strand, which is an interface to form homodimers of MerR family response regulators (6). MerR represses expression of the mer operon in the absence of Hg(II) but activates expression in the presence of Hg(II) (15). MerR is expressed irrespective of Hg(II) (15). In contrast, our results indicate that CadR represses its own expression in the absense of Cd(II), but it is induced in the presence of Cd(II). Induction of ZntR by cadmium or other metal ions has not been reported, to our knowledge. Also, in contrast to MerR and ZntR, CadR has an unusual histidine-rich C-terminal extension (HSHVGRSHGH) (Fig. 3). It will be interesting to test if the histidine-rich C-terminal of CadR contributes to the metal specificity of the cadmium operon in P. putida 06909.
FIG. 3.
Alignment of CadR of P. putida 06909 with the MerR family of activator-repressors, including the predicted product of gene PA3689 of P. aeruginosa (51), ZntR of E. coli (6), and MerR of Tn501 in pVS1 of P. aeruginosa (31). Identical bases are shown as white letters on a dark background. Asterisks are Hg(II)-binding cysteine residues of MerR (62), and numbers 1 to 7 are the heptad repeats that form helical structures to promote homodimer formation (6). The identities (ID) of the amino acid sequences to CadR are indicated.
Mutation of cadA and cadR and heavy metal sensitivity.
While the MIC of cadmium chloride for the wild-type P. putida 06909 was 1.7 mM, the MICs of cadmium chloride for the mutant P. putida 06909s22x (cadA::km) and 06909s23 (cadR::gm) were 0.05 and 1.3 mM, respectively (Table 2). The MIC of zinc sulfate was also different among mutant strains. The 06909s22x (cadA::km) mutant showed less resistance to zinc sulfate, although the MIC (7.0 mM) was still high. The mutant 06909s23 (cadR::gm) also showed only a slight decrease in its MIC (10.5 mM) of zinc sulfate from the wild-type MIC of 11.5 mM. A trivial decrease in the MIC of lead acetate was observed from the two mutant strains. The MIC to lead acetate was 2.4 mM for the wild type and 2.3 mM for both the cadA and the cadR mutants. However, the MICs of other heavy metals tested for mutant P. putida 06909s22x and 06909s23 strains were not different from that of wild-type P. putida 06909 (Table 2). The mutational analysis was consistent with cadA encoding a cadmium-specific transporting ATPase that is partially responsible for zinc resistance in P. putida 06909. cadR is also necessary for full resistance to zinc and cadmium. The MIC of cadmium chloride for E. coli K-12 strain carrying zntA was 0.1 mM on MGY plates (data not shown), while the MIC of cadmium acetate for this strain on LB medium was 1.5 mM (42). This indicated that cadA of P. putida 06909 is responsible for much higher resistance to Cd(II) than zntA of E. coli.
A LysR family response regulator was located immediately downstream from cadA with nine bases between the start codon of the LysR family response regulator and the stop codon of CadA. The Shine-Dalgarno box of the LysR regulator overlaps with the stop codon of cadA, which suggests that cadA-lysR may be part of a single transcriptional unit. However, disruption of the response regulator in the wild-type chromosome (06909s21x) did not affect cadmium resistance and showed the same resistance to all of the tested metals at the level of the wild-type strain P. putida 06909 (Table 2). Thus, the LysR response regulator does not appear to be involved in cadmium resistance in P. putida 06909, but the loss of LysR did impair bacterial growth rate and colony morphology (26).
Induction of the promoter region by cadmium and its specificity.
The 84-bp space between the divergently transcribed cadA and cadR genes was assumed to contain two promoters with different orientations (Fig. 1A). A transcriptional fusion vector, pRKL1, with a promoterless lacZ gene was constructed. Since the vector pRKL1 alone showed a low background level of β-galactosidase activity in the presence of various metals in our strain (data not shown), it was used to construct a transcriptional fusion between the cadA or cadR promoter and the promoterless lacZ.
