ABSTRACT
Comamonas spp. are Gram-negative bacteria that catabolize a wide range of organic and inorganic substrates. Comamonas spp. are abundant in aquatic and soil environments, including wastewater, and can cause opportunistic infections in humans. Because of their potential in wastewater bioaugmentation and bioremediation strategies, the identification of Comamonas species harboring genes encoding carbapenemases and other clinically important antibiotic resistance genes warrant further investigation. Here, we present an analysis of 39 whole-genome sequences comprising three Comamonas species from aquatic environments in South Australia that were recovered on media supplemented with carbapenems. The analysis includes a detailed description of 33 Comamonas denitrificans isolates, some of which carried chromosomally acquired blaGES-5, blaOXA, and aminoglycoside resistance (aadA) genes located on putative genomic islands (GIs). All blaGES-5- and blaOXA-containing GIs appear to be unique to this Australian collection of C. denitrificans. Notably, most open reading frames (ORFs) within the GIs, including all antimicrobial resistance (AMR) genes, had adjacent attC sites, indicating that these ORFs are mobile gene cassettes. One C. denitrificans isolate carried an IncP-1 plasmid with genes involved in xenobiotic degradation and response to oxidative stress. Our assessment of the sequences highlights the very distant nature of C. denitrificans to the other Comamonas species and its apparent disposition to acquire antimicrobial resistance genes on putative genomic islands.
IMPORTANCE Antimicrobial resistance (AMR) poses a global public health threat, and the increase in resistance to “last-resort drugs,” such as carbapenems, is alarming. Wastewater has been flagged as a hot spot for AMR evolution. Comamonas spp. are among the most common bacteria in wastewater and play a role in its bioaugmentation. While the ability of Comamonas species to catabolize a wide range of organic and inorganic substrates is well documented, some species are also opportunistic pathogens. However, data regarding AMR in Comamonas spp. are limited. Here, through the genomic analyses of 39 carbapenem-resistant Comamonas isolates, we make several key observations, including the identification of a subset of C. denitrificans isolates that harbored genomic islands encoding carbapenemase blaGES-5 or extended-spectrum β-lactamase blaOXA alleles. Given the importance of Comamonas species in potential wastewater bioaugmentation and bioremediation strategies, as well as their status as emerging pathogens, the acquisition of critically important antibiotic resistance genes on genomic islands warrants future monitoring.
KEYWORDS: Comamonas denitrificans, wastewater, whole-genome sequencing (WGS), genomic islands, carbapenemase, incP-1, antimicrobial resistance (AMR), Comamonas, bla GES-5
INTRODUCTION
Comamonas spp. are Gram-negative bacilli that reside within the family Comamonadaceae and the phylum Proteobacteria (1). Comamonas spp. are commonly found in a variety of environmental samples and are among the common bacteria in soil (2–4), wetlands (5–7), and wastewater (8, 9). The ability of Comamonas spp. to catabolize a wide range of organic and inorganic substrates, including amino acids, aromatic compounds, carboxylic acids, heavy metals, and steroids, has been extensively documented (10–12). Furthermore, several studies have documented the roles Comamonas spp. can play in wastewater bioaugmentation and bioremediation (13–15). Comamonas denitrificans was first identified because of its significant ability to grow in anoxic conditions and denitrify sludge (16, 17), a by-product during sewage treatment rich in antimicrobial residues, metals, pharmaceuticals, and drug-resistant microbial communities (18, 19). Unlike other species in the Comamonas genus, C. denitrificans can reduce nitrate to nitrogen gas in both aerobic and anaerobic conditions (16) and has a notable capacity of biofilm formation (20).
While Comamonas spp. regularly feature in environmental microbial communities, several species have been described as aggressive opportunistic pathogens capable of surviving in hospital environments (21). Reports of Comamonas spp. causing disease in humans have been increasing in multiple countries across six continents (22–28). Comamonas testosteroni is the species most frequently associated with human disease and is known to cause invasive infections such as cellulitis (29), peritonitis (30), endocarditis (31), meningitis (32), endophthalmitis (33), pneumonia (34), and appendicitis associated with bacteremia (25). However, other Comamonas species, such as Comamonas terrigena and Comamonas kerstersii, can also cause pathologies, including eye and intra-abdominal infections (35, 36). Due to the omnipresence of Comamonas spp. in the environment and its apparent capacity to cause opportunistic infection, it is important to understand whether members of the genus display resistance to antibiotics. Antimicrobial resistance (AMR) poses a global public health threat, and the increase in resistance to “last-resort drugs,” such as carbapenems, is alarming (37–39). The environment is an important reservoir of resistant bacteria, antimicrobial-resistant genes (ARGs) and antimicrobial residues, and there is mounting evidence suggesting the dispersal of environmental and clinical ARGs between wildlife, agriculture, and humans (40–42). Carbapenem-resistant bacteria have been isolated from natural water bodies (43, 44), wastewater (45–47), wildlife, and synanthropic species that frequent wastewater (42, 48). Airline waste (49) and wastewater in particular has been flagged as a hot spot for AMR evolution given the high abundance of bacteria combined with sublethal antibiotic concentrations derived from anthropogenic sources, including agriculture, industry, hospitals, and households (19, 47).
