ABSTRACT
Microplastics provide new microbial niches in aquatic environments. Nevertheless, information on the assembly processes and potential ecological mechanisms of bacterial communities on microplastics from reservoirs is lacking. Here, we investigated the assembly processes and potential ecological mechanisms of bacterial communities on microplastics through full-length 16S rRNA sequencing in the Three Gorges Reservoir area of the Yangtze River, compared to water and sediment. The results showed that the Burkholderiaceae were the dominant composition of bacterial communities in microplastics (9.95%), water (25.14%), and sediment (7.22%). The niche width of the bacterial community on microplastics was lower than those in water and sediment. For the microplastics and sediment, distance-decay relationship results showed that the bacterial community similarity was significantly decreased with increasing geographical distance. In addition, the spatial turnover rate of the bacterial community on microplastics along the ~662-km reaches of the Yangtze River in the Three Gorges Reservoir area was higher than that in sediment. Null model analysis showed that the assembly processes of the bacterial community on microplastics were also different from those in water and sediments. Dispersal limitation (52.4%) was the primary assembly process of the bacterial community on microplastics, but variable selection was the most critical assembly process of the bacterial communities in water (47.6%) and sediment (66.7%). Thus, geographic dispersal limitation dominated the assembly processes of bacterial communities on microplastics. This study can enhance our understanding of the assembly mechanism of bacterial communities caused by the selection preference for microplastics from the surrounding environment.
IMPORTANCE In river systems, microplastics create new microbial niches that significantly differ from those of the surrounding environment. However, the potential relationships between the biogeographic distribution and assembly processes of microbial communities on microplastics were still not well understood. This study could help us address the lack of knowledge about the assembly processes of bacterial communities on microplastics caused by selection from the surrounding environment. In this study, strong geographic dispersal limitation dominated assembly processes of bacterial communities on microplastics, compared to water and sediment, which may be responsible for the microplastic bacterial richness, and the niche distance was lower than those in water and sediment. In addition, sediment may be the main potential source of bacterial communities on microplastics in the Three Gorges Reservoir area, which makes higher community similarity between microplastics and sediment than between microplastics and water.
KEYWORDS: microplastics, dispersal limitation, community assembly, niche distance, full-length 16S rRNA
INTRODUCTION
Microplastics are an emerging pollutant of great public concern because they are considered a potential threat to aquatic biota and human health. Microplastics are ubiquitous in aquatic ecosystems and are well documented in oceans (1, 2), rivers (3, 4), and lakes (5). In particular, the lightweight and persistent nature of microplastics allows their dispersion across large distances throughout the aquatic ecosystem (6, 7). It has been proven that microplastics can transport microorganisms, persistent organic contaminants, or heavy metals from one ecosystem to another (8, 9). Besides, microplastics could selectively enrich microbial communities from the surrounding environment, creating new microbial niches that significantly differ from those of the surrounding environment (including water and sediment, etc.) (7, 10, 11). Although the microplastic biofilm is distinct, the formation process could not be completed without the surrounding environment (12, 13). The formation and development of the microbial community on microplastics to a large extent depend on polymer types, habitats, and time (7). However, to date, most studies reveal that habitat plays a more critical role than polymer type in shaping the bacterial community composition of microplastic biofilms (7, 14, 15).
Revealing the biogeographic patterns of river bacteria and their potential ecological assembly mechanisms is beneficial for us to understand the consequences of bacterial community changes on the functioning of the Earth’s ecosystems (16–18). Especially, the microbial community in the marine plastisphere had a biogeographic pattern, and biogeography rather than substrate type determined the bacterial colonization dynamics on plastic (19, 20). Furthermore, previous studies found that stochastic processes dominated by dispersal limitations played a dominant role in marine plastic microbial community assembly (21) and Hangzhou Bay (22). Stochastic processes involve species probabilistic dispersal, birth, death, extinction, and ecology drift that can drive microbial communities (23, 24), while determinism processes mainly cover ecological selection where abiotic and biotic factors represent distinct niches determining the fitness differences of microorganisms (25). Dispersal and selection have been collectively regarded as two principal forces driving microbial community distribution (26–28). However, the potential relationships between the biogeographic distribution and assembly processes of microbial communities on microplastics are still not well understood.
Variable selection, homogeneous selection, homogenizing dispersal, dispersal limitation, and undominated limitation have been identified as the main assembly processes for microbial communities in the environment (29–31). Stochastic and deterministic processes involved in these assembly processes provide a theoretical framework to better understand the spatial and temporal community dynamics (32, 33). Besides, environmental selection and dispersal were the most important assembly processes for microbial communities (29, 34). Understanding the contrasting roles of environmental selection and dispersal is especially important because dispersal can decrease the strength of selection, which can be well allocated by the null model based on their contributions (35, 36). Furthermore, the distance-decay relationship is one of the most common biogeographic models, indicating that community similarity decreases with increasing geographical distance (29, 37). The slope of the distance-decay relationship reflects the rate of species replacement at different locations (29, 38).
In lotic rivers, the longitudinal, lateral, and vertical movement of water promoted community exchange and evolutionary processes of microbial communities in microplastic biofilms with the surrounding environment (39, 40). This could lead to an exchange of bacteria on microplastics with those in water and sediment (41). In particular, the river ecosystems in the Three Gorges Reservoir have dual characteristics of the reservoir and river (42). Three Gorges dam may further enhance the exchange of microbial communities among microplastics, water, and sediment during water storage and flood discharge. However, little is known about the ecological assembly mechanism of microplastic bacterial communities in reservoirs and the contribution of the surrounding environment to their ecological assembly processes. Therefore, in this study, the assembly processes and ecological mechanisms of the bacterial community on microplastics were investigated through full-length 16S rRNA gene sequencing of samples from the Three Gorges Reservoir area along the Yangtze River compared to water and sediment (Fig. 1). This study could help us address the lack of knowledge about the assembly processes of bacterial communities on microplastics caused by selection from the surrounding environment.
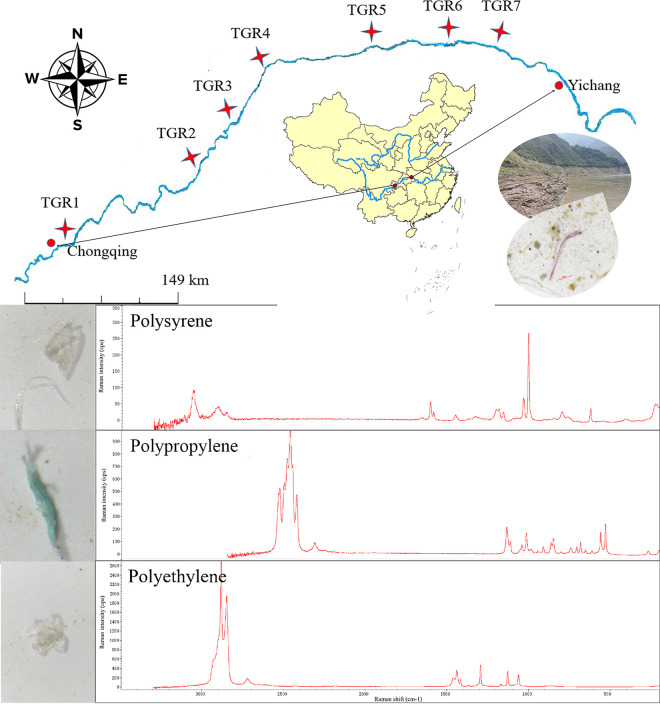
FIG 1.
Sites of sampling of microplastics, water, and sediment in the Three Gorges Reservoir area, China, and some main shapes and materials of microplastics detected in the Three Gorges Reservoir Region. (The map of China was created with ArcGIS.)