Both the cadA promoter (PcadA) and the cadR promoter (PcadR) were inducible by cadmium at subinhibitory concentrations in the wild-type strain 06909 background, but induction of cadA by Cd(II) was stronger than that of cadR (Table 3). Cd(II) was the most effective divalent cation for induction of cadA, but Pb(II) was also an effective inducer at a high concentration. Zn(II) and Hg(II) only induced cadA slightly. These results are similar to the induction of ZntA in E. coli, which was induced with the following order of effectiveness: Cd(II) > Pb(II) > Zn(II) (4). cadA of pI258 in S. aureus is induced with the following order of effectiveness: Pb(II) > Cd(II) > Zn(II) (41). However, only cadmium induced PcadR (Table 3). The lack of induction of cadR by lead and zinc also suggests differences in the transcriptional apparatus at the cadA and cadR promoters.
TABLE 3.
Specificity of expression of β-galactosidase activity from the promoter regions of cadA and cadR in wild-type 06909 with different metal ions
| Metal | Concn (μM) | β-Galactosidase activity (Miller units)a with plasmid:
|
||
|---|---|---|---|---|
| pRKL1 | pRCD31 (PcadA) | pRCD32 (PcadR) | ||
| No metal | 196 | 690 | 735 | |
| CdCl2 · 2.5H2O | 12.5 | 235 | 8,417 | 1,652 |
| CuSO4 · 5H2O | 25 | NT | 802 | 656 |
| ZnSO4 · 7H2O | 50 | NT | 1,015 | 745 |
| ZnSO4 · 7H2O | 500 | NT | 1,531 | NT |
| HgCl2 | 0.5 | NT | 1,007 | 592 |
| CoCl2 · 6H2O | 5 | NT | 709 | 493 |
| NiSO4 | 10 | NT | 683 | 639 |
| MnCl2 · 4H2O | 25 | NT | 761 | 605 |
| Pb(C2H3O2) · 3H2O | 10 | NT | 1,033 | 672 |
| Pb(C2H3O2) · 3H2O | 500 | NT | 3,250 | NT |
NT, not tested.
In the cadR mutant 06909s23 (cadR::gm) background, PcadA was still cadmium inducible, but PcadR was constitutive (Table 4). PcadA was induced more than 10-fold in the presence of 12.5 μM cadmium chloride both in the wild-type and in the cadR mutant background. However, the induction of PcadR by cadmium was only about 2.5-fold in the wild-type background. The constitutive expression of PcadR in the cadR mutant background suggests that CadR represses its own expression. However, it is not clear if CadR is a repressor for cadA expression. The cadR mutant 06909s23 may still produce a truncated CadR, which maintains the N-terminal DNA binding domain and the first Cys77 residue for cation binding but missing the C-terminal 50 amino acids. The mutated CadR may thus retain some activity as an activator of cadA. However, the helical domain to form homodimers of CadR should have been disrupted. Alternatively, another regulatory gene may contribute to cadA induction.
TABLE 4.
β-Galactosidase activity from promoter regions of cadA and cadR in wild-type strain 06909 and CadR− mutant 06909s23 backgrounds
| Plasmid (promoter) | β-Galactosidase activity (Miller units)a in strain:
|
|||
|---|---|---|---|---|
| 06909
|
06909s23
|
|||
| −Cd | +Cd | −Cd | +Cd | |
| pRKL1 (control) | 208 | 235 | 341 | 340 |
| pRCD31 (PcadA) | 690 | 8,417 | 636 | 8,129 |
| pRCD32 (PcadR) | 776 | 1,994 | 3,560 | 4,127 |
Activity was determined without (−) or with (+) cadmium (Cd).
Conservation of cadmium-transporting ATPase genes among Pseudomonas species.
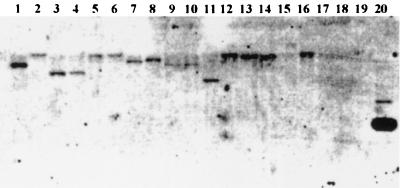
Southern hybridizations were carried out to determine whether the cadmium-specific P-type ATPase is widespread in various Pseudomonas species. The bacterial strains, their characteristics, and respective cadmium chloride MICs are summarized in Fig. 4. Most of the pseudomonads tested showed various degrees of hybridization signals with the cadmium-transporting ATPase of P. putida 06909 except Pseudomonas sp. strain 07887 isolated from tomato (Fig. 4). No hybridization was observed from cadmium-sensitive Xanthomonas axonopodis pv. vesicatoria, Agrobacterium radiobacter, or E. coli DH5α. Southern hybridizations suggested that the CadA ATPase is conserved among many Pseudomonas species and strains. It will be interesting to investigate whether variations in the CadA sequence or in its expression contribute to the observed variation in cadmium resistance among Pseudomonas species and strains.