To date, only one carbapenem resistance gene has been identified in any Comamonas spp. (blaIMP-8 in Comamonas thiooxydans) (50), and in general, data regarding ARG carriage in Comamonas are limited. Published case reports suggest that C. testosteroni is susceptible to common antibiotics such as aminoglycosides, fluoroquinolones, ceftazidime, carbapenems, and piperacillin-tazobactam (1, 51). However, in a severe meningitis case caused by C. testosteroni, the pathogen was resistant to broad spectrum cephalosporin but sensitive to carbapenem (32). C. denitrificans has been reported to survive high doses of amoxicillin (52), and a genomic analysis of a Comamonas aquatica isolate from the Daguan River, China, identified genes encoding resistance to several antibiotics, including cephalosporins, penicillins, and bacitracin; however, the genes that could account for these resistances were not reported (53).
Given the growing number of studies identifying carbapenem-resistant bacteria in water matrices and the abundance of Comamonas in the environment, this study sought to shed light on the potential for Comamonas species to acquire genes that encode resistance to critically important antibiotics in Australia. Here, we provide a whole-genome sequence (WGS) and phylogenetic analysis of 39 Comamonas spp. isolates that grew on Oxoid Brilliance CRE agar plates, including 33 diverse C. denitrificans isolates, the majority of which carried acquired chromosomal carbapenemase blaGES-5 and β-lactamase blaOXA genes.
RESULTS
From a total of 471 isolates cultured from CRE-agar plates inoculated with samples from aquatic environments in South Australia, including wastewater, wetland, and lake samples from 2018 to 2019, 39 isolates were Comamonas spp. Comamonas constituted the third most prevalent genus isolated from wastewater and were the subject of this study. Most Comamonas spp. were sourced from influent wastewater (n = 37; 94.8%) with single isolates from a lake (n = 1; 2.6%) and a wetland (n = 1; 2.6%). The associated metadata on all 39 isolates described here, as well as 48 Comamonas spp. sourced from GenBank, are available in Data Set S1.
Genome assembly.
Draft genomes were assembled using Shovill version 1.0.4. Consistent with an earlier whole-genome sequencing study (27), genome sizes ranged between 2,782,493 and 4,055,492 bp with an average size of 3,101,902 bp. The number of scaffolds per genome ranged from 29 to 321, with a mean of 128.9. Full assembly statistics can be viewed in Data Set S2. Draft genomes have been deposited in the NCBI database under the accession numbers SAMN25632339 to SAMN25632377 and BioProject PRJNA803140.
Identification of Comamonas species.
Identification of Comamonas isolates to the species level varied between the typing techniques used (Table 1). Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) of the 39 isolates sourced from Australian aquatic environments predicted Comamonas sp. for 25 (62.5%) and one C. aquatica. Kraken2 predicted 28 isolates as Comamonas species (72%), but some were misidentified as Pseudomonas aeruginosa (n = 1) and Acidovorax spp. (n = 10). Using our phylogenetic approach, all 39 isolates were identified as Comamonas spp., specifically 33 C. denitrificans (84.6%), 5 C. testosteroni (12.5%), and 1 C. aquatica (2.5%) isolate.
TABLE 1.
Comamonas identification to the species level
| Species identification | MALDI-TOF MS | Kraken2a | WGS phylogenetic characterization |
|---|---|---|---|
| C. aquatic | 1 (2.6%) | 12 (30.7%) | 1 (2.6%) |
| C. testosterone | 0 | 6 (15.4%) | 5 (12.8%) |
| C. denitrificans | 0 | 0 | 33 (84.6%) |
| C. kerstersii | 0 | 8 (20.5%) | 0 |
| Comamonas sp. | 25 (64.1%) | 2 (5.1%) | 0 |
| Nonreliable identification | 13 (33.3%) | 0 | 0 |
aKraken2 typing also resulted in P. aeruginosa (n = 1; 2.6%), Acidovorax carolinensis (n = 2; 5.1%), and Acidovorax spp. (n = 8; 20.5%).
Phylogenetic analysis.