RESULTS
Diversity and composition of bacterial communities in different habitats.
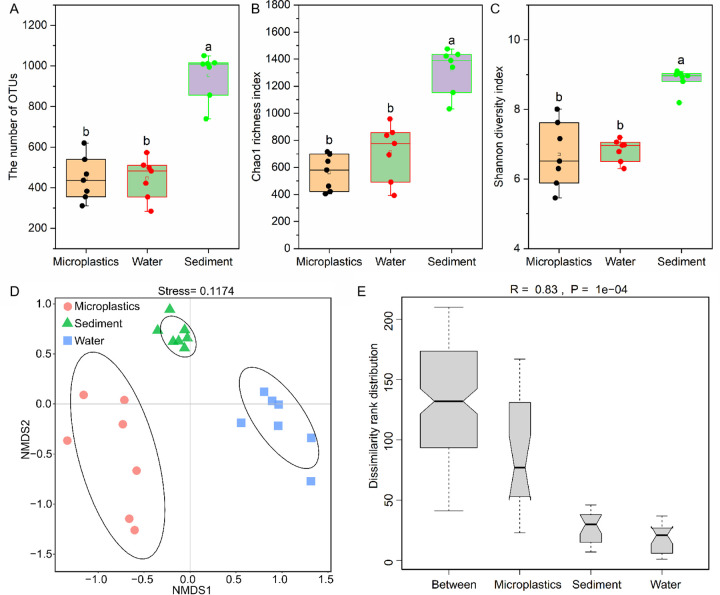
After quality filtering and rarefaction, microplastics, water, and sediment all present rarefaction curves with a stationary phase, suggesting the sufficient depth and accuracy of sequencing for performing bacterial community analysis (see Fig. S1 in the supplemental material). The high-quality sequences were clustered into 2,256 operational taxonomic units (OTUs), and the numbers of OTUs on microplastics (mean = 444) and in water (mean = 446) were significantly lower than that in sediment (mean = 953) (Fig. 2A). The Chao1 richness values of bacterial communities on microplastics (mean = 561) and in water (mean = 715) were significantly lower than that in sediment (mean = 1,321) (Fig. 2B). The Shannon diversity indices of bacterial communities on microplastics (mean = 6.71) and in water (mean = 6.83) were also significantly lower than that in sediment (mean = 8.86) (Fig. 2C). Nonmetric multidimensional scaling (NMDS) analysis showed visible differences in bacterial communities among the three habitats at the OTU level (Fig. 2D). The proximal bacterial communities on microplastics and in sediment were more closely aligned but not for water and microplastics. Furthermore, the NMDS analysis found that the aggregation degree of bacteria on microplastics was relatively dispersed. Analysis of similarity (ANOSIM) showed that the differences in bacterial communities among the three habitats were significantly higher than those in the bacterial communities within the same habitats (R = 0.83; P < 0.01), and the differences among bacterial communities on microplastics were also significantly higher than those in water and sediment (Fig. 2E).
FIG 2.
Difference analysis of bacterial communities among microplastics, water, and sediment based on α- and β-diversity. (A to C) Differences in the bacterial communities’ α-diversity among microplastics, water, and sediment determined by one-way analysis of variance (ANOVA). Different lowercase letters indicate significant differences at the 0.05 level by Duncan’s test. The hollow squares in the box plots show the average values of community similarity, and the lines in the box plots are the median values. (D and E) NMDS analysis and ANOSIM of the bacterial communities’ β-diversity among microplastics, water, and sediment based on Bray-Curtis distance.
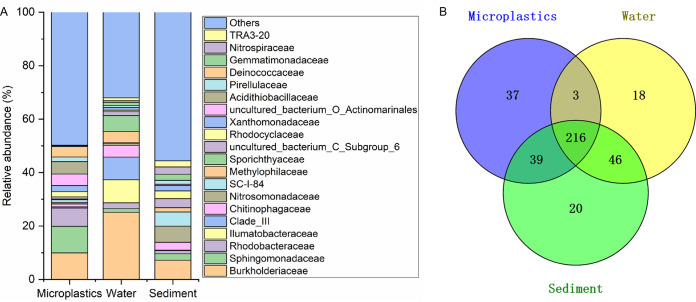
Furthermore, full-length 16S rRNA sequencing provided more detailed and accurate taxon resolution of the bacterial communities on microplastics, in water, and in sediment. A total of 47,376 OTUs from all microplastic, water, and sediment samples were classified into 377 family taxonomic taxa at the 97% similarity level, which accounted for 99.1% of the total OTUs. The microplastic bacterial community was composed mainly of Burkholderiaceae (9.95%), Sphingomonadaceae (9.93%), and Rhodobacteraceae (6.85%), whereas the bacterial communities in water were composed mainly of Burkholderiaceae (25.14%), Ilumatobacteraceae (8.63%), and clade III (8.47%) (Fig. 3A). The sedimentary bacterial community was composed mainly of Burkholderiaceae (7.22%), Nitrosomonadaceae (6.03%), and SC-I-84 (5.32%) (Fig. 3A). limma voom analysis showed that microplastics had significant enrichment for Sphingomonadaceae, Hyphomicrobiaceae, Rhizobiaceae, uncultured bacterium o_Saccharimonadales, Trueperaceae, and uncultured bacterium o_Elsterales from the surrounding environment (water and sediment) (see Tables S2 and S3 in the supplemental material). These results indicated that the composition of the bacterial community on microplastics was significantly different from those in water and sediment. Microplastics provided a microbial niche different from those of water and sediment.
FIG 3.
(A) Main composition of bacterial communities on microplastics, in water, and in sediment at the family taxonomic level. (B) Numbers of unique and shared bacterial taxa at the family taxonomic level described by a Venn diagram.
Moreover, a Venn diagram was applied to identify the unique and shared bacterial taxa among microplastics, water, and sediment (Fig. 3B). A total of 73.2% (216 families) of microplastic bacteria (at the taxonomic level of family) were shared by water and sediments, indicating that microplastic bacteria from the Three Gorges Reservoir area may be mainly from water and sediment. However, 46 bacterial families (e.g., Armatimonadaceae, Cyclobacteriaceae, and Gallionellaceae) were shared in water and sediment but were not detected on microplastics (see Table S4 in the supplemental material), showing that microplastics can selectively enrich bacteria. More bacteria were shared between microplastics and sediment (39 families) than between microplastics and water (3 families), suggesting that sediment may be a major source of bacteria on microplastics, or they are just adapted for a surface-attached/biofilm lifestyle. However, 37 families of bacteria on microplastics were not detected in water and sediment, indicating that bacteria colonizing microplastics in rivers are not necessarily from water and sediment and may be related to the original source of the microplastics.
Geographic distribution of bacterial communities in different habitats.
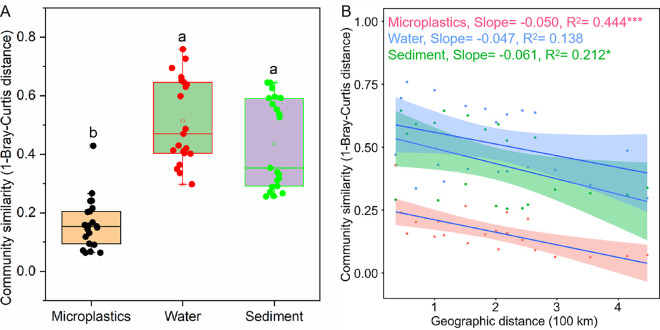
Community similarity (1-Bray-Curtis distance) analysis at the OTU level showed that the community similarity on microplastics (0.16) was significantly lower than those in water (0.51) and sediment (0.44) (Fig. 4A). Thereafter, we further evaluated the relationship between bacterial community similarity and distance between sampling sites (Fig. 4B). The results showed that the bacterial community similarity on microplastics (R2 = 0.444; P = 0.001) and in sediment (R2 = 0.212; P = 0.036) decreased significantly with increasing geographic distance between the sampling sites. Furthermore, the R2 value on microplastics was higher than that in sediment, indicating that the bacterial community on microplastics showed a stronger distance attenuation pattern than the bacterial community in sediment. However, the slope for sediment (0.06) was higher than that for microplastics (0.05). The bacterial community similarity in water had no significant relationship with the distance between the sampling sites (R2 = 0.138; P = 0.098).