FIG. 4.
Southern blot hybridization of the cadA gene of P. putida 06909 to total DNA from cadmium-resistant Pseudomonas species and other gram-negative bacteria. The MICs (mM) of cadmium chloride for each strain are indicated in parentheses with the original strain reference in brackets as follows: lane 1, P. putida 06909 (1.7 [59]); lane 2, P. putida 08891 (4.0 [9]); lane 3, P. fluorescens 09906 (0.7 [58]); lane 4, P. fluorescens 2-79 (0.7 [55]); lane 5, P. fluorescens 08908 (>4.0 [8]); lane 6, P. fluorescens 0785-17 (1.0 [30]); lane 7, P. fluorescens 08892 (0.1 [9]); lane 8, P. fluorescens 513 (0.1); lane 9, P. aeruginosa PAO1 (4.0 [17]); lane 10, P. stutzeri ATCC 17588 (4.0); lane 11, P. cichorii 07881 (0.1 [9]); lane 12, P. syringae pv. syringae PS61 (0.5 [3]); lane 13, P. syringae pv. tomato (0.4 [2]); lane 14, Pseudomonas sp. strain 02894 (2.0 [8]); lane 15, Pseudomonas sp. strain 07887 (1.0 [8]); lane 16, Pseudomonas sp. strain 07888 (1.2 [8]); lane 17, Xanthomonas axonopodis pv. vesicatoria 07882 (0.2 [9]); lane 18, Agrobacterium radiobacter K84 (0.05 [34]); lane 19, E. coli DH5α (0.2 [44]); and lane 20, plasmid DNA of pUIVS22 carrying the cadA gene. The DNA in each lane was completely digested with BamHI, and a total of 2 μg of DNA per lane was loaded for each sample. A total of 100 ng of plasmid DNA was loaded for the plasmid in the lane 20.
ACKNOWLEDGMENTS
We thank Caroline S. Harwood (University of Iowa) for providing plasmids and helpful information. Preliminary sequence data was obtained from The Institute for Genomic Research website at http://www.tigr.org.
This work was supported by U.S. Department of Agriculture/National Research Initiative competitive grant 93-37303-9227 to D. A. Cooksey.
REFERENCES
- 1.Alonso A, Sanchez P, Martinez J L. Stenotrophomonas maltophilia D457R contains a cluster of genes from gram-positive bacteria involved in antibiotic and heavy metal resistance. Antimicrob Agents Chemother. 2000;44:1778–1782. doi: 10.1128/aac.44.7.1778-1782.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bender C L, Cooksey D A. Indigenous plasmids in Pseudomonas syringae pv. tomato: conjugative transfer and role in copper resistance. J Bacteriol. 1986;165:534–541. doi: 10.1128/jb.165.2.534-541.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender C L, Cooksey D A. Molecular cloning of copper resistance genes from Pseudomonas syringae pv. tomato. J Bacteriol. 1987;169:470–474. doi: 10.1128/jb.169.2.470-474.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binet M R B, Poole R K. Cd(II), Pb(II), and Zn(II) ions regulate expression of the metal-transporting P-type ATPase ZntA in Escherichia coli. FEBS Lett. 2000;473:67–70. doi: 10.1016/s0014-5793(00)01509-x. [DOI] [PubMed] [Google Scholar]
- 5.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli J. Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 6.Brocklehurst K R, Hobman J L, Lawley B, Blank L, Marshall S J, Brown N L, Morby A P. ZntR is a Zn(II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli. Mol Microbiol. 1999;31:893–902. doi: 10.1046/j.1365-2958.1999.01229.x. [DOI] [PubMed] [Google Scholar]
- 7.Chaouni L B-A, Etienne J, Greenland T, Vandenesch F. Nucleic acid sequence and affiliation of pLUG10 of a novel cadmium resistance plasmid from Staphylococcus lugdunensis. Plasmid. 1996;36:1–8. doi: 10.1006/plas.1996.0025. [DOI] [PubMed] [Google Scholar]