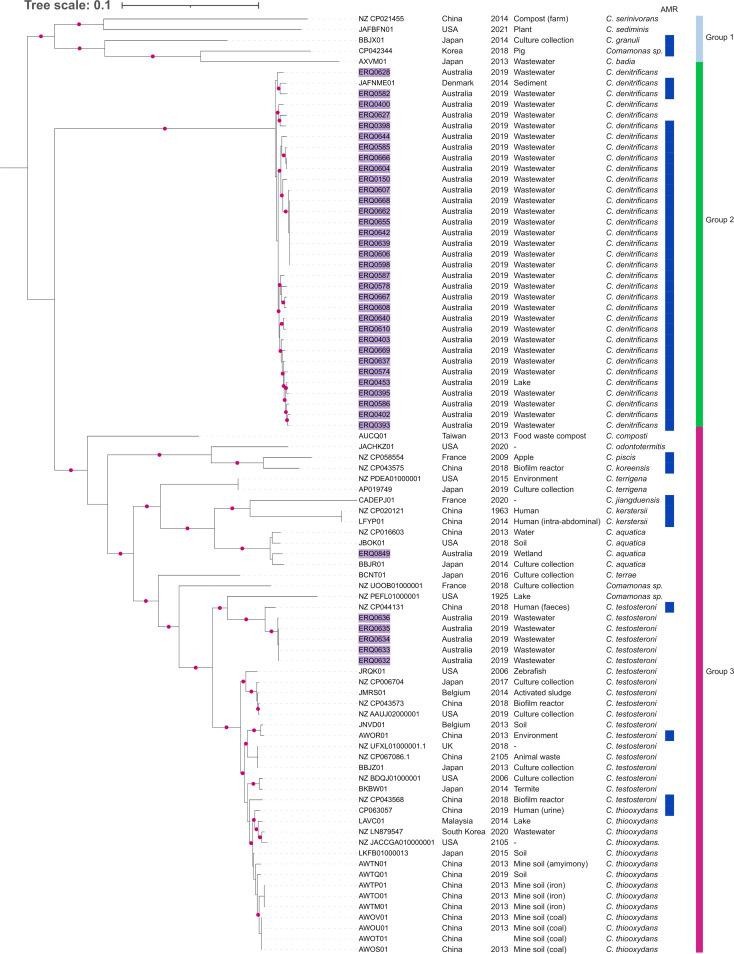
A phylogenetic tree comprising 88 Comamonas spp. genomes was constructed using Phylosift (Fig. 1) with 39 isolates from the Australian aquatic environment in this collection and 49 global strains available from GenBank. The isolates were derived from diverse sources, primarily environmental samples (n = 75), humans (n = 5), and various animal waste (n = 5). The count of species postphylogeny were as follows: C. denitrificans (n = 34), C. testosteroni (n = 19), Comamonas thiooxydens (n = 14), C. aquatica (n = 4), C. kerstersii (n = 2), Comamonas terrigena (n = 2), and one isolate each of Comamonas badia, Comamonas terrae, Comamonas composti, Comamonas granuli, Comamonas jiangduensis, Comamonas koreensis, Comamonas odontotermitis, Comamonas piscis, Comamonas sediminis, and Comamonas serinivorans. Three isolates could not be characterized to the species level.
FIG 1.
Comamonas phylogeny. Midpoint-rooted maximum-likelihood phylogeny and associated metadata of 88 Comamonas species using Phylosift. Isolates from this collection are colored in purple. Bootstrap values greater than 0.9 are shown as pink dots. The blue strip designates isolates carrying antimicrobial-resistant genes (ARGs). AMR, antimicrobial resistance.
Based on the distribution of sequence alignments by PhyloSift, Comamonas species were separated into three primary clusters, with a clear separation between species in all cases except C. testosteroni and C. thiooxydans, which were closely related. C. denitrificans was notably separated from other Comamonas species, mirroring prior analyses of RNA sequences (54).
Comamonas group 1 (Fig. 1) comprised five species: C. serinivorans, C. sediminis, C. granuli, C. badia, and the undefined Comamonas sp. strain NLF 7-7 (Sus scrofa, South Korea, CP042344). Group 2 was defined as C. denitrificans only, including the environmental/wastewater isolates from this study plus the only available C. denitrificans genome sequence from GenBank (JAFNME01, from sediment, Denmark). Group 3 constituted isolates across at least 11 species based on current nomenclature, with clade structure suggesting that up to 13 species may be present. Our five C. testosteroni isolates were most closely related to C. testosteroni strain NFYY023 (NZ_CP044131) from human feces, which sits apart from the C. testosteroni/C. thiooxydans subclade; however, C. testosteroni strain NFYY023 was ~90,000 single-nucleotide polymorphisms (SNPs) from the closest C. testosteroni from this collection. It is likely that our five isolates constitute a closely related yet distinct species to C. testosteroni. Using the NCBI C. testosteroni representative strain G1 (NZ_CP067086) as a reference, we performed average nucleotide identity BLASTn (ANIb), ANI average nucleotide identity MUMer (ANIm), and in silico DNA-DNA hybridization (DDH) analyses to determine whether the isolates belong to C. testosteroni. All five isolates had ANIb and ANIm values of 83 and 87%, respectively, which are below the 95% cutoff values for species delimitation, and the in silico DDH analysis returned a 0.04% probability of being the same species as NZ_CP067086 (Data Set S3). Definitive novel species characterizations will be the subject of future studies.