FIG 4.
Difference in bacterial community similarity among microplastics, water, and sediment (A) and its attenuation relationship with geographical distance (B). In panel A, differences in bacterial community similarity among microplastics, water, and sediment were determined by one-way ANOVA. Different lowercase letters indicate a significant difference among different media (P < 0.05). The hollow squares in the box plots show the average values of community similarity, and the lines in the box plots are the median values. In panel B, asterisks denote a significant correlation (*, P < 0.05; ***, P < 0.001).
Assembly processes of bacterial communities in different habitats.
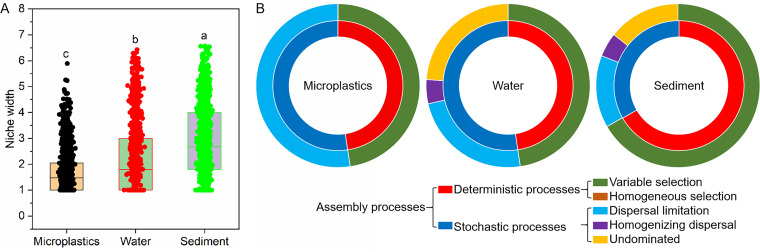
The average niche width of bacterial communities on microplastics (1.72) was significantly lower than those in water (2.20) and sediment (2.85) (Fig. 5A), indicating that there may be different assembly patterns in bacterial communities among microplastics, water, and sediment. Null model analyses revealed that dispersal limitation (52.4%) was the main assembly process of the bacterial community on microplastics, while variable selection was the main assembly process of the bacterial communities in water (47.6%) and sediment (66.7%) (Fig. 5B). Besides, the assembly processes of bacterial communities for both microplastics (47.6%) and water (47.6%) were dominated by stochastic processes, while the assembly processes of bacterial communities in sediment were dominated by deterministic processes (66.7%) (Fig. 5B).
FIG 5.
(A) Niche distances of bacterial communities in microplastics, water, and sediment. Different lowercase letters indicate that there are significant differences at the 0.05 level via Duncan’s test. The hollow squares in the box plots show the average values of community similarity, and the lines in the box plots are the median values. (B) Relative contributions of different ecological processes to the bacterial community assembly in these three habitats based on null model analysis.
Co-occurrence patterns of bacterial communities in different habitats.
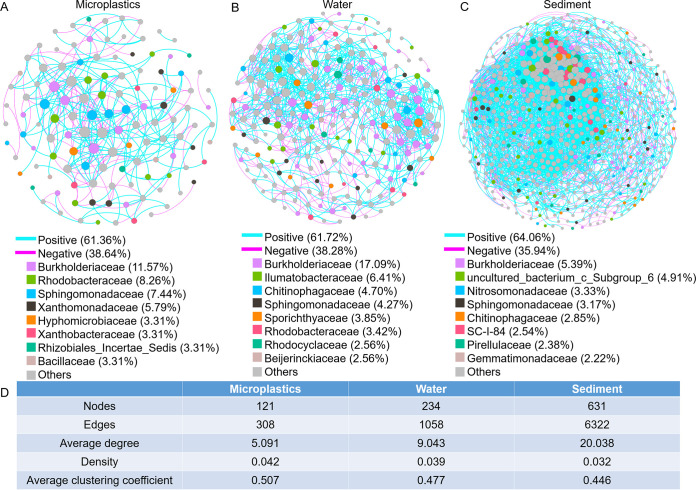
Co-occurrence networks were constructed to disentangle the potential associations among bacterial communities of microplastics, water, and sediment from the Three Gorges Reservoir area (Fig. 6). The network results showed that the microplastic network consisted of 121 nodes and 308 edges, which were substantially lower than those for water (234 nodes and 1,058 edges) and sediment (631 nodes and 6,322 edges). The negative correlation percentage for the microplastic bacteria (38.64%) was higher than those for water (38.28%) and sediment (35.94%). Burkholderiaceae, Rhodobacteraceae, and Sphingomonadaceae were vital participants in the co-occurrence network of the microplastic bacteria. In addition, Burkholderiaceae, Ilumatobacteraceae, and Chitinophagaceae were vital bacterial participants in the co-occurrence network in water, while Burkholderiaceae, uncultured bacterium c_Subgroup_6, and Nitrosomonadaceae were vital bacterial participants in the co-occurrence network in sediment. Thus, the critical bacterial taxa at the family level in the different co-occurrence networks depended on the different habitats. Topological features revealed that the average degree of microplastics was lower than those of water and sediment, while the density and average clustering coefficient of microplastics were higher than those of water and sediment.
FIG 6.
(A to C) Co-occurrence network patterns of bacterial communities in microplastics (A), water (B), and sediment (C) at the OTU level based on Spearman correlation analysis (|r| > 0.8; P < 0.01). The percentages next to family names are the percentages of the number of OTUs accounted for by this family out of the total number of OTUs in the networks. (D) Major topological properties of the co-occurrence network of the bacterial communities in microplastics, water, and sediment.
DISCUSSION
Microplastics enter aquatic ecosystems, and the bacteria from the surrounding environment could selectively colonize and propagate on the microplastic surface, forming a new microbial ecological niche (7, 10, 43, 44). This study found that the α-diversity of bacterial communities on microplastics from the Three Gorges Reservoir area was lower than that in sediment, but there was no significant difference from that in water. This may be because water and microplastics have limited nutrient contents, so their microbial diversities are lower than that in sediment and about equal to each other. However, the composition of the bacterial community on microplastics was significantly different from those in water and sediment. The reason for this may be due to the different physical and chemical properties inherent in different habitats, which lead to differences in the compositions and structures of the bacterial communities (7, 45, 46). Specifically, Sphingomonadaceae were not only abundant bacteria on microplastics but also showed a significant enrichment on microplastics. Sphingomonadaceae were also the major inhabitants of microplastics from Italian lakes (6), which was consistent with the findings of this study. Nitrosomonadaceae play a major role in controlling the nitrogen cycle in terrestrial and aquatic ecosystems (47). In this study, Nitrosomonadaceae were the second most abundant bacteria in sediment and were significantly enriched, while only Nitrosomonadaceae with low relative abundances were detected in microplastic biofilms and water. Oligotrophic bacteria of clade III (affiliated with the SAR11 clade of the phylum Proteobacteria) were the main components in water, while clade III was detected at a low abundance in only one microplastic sample and was not detected in any sediment sample. These results further proved that microplastics in the Three Gorges Reservoir area provide a microbial niche different from those in the surrounding habitats.