- 8.Cooksey D A, Azad H R. Accumulation of copper and other metals by copper-resistant plant-pathogenic and saprophytic pseudomonads. Appl Environ Microbiol. 1992;58:274–278. doi: 10.1128/aem.58.1.274-278.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooksey D A, Azad H R, Cha J-S, Lim C-K. Copper resistance gene homologs in pathogenic and saprophytic bacterial species from tomato. Appl Environ Microbiol. 1990;56:431–435. doi: 10.1128/aem.56.2.431-435.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Courvalin P. Transfer of antibiotic resistance gene between gram-positive and gram-negative bacteria. Antimicrob Agents Chemother. 1994;38:1447–1451. doi: 10.1128/aac.38.7.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crupper S S, Worrell V, Stewart G C, Iandolo J J. Cloning and expression of cadD, a new cadmium resistance gene of Staphylococcus aureus. J Bacteriol. 1999;181:4071–4075. doi: 10.1128/jb.181.13.4071-4075.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Endo G, Silver S. CadC, the transcriptional regulatory protein of the cadmium resistance system of Staphylococcus aureus plasmid pI258. J Bacteriol. 1995;177:4437–4441. doi: 10.1128/jb.177.15.4437-4441.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hassan M E T, van der Lelie D, Springael D, Römling U, Ahmed N, Mergeay M. Identification of a gene cluster, czr, involved in cadmium and zinc resistance in Pseudomonas aeruginosa. Gene. 1999;238:417–425. doi: 10.1016/s0378-1119(99)00349-2. [DOI] [PubMed] [Google Scholar]
- 15.Heltzel A, Lee I W, Totis P A, Summers A O. Activator-dependent preinduction binding of ς-70 RNA polymerase at the metal regulated mer promoter. Biochemistry. 1990;29:9572–9584. doi: 10.1021/bi00493a011. [DOI] [PubMed] [Google Scholar]
- 16.Herrmann L, Schwan D, Garner R, Mobley H L T, Haas R, Schafer K P, Melchers K. Helicobacter pylori cadA encodes an essential Cd(II)-Zn(II)-Co(II) resistance factor influencing urease activity. Mol Microbiol. 1999;33:524–536. doi: 10.1046/j.1365-2958.1999.01496.x. [DOI] [PubMed] [Google Scholar]
- 17.Holloway B W, Krishnapillai V, Morgan A F. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979;43:73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horitsu H, Yamamoto K, Wachi S, Kawai K, Fukuchi A. Plasmid-determined cadmium resistance in Pseudomonas putida GAM-1. J Bacteriol. 1986;165:33–335. doi: 10.1128/jb.165.1.334-335.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivey D M, Guffanti A A, Shen Z, Kudyan N, Krulwich T A. The cadC gene product of alkaliphilic Bacillus firmus OF4 partially restores Na+ resistance to an Escherichia coli strain lacking an Na+/H+ antiporter (NhaA) J Bacteriol. 1992;174:4878–4884. doi: 10.1128/jb.174.15.4878-4884.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keane P J, Kerr A, New P B. Crown gall of stone fruit. II. Identification and nomenclature of Agrobacterium isolates. Aust J Biol Sci. 1970;23:585–595. [Google Scholar]
- 21.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 22.Laddaga R A, Silver S. Cadmium uptake in Escherichia coli K-12. J Bacteriol. 1985;162:1100–1105. doi: 10.1128/jb.162.3.1100-1105.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lebrun M, Audurier A, Cossart P. Plasmid-borne cadmium resistance genes in Listeria monocytogenes are present on Tn5422, a novel transposon closely related to Tn917. J Bacteriol. 1994;176:3049–3061. doi: 10.1128/jb.176.10.3049-3061.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lebrun M, Loulergue J, Chaslus-Dancla E, Audurier A. Plasmids in Listeria monocytogenes in relation to cadmium resistance. Appl Environ Microbiol. 1992;58:3183–3186. doi: 10.1128/aem.58.9.3183-3186.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee S-W, Cooksey D A. Genes expressed in Pseudomonas putida during colonization of a plant-pathogenic fungus. Appl Environ Microbiol. 2000;66:2764–2772. doi: 10.1128/aem.66.7.2764-2772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee S-W. Identification of chromosomal loci of Pseudomonas putida Induced during colonization of Phytophthora parasitica. Ph.D. thesis. Riverside: University of California; 1999. [Google Scholar]