Subsequently, the differences among the bacterial communities on microplastics, water, and sediments were further analyzed. NMDS and ANOSIM based on Bray-Curtis distance showed that the differences among the bacterial communities on microplastics were higher than those in water and sediment (Fig. 2D and E). The bacterial community similarity on microplastics was also significantly lower than those in water and sediment (Fig. 4A). This may be due to various polymer types of microplastics from different sampling sites (48, 49). In this study, the bacterial communities on microplastics at each sampling site come from no fewer than five microplastics, which included polystyrene, polypropylene, and polyethylene (Fig. 1). Previous studies also confirmed that different composition patterns of microplastic types were found along the reaches of the Three Gorges Reservoir area (3, 50), which provides more favorable evidence for our inference. Furthermore, the narrower niche distance of the bacterial community was more influenced by environmental factors (37, 51). We found that the niche distance of the bacterial community on microplastics was lower than those in water and sediment (Fig. 6). The small size and limited nutrients of microplastics may lead to competition and selection of bacteria from the surrounding environment. Thus, many microbial communities colonize in limited microplastic spaces without adequate nutrient support compared to water and sediment, limiting their niche width.
Geographical distance is an important factor shaping the biogeography patterns and assembly mechanisms of planktonic and sedimentary microbial communities in rivers such as the Yangtze River (52) and Jinsha River (53). The distance-decay relationship was one of the most common biogeographic patterns, referring to the decreasing similarity of the bacterial community and geographic distance (34, 54). Although the geographical factor was suggested to influence the bacterial community composition on microplastics collected from brackish ecosystems (55) and ocean ecosystems (56, 57), the potential correlation of bacterial community composition on microplastics with their geographical distribution was not clear. In this study, the bacterial community similarity on microplastics decreased significantly with geographic distance along the ~662-km reach of the Yangtze River, indicating that geographic distance limits were another significant factor influencing the composition of the bacterial community on microplastics.
The null model also suggested that the assembly processes of bacterial communities on microplastics were different from those in water and sediments. Dispersal limitation was the main assembly process of the bacterial community on microplastics (Fig. 5), which was consistent with the assembly process of the bacterial community on microplastics from Hangzhou Bay (22). Thus, geographic dispersal limitation dominated the assembly processes of bacterial communities on microplastics. Furthermore, selection (including variable selection and homogeneous selection) was recognized as the ecological force that alters the community due to fitness differences among organisms and environmental heterogeneity among regional pools, which was a determinism process in the absolute sense (33). In this study, variable selection was the most critical assembly process of the bacterial community, indicating that deterministic processes play the most important role in the assembly of sediment bacterial communities (Fig. 5). A previous study demonstrated that more interconnected microbial communities experienced lower turnover and susceptibility to homogenizing selection in unperturbed aquifers (58). This study found that dispersal limitation had a weak contribution to the assembly of the bacterial community in water, which was similar to the distance-decay relationship results (Fig. 4 and 5). Meanwhile, undominated and dispersal limitations contributed equally to the assembly of the bacterial community in the water. So the assembly pattern of the bacterial community in water was dominated by the stochastic process. This also indicates that the selective enrichment of the bacterial community by microplastics shapes its unique community assembly processes.
The assembly of microbial communities may involve changes in environmental factors but also interactions between species (18). Co-occurrence network analysis can help us better understand the potential interaction and niche spaces shared within the bacterial community (59). This study found that the structure and topological properties of the co-occurrence network were different among microplastics, water, and sediment, indicating that the potential interaction and niche spaces shared within microplastic bacterial communities were different from those of their surroundings (Fig. 5). More OTUs in the network of microplastics were affiliated with Burkholderiaceae, Rhodobacteraceae, and Sphingomonadaceae, indicating that these bacteria have a vital control potential within the microplastic bacterial community assembly process, which is a vital status. Previous studies have shown that Rhodobacteraceae and Sphingomonadaceae are the initial membrane colonists and maintain a preponderance in biofilm formation (60). Besides, Rhodobacteraceae and Sphingomonadaceae were also known as “initial colonizers” in the plastisphere (6). To initiate biofilm formation, pioneer microorganisms would be able to adhere to the bare substratum and then coaggregate by producing exopolysaccharides and surface adhesion proteins, which may facilitate the colonization process (6, 60). Thus, perhaps the preferential colonization effect of Rhodobacteraceae and Sphingomonadaceae on microplastics may mediate the colonization of other bacteria, leading to their vital status in the microplastic bacterial community.
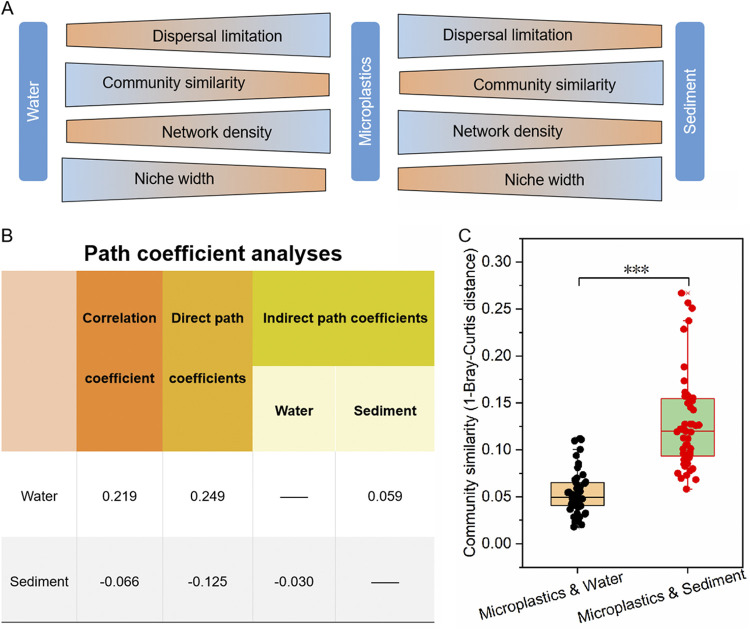
Overall, in this study, a conceptual paradigm could help us understand the relationship between bacterial community assembly processes on microplastics and those in water and sediment (Fig. 7A). The niche width and community similarity of the bacterial community on microplastics were lower than those in water and sediment. The dispersal limitation assembly process of the bacterial community on microplastics was higher than those in sediment and water, but the homogenizing dispersal assembly process of the bacterial community on microplastics was lower than those in sediment and water. Furthermore, dispersal limitation was the main assembly process of the bacterial community on microplastics, and the bacterial community similarity on microplastics and sediment significantly decreased with increasing geographical distance. Thus, geographic dispersal limitation dominated the assembly processes of bacterial communities on microplastics. Moreover, the bacterial communities’ assembly on microplastics may also be affected by their surrounding environment. This study found that the 216 families of microplastic bacteria were shared by water and sediments, indicating that the bacteria colonizing microplastics were highly dependent on the surrounding environment. However, some bacteria (e.g., Armatimonadaceae, Cyclobacteriaceae, and Gallionellaceae) were detected in both water and sediment but not on microplastics, indicating that microplastics are selective for the formation of bacterial communities on their surfaces. Microplastics collected from rivers (43), marine habitats (56), and a coastal bay close to Stockholm, Sweden (61), also showed selective enrichment of microbial communities, which was consistent with the results of this study. Surprisingly, some bacteria (Ruminococcaceae, Leptolyngbyaceae, and Phormidiaceae, etc.) were detected on microplastics but were not detected in water and sediments. These bacteria may have come from terrestrial ecosystems or other sources and colonized microplastics before the microplastics entered the aquatic environment (62, 63). These studies have described the potential sources of bacterial communities on microplastics from a quantified perspective, but there is currently a lack of quantified analyses of the contribution of surrounding habitats to colonization by microbial communities on microplastics. The path coefficient analysis showed that the direct effect of the sediment bacterial community (−0.13) on the microplastic bacterial community was lower than that of the water bacterial community (0.25). Besides, water also contributed to the accumulation of bacterial communities on microplastics through sediment, which may be related to the lotic characteristics of the river in the Three Gorges Reservoir area (39, 40). The freshwater ecosystems in the Three Gorges Reservoir have the dual characteristics of the reservoir and the river; the dam’s capacity to hold water makes its water flow backward, downstream, or relatively static (64, 65). Under the dynamic action of water, the particle-associated bacteria in the sediment could come into contact with the floating microplastics in water, resulting in the exchange of bacterial communities. Frère et al. (66) found that microplastic bacterial communities in the water had a strong similarity to particle-associated bacteria, which also supported our interpretation. This may be the reason why the composition of the core groups of suspended microplastics in water more closely resembles that of sediment than that of water.
FIG 7.
(A) Conceptual paradigm showing the relationship of bacterial community assembly processes on microplastics compared to those in water and sediment. (B) The direct and indirect effects of the bacterial community composition in water and sediment on the bacterial community assembly on microplastics were analyzed by path coefficient analysis. Among the direct effects of water and sediment on the microplastic bacterial communities, water was the largest (0.219), followed by sediment (−0.066). From the indirect effect, it was found that water also strongly contributed to the accumulation of bacterial communities on microplastics through sediment (0.059). (C) Bacterial community similarity between microplastics and water and between microplastics and sediment based on Bray-Curtis distance. The hollow squares in the box plots show the average values of community similarity, and the lines in the box plots are the median values.
Conclusion.
This study provides empirical support for the assembly processes and potential ecological mechanisms of bacterial communities on microplastics in aquatic ecosystems. Microplastics have been proven to be a new microbial niche in the aquatic environment. In this study, we further found that the bacterial community assembly on microplastics was different from that in water and sediments, which was dominated by dispersal limitation. Besides, the bacterial community similarity on microplastics significantly decreased with increasing geographical distance. Furthermore, the bacterial community niche width on microplastics was lower than those in water and sediment. In brief, geographic dispersal limitation dominated the assembly processes of bacterial communities on microplastics.
MATERIALS AND METHODS
Sample collection.
The Three Gorges Reservoir area is located in the upstream reach of the Yangtze River, China, with a length of 662 km, and includes a total water surface area of 1,080 km2 and surrounding areas of the reservoir of about 5.8 × 104 km2 (67, 68), which not only is the largest human-made reservoir in the world but also provides important ecosystem services (42). Unfortunately, this region is found to have a large number of microplastics in the surface water, with an abundance in the range of 1,597 to 12,611 items/m3 (3, 50, 69). Thus, in this study, we collected microplastic, water, and sediment samples at 7 sites along the mainstream of Yangtze River (length of 662 km) in the Three Georges Reservoir area, China (Fig. 1). At each site, the surficial sediment (0 to 5 cm) was sampled with a stainless steel core sampler, and three subsamples were collected and mixed as one sample. Microplastic samples were collected from surface water (0 to 20 cm) using a handheld portable microplastic collection device. Briefly, the surface water passed through a 5-mm stainless steel filter, the filtered water passed through a 0.05-mm stainless steel filter, and the microplastic samples enriched on the 0.05-mm stainless steel screen were then collected. Meanwhile, a 1-L water sample was collected and filtered through a 0.22-μm via a portal vacuum pump. Next, microplastic samples were collected from water and rinsed onto a 50-μm sterile sieve and a sterile forceps clip into 10-mL sterile specimen containers. All collected microplastics, filter membranes, and sediment samples were placed into a 10-mL sterile specimen container and stored in liquid nitrogen. Furthermore, the random selection of microplastics was detected by micro-Raman spectroscopy (DXR2; Thermo Fisher Scientific) (532-nm laser and Raman shift of 50 to 3,500 cm−1) according to the method described previously by Di and Wang (3), and the chemical composition of the particles was determined by comparing the obtained spectra with the database on the instrument. The results showed that the microplastics of fiber and fragments that were collected from the Three Gorges Reservoir area surface water were mainly polystyrene, polypropylene, and polyethylene (Fig. 1). However, due to the limited collection and identification of microplastics in field sampling, at least five microplastic particles were picked from each site, which led to not having all microplastic polymer types being collected at each sampling site.
DNA extraction and full-length 16S rRNA sequencing.
The bacterial DNA from each sample was extracted by using the DNeasy PowerSoil kit according to the manufacturer’s guidelines (Qiagen). After the total DNA was extracted from all samples, the primers 27F (5′-AGRGTTYGATYMTGGCTCAG-3′) and 1492R (5′-RGYTACCTTGTTACGACTT-3′), designed for the full-length 16S rRNA V1-V9 region, were synthesized with Barcode (70). The single-molecule real-time (SMRT) cell method was used to sequence marker genes (71, 72), and circular consensus sequencing (CCS) data were filtered to obtain optimized CCS for operational taxonomic unit (OTU) clustering and taxonomic annotation. The specific methods were described in detail in our previous study (41). In brief, the sequences were clustered at the level of 97% similarity (USEARCH, version 10.0) (73), and 0.005% of the number of sequenced sequences were used as the threshold to filter the OTUs of each sample (74). The taxonomic annotation of OTUs was carried out using the Silva (bacteria) taxonomic database (release 132; http://www.arb-silva.de) (75), and the confidence threshold was 0.8 (76). For comparisons across different habitats, the sequencing depth of samples was normalized to 4,100 reads via rarefaction (see Table S1 in the supplemental material).
Statistical analysis.
To determine the significance of the differences in community composition among the three habitats, we used the Bray-Curtis distance to calculate the NMDS plot and perform ANOSIM. Besides, the bacterial community similarities at the OTUs level were estimated based on 1-Bray-Curtis distance between samples using the vegan package (version 2.5-7) in R. Meanwhile, the geographic distance between the sampling sites was determined based on the geographic coordinates, and the correlation between community similarity and geographical distance was then evaluated via the ggplot 2 package (version 3.3.5) in R. Previous studies have found that limma voom has good performance in composition difference analyses (77, 78), so the composition differences in bacterial communities among the microplastics, water, and sediment were described via limma Bioconductor packages (version 3.38.3) in this study.
The null model was applied via the picante package (version 1.8.2) in R to analyze phylogenetic and taxonomic β-diversity metrics (β-nearest taxon index, βNTI and value of Bray–Curtis-based Raup–Crick, RCBray) of bacterial communities on microplastics, in water, and in sediment, according to methods described in a previous study (29). A βNTI of greater than 2 was defined as variable selection (more phylogenetic turnover than expected), and a βNTI of less than −2 was defined as homogeneous selection (less phylogenetic turnover than expected). Furthermore, an absolute value of βNTI (|βNTI|) more than 2 expressed the dominance of selection processes. Subsequently, RCBray was used to further partition |βNTI| values of <2. A |βNTI| of less than 2 and an RCBray of greater than 0.95 were defined as dispersal limitation, a |βNTI| of less than 2 and an RCBray of less than −0.95 were defined as homogenizing dispersal, and a |βNTI| of less than 2 and an |RCBray| of less than 0.95 were defined as undominated (consisting mostly of weak selection, weak dispersal, diversification, and/or drift) (29, 31). Besides, to help reveal the effects of habitats and transmission constraints on bacterial communities, the niche distance of bacterial communities was evaluated via the spaa package (version 0.2.2) in R. Meanwhile, the direct or indirect effects of sediment and water bacterial communities on the assembly of bacterial communities on microplastics were described based on path coefficient analysis via SPSS (version 19.0). Co-occurrence networks of bacterial communities in different habitats were constructed based on a strong (|r| > 0.8) and significant (P < 0.001) Spearman correlation at the OTU level. The co-occurrence networks of bacterial communities in different habitats (microplastics, water, and sediment) were visualized via Gephi (version 0.9.2), and the topological properties (including average weighted degree, average path length, average clustering coefficient, modularity, and number of modules) of the co-occurrence network were also calculated via Gephi (version 0.9.2).