- 27.Lund P A, Ford S J, Brown N L. Transcriptional regulation of the mercury-resistance genes of transposon Tn501. J Gen Microbiol. 1986;132:465–480. doi: 10.1099/00221287-132-2-465. [DOI] [PubMed] [Google Scholar]
- 28.Malik A, Jaiswal R. Metal resistance in Pseudomonas strains isolated from soil treated with industrial wastewater. World J Microbiol Biotech. 2000;16:177–182. [Google Scholar]
- 29.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 30.Mills S D, Jasalavich C A, Cooksey D A. A two-component regulatory system required for copper-inducible expression of the copper resistance operon of Pseudomonas syringae. J Bacteriol. 1993;175:1656–1664. doi: 10.1128/jb.175.6.1656-1664.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Misra T K, Brown N L, Fritzinger D C, Pridmore R D, Barnes W M, Haberstroh L, Silver S. Mercuric ion-resistance operons of plasmid R100 and transposon Tn501: the beginning of the operon including the regulatory region and the first two structural genes. Proc Natl Acad Sci USA. 1984;81:5975–5979. doi: 10.1073/pnas.81.19.5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murray K, Duggleby C J, Sala-Trepat J M, Williams P A. The metabolism of benzoate and methylbenzoates via the meta-cleavage pathway by Pseudomonas arvilla mt-2. Eur J Biochem. 1972;28:301–310. doi: 10.1111/j.1432-1033.1972.tb01914.x. [DOI] [PubMed] [Google Scholar]
- 33.Murillo J, Shen H, Gerhold D, Sharma A, Cooksey D A, Keen N T. Characterization of pPT23B, the plasmid involved in syringolide production by Pseudomonas syringae pv. tomato PT23. Plasmid. 1994;31:275–287. doi: 10.1006/plas.1994.1029. [DOI] [PubMed] [Google Scholar]
- 34.New P B, Kerr A. Biological control of crown gall: field measurements and glasshouse experiments. J Appl Bacteriol. 1972;35:279–287. [Google Scholar]
- 35.Nies D H, Nies A, Chu L, Silver S. Expression and nucleotide sequence of a plasmid-determined divalent cation efflux system from Alcaligenes eutrophus. Proc Natl Acad Sci USA. 1989;86:7351–7355. doi: 10.1073/pnas.86.19.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nucifora G, Chu L, Misra T K, Silver S. Cadmium resistance from Staphylococcus aureus plasmid pI258 cadA gene results from a cadmium-efflux ATPase. Proc Natl Acad Sci USA. 1989;86:3544–3548. doi: 10.1073/pnas.86.10.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Outten C E, Outten F W, O'Halloran T V. DNA distortion mechanism for transcriptional activation by ZntR, a Zn(II)-responsive MerR homologue in Escherichia coli. J Biol Chem. 1999;274:37517–37524. doi: 10.1074/jbc.274.53.37517. [DOI] [PubMed] [Google Scholar]
- 38.Parales R E, Harwood C S. Construction and use of a new broad-host-range lacZ transcriptional fusion vector, pHRP309, for gram− bacteria. Gene. 1993;133:23–30. doi: 10.1016/0378-1119(93)90220-w. [DOI] [PubMed] [Google Scholar]
- 39.Perry R D, Silver S. Cadmium and manganese transport in Staphylococcus aureus membrane vesicles. J Bacteriol. 1982;150:973–976. doi: 10.1128/jb.150.2.973-976.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rensing C, Ghosh M, Rosen B P. Families of soft-metal-ion-transporting ATPases. J Bacteriol. 1999;181:5891–5897. doi: 10.1128/jb.181.19.5891-5897.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rensing C, Sun Y, Mitra B, Rosen B P. Pb(II)-translocating P-type ATPases. J Biol Chem. 1998;273:32614–32617. doi: 10.1074/jbc.273.49.32614. [DOI] [PubMed] [Google Scholar]
- 42.Rensing C, Mitra B, Rosen B P. The zntA gene of Escherichia coli encodes a Zn(II)-translocating P-type ATPase. Proc Natl Acad Sci USA. 1997;94:14326–14331. doi: 10.1073/pnas.94.26.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roane T M, Pepper I L. Microbial responses to environmentally toxic cadmium. Microb Ecol. 2000;38:358–364. doi: 10.1007/s002489901001. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 45.Sharma R, Rensing C, Rosen B P, Mitra B. The ATP hydrolytic activity of purified ZntA, a Pb(II)/Cd(II)/Zn(II)-translocating ATPase from Escherichia coli. J Biol Chem. 2000;275:3873–3878. doi: 10.1074/jbc.275.6.3873. [DOI] [PubMed] [Google Scholar]