Data availability.
The raw full-length 16S rRNA gene sequences generated in the present study were deposited in the Sequence Read Archive (SRA) database (https://submit.ncbi.nlm.nih.gov/subs/sra/) under accession number PRJNA647658.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grant number 32071614); the Starting Research Fund and Opening Research Fund from the Key Laboratory of Aquatic Botany and Watershed Ecology, Chinese Academy of Sciences (grant numbers Y9519802 and E0520202); and the Funding Project of the Sino-Africa Joint Research Center, Chinese Academy of Sciences (grant number E0291P0101).
We declare that we have no competing interests.
Weihong Zhang and Yuyi Yang, conceptualization, methodology, and writing – original draft preparation; Lu Chen, visualization and software; Wenzhi Liu, software and validation; Haiyang Chen and Yuyi Yang, writing – reviewing and editing.
Footnotes
Supplemental material is available online only.
Contributor Information
Yuyi Yang, Email: yangyy@wbgcas.cn.
Laura Villanueva, Royal Netherlands Institute for Sea Research.
REFERENCES
- 1.Amon DJ, Kennedy BRC, Cantwel K, Suhre K, Glickson D, Shank TM, Rotjan RD. 2020. Deep-sea debris in the Central and Western Pacific Ocean. Front Mar Sci 7:369. 10.3389/fmars.2020.00369. [DOI] [Google Scholar]
- 2.Kelly A, Lannuzel D, Rodemann T, Meiners KM, Auman HJ. 2020. Microplastic contamination in East Antarctic sea ice. Mar Pollut Bull 154:111130. 10.1016/j.marpolbul.2020.111130. [DOI] [PubMed] [Google Scholar]
- 3.Di M, Wang J. 2018. Microplastics in surface waters and sediments of the Three Gorges Reservoir, China. Sci Total Environ 616–617:1620–1627. 10.1016/j.scitotenv.2017.10.150. [DOI] [PubMed] [Google Scholar]
- 4.Xiong X, Wu C, Elser JJ, Mei Z, Hao Y. 2019. Occurrence and fate of microplastic debris in middle and lower reaches of the Yangtze River—from inland to the sea. Sci Total Environ 659:66–73. 10.1016/j.scitotenv.2018.12.313. [DOI] [PubMed] [Google Scholar]
- 5.Yuan W, Liu X, Wang W, Di M, Wang J. 2019. Microplastic abundance, distribution and composition in water, sediments, and wild fish from Poyang Lake, China. Ecotoxicol Environ Saf 170:180–187. 10.1016/j.ecoenv.2018.11.126. [DOI] [PubMed] [Google Scholar]
- 6.Di Pippo F, Venezia C, Sighicelli M, Pietrelli L, Di Vito S, Nuglio S, Rossetti S. 2020. Microplastic-associated biofilms in lentic Italian ecosystems. Water Res 187:116429. 10.1016/j.watres.2020.116429. [DOI] [PubMed] [Google Scholar]
- 7.Yang Y, Liu W, Zhang Z, Grossart H, Gadd G. 2020. Microplastics provide new microbial niches in aquatic environments. Appl Microbiol Biotechnol 104:6501–6511. 10.1007/s00253-020-10704-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckert E, Di Cesare A, Kettner M, Arias-Andres M, Fontaneto D, Grossart HP, Corno G. 2018. Microplastics increase impact of treated wastewater on freshwater microbial community. Environ Pollut 234:495–502. 10.1016/j.envpol.2017.11.070. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Huang M, Wang Q, Sun Y, Zhao Y, Huang Y. 2020. LDPE microplastics significantly alter the temporal turnover of soil microbial communities. Sci Total Environ 726:138682. 10.1016/j.scitotenv.2020.138682. [DOI] [PubMed] [Google Scholar]
- 10.Jiang P, Zhao S, Zhu L, Li D. 2018. Microplastic-associated bacterial assemblages in the intertidal zone of the Yangtze Estuary. Sci Total Environ 624:48–54. 10.1016/j.scitotenv.2017.12.105. [DOI] [PubMed] [Google Scholar]
- 11.McCormick A, Hoellein TJ, Mason SA, Schluep J, Kelly JJ. 2014. Microplastic is an abundant and distinct microbial habitat in an urban river. Environ Sci Technol 48:11863–11871. 10.1021/es503610r. [DOI] [PubMed] [Google Scholar]
- 12.Gong M, Yang G, Zhuang L, Zeng EY. 2019. Microbial biofilm formation and community structure on low-density polyethylene microparticles in lake water microcosms. Environ Pollut 252:94–102. 10.1016/j.envpol.2019.05.090. [DOI] [PubMed] [Google Scholar]
- 13.Wu X, Pan J, Li M, Li Y, Bartlam M, Wang Y. 2019. Selective enrichment of bacterial pathogens by microplastic biofilm. Water Res 165:114979. 10.1016/j.watres.2019.114979. [DOI] [PubMed] [Google Scholar]
- 14.Li J, Huang W, Jiang R, Han X, Zhang D, Zhang C. 2020. Are bacterial communities associated with microplastics influenced by marine habitats? Sci Total Environ 733:139400. 10.1016/j.scitotenv.2020.139400. [DOI] [PubMed] [Google Scholar]
- 15.Oberbeckmann S, Löder MGJ, Labrenz M. 2015. Marine microplastic-associated biofilms—a review. Environ Chem 12:551–562. 10.1071/EN15069. [DOI] [Google Scholar]
- 16.Rivett D, Bell T. 2018. Abundance determines the functional role of bacterial phylotypes in complex communities. Nat Microbiol 3:767–772. 10.1038/s41564-018-0180-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan Y, Kuramae E, de Hollander M, Klinkhamer P, van Veen J. 2017. Functional traits dominate the diversity-related selection of bacterial communities in the rhizosphere. ISME J 11:56–66. 10.1038/ismej.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang W, Wan W, Lin H, Pan X, Lin L, Yang Y. 2022. Nitrogen rather than phosphorus driving the biogeographic patterns of abundant bacterial taxa in a eutrophic plateau lake. Sci Total Environ 806:150947. 10.1016/j.scitotenv.2021.150947. [DOI] [PubMed] [Google Scholar]
- 19.Amaral-Zettler L, Zettler E, Slikas B, Boyd G, Melvin D, Morrall C, Proskurowski G, Mincer T. 2015. The biogeography of the plastisphere: implications for policy. Front Ecol Environ 13:541–546. 10.1890/150017. [DOI] [Google Scholar]
- 20.Coons A, Busch K, Lenz M, Hentschel U, Borchert E. 2021. Biogeography rather than substrate type determines bacterial colonization dynamics of marine plastics. PeerJ 9:e12135. 10.7717/peerj.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang S, Zeng Y, Zhu J, Cai Z, Zhou J. 2022. The structure and assembly mechanisms of plastisphere microbial community in natural marine environment. J Hazard Mater 421:126780. 10.1016/j.jhazmat.2021.126780. [DOI] [PubMed] [Google Scholar]
- 22.Sun Y, Zhang M, Duan C, Cao N, Jia W, Zhao Z, Ding C, Huang Y, Wang J. 2021. Contribution of stochastic processes to the microbial community assembly on field-collected microplastics. Environ Microbiol 23:6707–6720. 10.1111/1462-2920.15713. [DOI] [PubMed] [Google Scholar]
- 23.Chave J. 2004. Neutral theory and community ecology. Ecol Lett 7:241–253. 10.1111/j.1461-0248.2003.00566.x. [DOI] [Google Scholar]
- 24.Hubbell SP. 2005. Neutral theory in community ecology and the hypothesis of functional equivalence. Funct Ecol 19:166–172. 10.1111/j.0269-8463.2005.00965.x. [DOI] [Google Scholar]