- 46.Silver S. Bacterial resistances to toxic metal ions: a review. Gene. 1996;179:9–19. doi: 10.1016/s0378-1119(96)00323-x. [DOI] [PubMed] [Google Scholar]
- 47.Silver S, Phung L T. Bacterial heavy metal resistance: new surprises. Annu Rev Microbiol. 1996;50:753–789. doi: 10.1146/annurev.micro.50.1.753. [DOI] [PubMed] [Google Scholar]
- 48.Silver S, Nucifora G, Phung L T. Human Menkes X-chromosome disease and the staphylococcal cadmium-resistance ATPase: a remarkable similarity in protein sequences. Mol Microbiol. 1993;10:7–12. doi: 10.1111/j.1365-2958.1993.tb00898.x. [DOI] [PubMed] [Google Scholar]
- 49.Silver S, Walderhaug M. Gene regulation of plasmid- and chromosome-determined inorganic ion transport in bacteria. Microbiol Rev. 1992;56:195–228. doi: 10.1128/mr.56.1.195-228.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stohs S J, Bagchi D. Oxidative mechanisms in the toxicity of metal-ions. Free Radic Biol Med. 1995;18:321–336. doi: 10.1016/0891-5849(94)00159-h. [DOI] [PubMed] [Google Scholar]
- 51.Stover C K, Pham X Q, Erwin A L, Mizoguchi S D, Warrener P, Hickey M J, Brinkman F S L, Hufnagle W O, Kowalik D J, Lagrou M, Garber R L, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody L L, Coulter S N, Folger K R, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong G K-S, Wu Z, Paulsen I T, Reizer J, Saier M H, Hancock R E W, Lory S, Olson M V. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 52.Summers A O. Untwist and shout: a heavy metal-responsive transcriptional regulator. J Bacteriol. 1992;174:551–557. doi: 10.1128/jb.174.10.3097-3101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tynecka Z, Gos Z, Zajac J. Energy-dependent efflux of cadmium coded by a plasmid resistance determinant in Staphylococcus aureus. J Bacteriol. 1981;147:305–312. doi: 10.1128/jb.147.2.313-319.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vallee B L, Ulmer D D. Biochemical effects of mercury, cadmium and lead. Annu Rev Biochem. 1972;41:91–128. doi: 10.1146/annurev.bi.41.070172.000515. [DOI] [PubMed] [Google Scholar]
- 55.Weller D M, Cook R J. Suppression of take-all of wheat by seed treatments with fluorescent pseudomonads. Phytopathology. 1983;73:463–469. [Google Scholar]
- 56.Witte W, Green L, Misra T K, Silver S. Resistance to mercury and to cadmium in chromosomally resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1986;29:663–669. doi: 10.1128/aac.29.4.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang C-H, Azad H R, Cooksey D A. A chromosomal locus required for copper resistance, competitive fitness, and cytochrome c biogenesis in Pseudomonas fluorescens. Proc Natl Acad Sci USA. 1996;93:7315–7320. doi: 10.1073/pnas.93.14.7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang C-H, Menge J A, Cooksey D A. Role of copper resistance in competitive survival of Pseudomonas fluorescens in soil. App Environ Microbiol. 1993;59:580–584. doi: 10.1128/aem.59.2.580-584.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang C-H, Menge J A, Cooksey D A. Mutations affecting hyphal colonization and pyoverdine production in pseudomonads antagonistic toward Phytophthora parasitica. Appl Environ Microbiol. 1994;60:473–481. doi: 10.1128/aem.60.2.473-481.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 61.Yoon K P. Isolation and characterization of Pseudomonas sp. KM10, a cadmium- and mercury-resistant, and phenol-degrading bacterium. J Microbiol Biotech. 1998;8:388–398. [Google Scholar]
- 62.Zeng Q C, Stalhandske C, Anderson M C, Scott R A, Summers A O. The core metal-recognition domain of MerR. Biochemistry. 1998;37:15885–15895. doi: 10.1021/bi9817562. [DOI] [PubMed] [Google Scholar]