- 25.Tilman D. 2004. Niche tradeoffs, neutrality, and community structure: a stochastic theory of resource competition, invasion, and community assembly. Proc Natl Acad Sci USA 101:10854–10861. 10.1073/pnas.0403458101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chu H, Sun H, Tripathi BM, Adams JM, Huang R, Zhang Y, Shi Y. 2016. Bacterial community dissimilarity between the surface and subsurface soils equals horizontal differences over several kilometers in the western Tibetan Plateau. Environ Microbiol 18:1523–1533. 10.1111/1462-2920.13236. [DOI] [PubMed] [Google Scholar]
- 27.Martiny J, Bohannan B, Brown J, Colwell R, Fuhrman J, Green J, Horner-Devine M, Kane M, Krumins J, Kuske C, Morin P, Naeem S, Øvreås L, Reysenbach A, Smith V, Staley J. 2006. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol 4:102–112. 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- 28.Zhang K, Adams JM, Shi Y, Yang T, Sun R, He D, Ni Y, Chu H. 2017. Environment and geographic distance differ in relative importance for determining fungal community of rhizosphere and bulk soil. Environ Microbiol 19:3649–3659. 10.1111/1462-2920.13865. [DOI] [PubMed] [Google Scholar]
- 29.Jiao S, Yang Y, Xu Y, Zhang J, Lu Y. 2020. Balance between community assembly processes mediates species coexistence in agricultural soil microbiomes across eastern China. ISME J 14:202–216. 10.1038/s41396-019-0522-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ning D, Yuan M, Wu L, Zhang Y, Guo X, Zhou X, Yang Y, Arkin A, Firestone M, Zhou J. 2020. A quantitative framework reveals ecological drivers of grassland microbial community assembly in response to warming. Nat Commun 11:4717. 10.1038/s41467-020-18560-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stegen JC, Lin X, Fredrickson JK, Konopka AE. 2015. Estimating and mapping ecological processes influencing microbial community assembly. Front Microbiol 6:370. 10.3389/fmicb.2015.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stegen JC, Lin X, Konopka AE, Fredrickson JK. 2012. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J 6:1653–1664. 10.1038/ismej.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou J, Ning D. 2017. Stochastic community assembly: does it matter in microbial ecology? Microbiol Mol Biol Rev 81:e00002-17. 10.1128/MMBR.00002-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zinger L, Boetius A, Ramette A. 2014. Bacterial taxa-area and distance-decay relationships in marine environments. Mol Ecol 23:954–964. 10.1111/mec.12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fodelianakis S, Lorz A, Valenzuela-Cuevas A, Barozzi A, Booth JM, Daffonchio D. 2019. Dispersal homogenizes communities via immigration even at low rates in a simplified synthetic bacterial metacommunity. Nat Commun 10:1314. 10.1038/s41467-019-09306-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zha Y, Lindstrom ES, Eiler A, Svanback R. 2020. Different roles of environmental selection, dispersal, and drift in the assembly of intestinal microbial communities of freshwater fish with and without a stomach. Front Ecol Evol 8:152. 10.3389/fevo.2020.00152. [DOI] [Google Scholar]
- 37.Wu W, Lu HP, Sastri A, Yeh YC, Gong GC, Chou WC, Hsie CH. 2018. Contrasting the relative importance of species sorting and dispersal limitation in shaping marine bacterial versus protist communities. ISME J 12:485–494. 10.1038/ismej.2017.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X, Lu X, Yao J, Wang Z, Deng Y, Cheng W, Zhou J, Han X. 2017. Habitat-specific patterns and drivers of bacterial beta-diversity in China’s drylands. ISME J 11:1345–1358. 10.1038/ismej.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gweon H, Bowes M, Moorhouse H, Oliver A, Bailey M, Acreman M, Read D. 2021. Contrasting community assembly processes structure lotic bacteria metacommunities along the river continuum. Environ Microbiol 23:484–498. 10.1111/1462-2920.15337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malki K, Sawaya N, Tisza M, Coutinho F, Rosario K, Szekely A, Breitbart M. 2021. Spatial and temporal dynamics of prokaryotic and viral community assemblages in a lotic system (Manatee Springs, Florida). Appl Environ Microbiol 87:e00646-21. 10.1128/AEM.00646-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang W, Yuan W, Chen L, Ye C, Jiang Y, Yang Y. 2021. Uniqueness and dependence of bacterial communities on microplastics: comparison with water, sediment, and soil. Microb Ecol 10.1007/s00248-021-01919-0. [DOI] [PubMed]
- 42.Zhang P, Tan Z, Hu Q, Liu J. 2019. Geological impact of the Three Gorges Reservoir on the Yangtze River in China. Environ Earth Sci 78:443. 10.1007/s12665-019-8461-3. [DOI] [Google Scholar]
- 43.Wang J, Qin X, Guo J, Jia W, Wang Q, Zhang M, Huang Y. 2020. Evidence of selective enrichment of bacterial assemblages and antibiotic resistant genes by microplastics in urban rivers. Water Res 183:116113. 10.1016/j.watres.2020.116113. [DOI] [PubMed] [Google Scholar]
- 44.Zettler ER, Mincer TJ, Amaral-Zettler LA. 2013. Life in the “plastisphere”: microbial communities on plastic marine debris. Environ Sci Technol 47:7137–7146. 10.1021/es401288x. [DOI] [PubMed] [Google Scholar]
- 45.Rummel CD, Jahnke A, Gorokhova E, Kühnel D, Schmitt-Jansen M. 2017. Impacts of biofilm formation on the fate and potential effects of microplastic in the aquatic environment. Environ Sci Technol Lett 4:258–267. 10.1021/acs.estlett.7b00164. [DOI] [Google Scholar]
- 46.Sun R, Tu Z, Fan L, Qiao Z, Liu X, Hu S, Zheng G, Wu Y, Wang R, Mi X. 2020. The correlation analyses of bacterial community composition and spatial factors between freshwater and sediment in Poyang Lake wetland by using artificial neural network (ANN) modeling. Braz J Microbiol 51:1191–1207. 10.1007/s42770-020-00285-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prosser JI, Head IM, Stein LY. 2014. The family Nitrosomonadaceae, p 901–918. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes: alphaproteobacteria and betaproteobacteria, 4th ed. Springer, Berlin, Germany. 10.1007/978-3-642-30197-1_372. [DOI] [Google Scholar]
- 48.Jacquin J, Cheng J, Odobel C, Pandin C, Conan P, Pujo-Pay M, Barbe V, Meistertzheim A-L, Ghiglione J-F. 2019. Microbial ecotoxicology of marine plastic debris: a review on colonization and biodegradation by the “Plastisphere.” Front Microbiol 10:865. 10.3389/fmicb.2019.00865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oberbeckmann S, Kreikemeyer B, Labrenz M. 2018. Environmental factors support the formation of specific bacterial assemblages on microplastics. Front Microbiol 8:2709. 10.3389/fmicb.2017.02709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang K, Xiong X, Hu H, Wu C, Bi Y, Wu Y, Zhou B, Lam PKS, Liu J. 2017. Occurrence and characteristics of microplastic pollution in Xiangxi Bay of Three Gorges Reservoir, China. Environ Sci Technol 51:3794–3801. 10.1021/acs.est.7b00369. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y, Li D, Qi J, Peng Z, Chen W, Wei G, Jiao S. 2021. Stochastic processes shape the biogeographic variations in core bacterial communities between aerial and belowground compartments of common bean. Environ Microbiol 23:949–964. 10.1111/1462-2920.15227. [DOI] [PubMed] [Google Scholar]
- 52.Liu T, Zhang AN, Wang J, Liu S, Jiang X, Dang C, Ma T, Liu S, Chen Q, Xie S, Zhang T, Ni J. 2018. Integrated biogeography of planktonic and sedimentary bacterial communities in the Yangtze River. Microbiome 6:16. 10.1186/s40168-017-0388-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen J, Wang P, Wang C, Wang X, Miao L, Liu S, Yuan Q, Sun S. 2020. Fungal community demonstrates stronger dispersal limitation and less network connectivity than bacterial community in sediments along a large river. Environ Microbiol 22:832–849. 10.1111/1462-2920.14795. [DOI] [PubMed] [Google Scholar]
- 54.Morlon H, Chuyong G, Condit R, Hubbell S, Kenfack D, Thomas D, Valencia R, Green JL. 2008. A general framework for the distance-decay of similarity in ecological communities. Ecol Lett 11:904–917. 10.1111/j.1461-0248.2008.01202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kettner MT, Oberbeckmann S, Labrenz M, Grossart HP. 2019. The eukaryotic life on microplastics in brackish ecosystems. Front Microbiol 10:538. 10.3389/fmicb.2019.00538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oberbeckmann S, Labrenz M. 2020. Marine microbial assemblages on microplastics: diversity, adaptation, and role in degradation. Annu Rev Mar Sci 12:209–232. 10.1146/annurev-marine-010419-010633. [DOI] [PubMed] [Google Scholar]
- 57.Pinto M, Langer TM, Hüffer T, Hofmann T, Herndl GJ. 2019. The composition of bacterial communities associated with plastic biofilms differs between different polymers and stages of biofilm succession. PLoS One 14:e0217165. 10.1371/journal.pone.0217165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Danczak RE, Johnston MD, Kenah C, Slattery M, Wilkins MJ. 2018. Microbial community cohesion mediates community turnover in unperturbed aquifers. mSystems 3:e00066-18. 10.1128/mSystems.00066-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma B, Wang H, Dsouza M, Lou J, He Y, Dai Z, Brookes PC, Xu J, Gilbert JA. 2016. Geographic patterns of co-occurrence network topological features for soil microbiota at continental scale in eastern China. ISME J 10:1891–1901. 10.1038/ismej.2015.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Vries HJ, Beyer F, Jarzembowska M, Lipińska J, van den Brink P, Zwijnenburg A, Timmers PHA, Stams AJM, Plugge CM. 2019. Isolation and characterization of Sphingomonadaceae from fouled membranes. NPJ Biofilms Microbiomes 5:6. 10.1038/s41522-018-0074-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ogonowski M, Motiei A, Ininbergs K, Hell E, Gerdes Z, Udekwu KI, Bacsik Z, Gorokhova E. 2018. Evidence for selective bacterial community structuring on microplastics. Environ Microbiol 20:2796–2808. 10.1111/1462-2920.14120. [DOI] [PubMed] [Google Scholar]
- 62.Bellasi A, Binda G, Pozzi A, Galafassi S, Volta P, Bettinetti R. 2020. Microplastic contamination in freshwater environments: a review, focusing on interactions with sediments and benthic organisms. Environments 7:30. 10.3390/environments7040030. [DOI] [Google Scholar]
- 63.Mammo F, Amoah ID, Gani KM, Pillay L, Ratha SK, Bux F, Kumari S. 2020. Microplastics in the environment: interactions with microbes and chemical contaminants. Sci Total Environ 743:140518. 10.1016/j.scitotenv.2020.140518. [DOI] [PubMed] [Google Scholar]
- 64.Li Y, Shang Q, Jiang J, Da B, Gao Y. 2019. Study on processes of Changshou Waterway after 175m experimental impoundment of the Three Gorges Reservoir. IOP Conf Ser Earth Environ Sci 304:e022025. 10.1088/1755-1315/304/2/022025. [DOI] [Google Scholar]
- 65.Li Y, Zhang Q, Werner AD, Yao J, Ye X. 2017. The influence of river-to-lake backflow on the hydrodynamics of a large floodplain lake system (Poyang Lake, China). Hydrol Process 31:117–132. 10.1002/hyp.10979. [DOI] [Google Scholar]
- 66.Frère L, Maignien L, Chalopin M, Huvet A, Rinnert E, Morrison H, Kerninon S, Cassone A-L, Lambert C, Reveillaud J, Paul-Pont I. 2018. Microplastic bacterial communities in the Bay of Brest: influence of polymer type and size. Environ Pollut 242:614–625. 10.1016/j.envpol.2018.07.023. [DOI] [PubMed] [Google Scholar]
- 67.Bao Y, Gao P, He X. 2015. The water-level fluctuation zone of Three Gorges Reservoir—a unique geomorphological unit. Earth Sci Rev 150:14–24. 10.1016/j.earscirev.2015.07.005. [DOI] [Google Scholar]
- 68.Zhang Q, Lou Z. 2011. The environmental changes and mitigation actions in the Three Gorges Reservoir region, China. Environ Sci Policy 14:1132–1138. 10.1016/j.envsci.2011.07.008. [DOI] [Google Scholar]
- 69.Zhang K, Shi H, Peng J, Wang Y, Xiong X, Wu C, Lam PK. 2018. Microplastic pollution in China’s inland water systems: a review of findings, methods, characteristics, effects, and management. Sci Total Environ 630:1641–1653. 10.1016/j.scitotenv.2018.02.300. [DOI] [PubMed] [Google Scholar]
- 70.Numberger D, Ganzert L, Zoccarato L, Mühldorfer K, Sauer S, Grossart H-P, Greenwood AD. 2019. Characterization of bacterial communities in wastewater with enhanced taxonomic resolution by full-length 16S rRNA sequencing. Sci Rep 9:9673. 10.1038/s41598-019-46015-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Burke CM, Darling AE. 2016. A method for high precision sequencing of near full-length 16S rRNA genes on an Illumina MiSeq. PeerJ 4:e2492. 10.7717/peerj.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cabello-Yeves PJ, Rodriguez-Valera F. 2019. Marine-freshwater prokaryotic transitions require extensive changes in the predicted proteome. Microbiome 7:117. 10.1186/s40168-019-0731-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bokulich N, Subramanian S, Faith J, Gevers D, Gordon J, Knight R, Mills D, Caporaso JD. 2013. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10:57–59. 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and Web-based tools. Nucleic Acids Res 41:D590–D596. 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nearing JT, Douglas GM, Hayes MG, MacDonald J, Desai DK, Allward N, Jones CMA, Wright RJ, Dhanani AS, Comeau AM, Langille MGI. 2022. Microbiome differential abundance methods produce disturbingly different results across 38 datasets. Nat Commun 13:342. 10.1038/s41467-022-28034-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Calgaro M, Romualdi C, Waldron L, Risso D, Vitulo N. 2020. Assessment of statistical methods from single cell, bulk RNA-seq, and metagenomics applied to microbiome data. Genome Biol 21:191. 10.1186/s13059-020-02104-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and descriptions of Tables S1 to S4. Download aem.00482-22-s0001.pdf, PDF file, 0.4 MB (368.9KB, pdf)
Tables S1 to S4. Download aem.00482-22-s0002.xlsx, XLSX file, 0.3 MB (257.4KB, xlsx)
Data Availability Statement
The raw full-length 16S rRNA gene sequences generated in the present study were deposited in the Sequence Read Archive (SRA) database (https://submit.ncbi.nlm.nih.gov/subs/sra/) under accession number PRJNA647658.