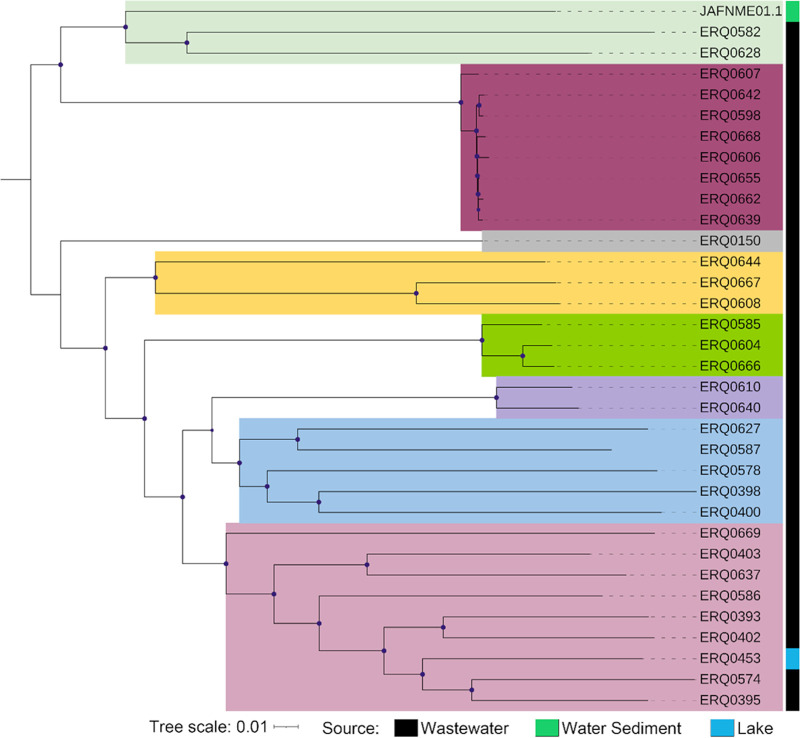
To illustrate the more nuanced distribution of C. denitrificans in our study, particularly against the representative strain from Denmark, a Parsnp tree was constructed (Fig. 2). First, despite the majority coming from the same Australian wastewater source, we noted a diverse set of clades that generally mirror the clade structure shown in the PhyloSift distribution, including several small subclades of only one to three representatives. Notably, one cluster of isolates (maroon) sat as a very closely related subclade (as low as seven SNPs between eight isolates). The external international reference sat most distant in the distribution, at approximately 6,000 SNPs from both its closest neighbors, suggesting significantly diverged lineages of the species.
FIG 2.
C. dentrificans phylogeny. Phylogenetic relationships of Australian C. denitrificans. The midpoint-rooted maximum-likelihood tree was generated from a single-nucleotide polymorphism (SNP)-based alignment via Parsnp, with C. denitrificans JAFNM01 used as a reference. Clades are colored in descending order: light green, maroon, gray, yellow, green, purple, blue, and pink.
Pangenome analysis of C. denitrificans.
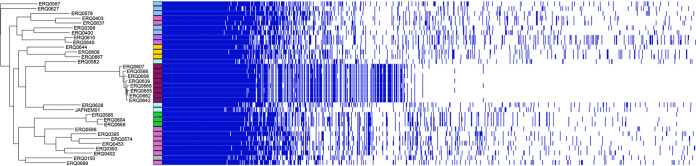
An analysis of the C. denitrificans pangenome supported the subclade associations as determined by the core-genome phylogeny and the high genetic diversity of the species (Fig. 3). The C. denitrificans pangenome consisted of 7,219 genes, with a core genome of only 946 genes and an accessory genome of 5,173 genes (147 soft-core, 2,637 shell, and 2,389 cloud genes). The distribution of genes generally matched the phylogenetic analysis presented in Fig. 2, with notable divergence in pangenome data for the top-most (light green) and lower two (blue and pink) clades, which had some isolates with distinct genotypes distant from the closest evolutionary neighbors. This comparison aligns with the highly variant nature of the isolates presented here, suggesting that up to 25 distinct lineages of C. denitrificans are now available as WGS.
FIG 3.
C. denitrificans pangenome. Pangenome analyses of 33 C. denitrificans from Australian aquatic environments and C. denitrificans JAFNEM01 sourced from GenBank. Color groupings match the clades designated in Fig. 2. Phylogenetic clustering is by accessory genomes.
Antimicrobial resistance gene (ARG) identification.
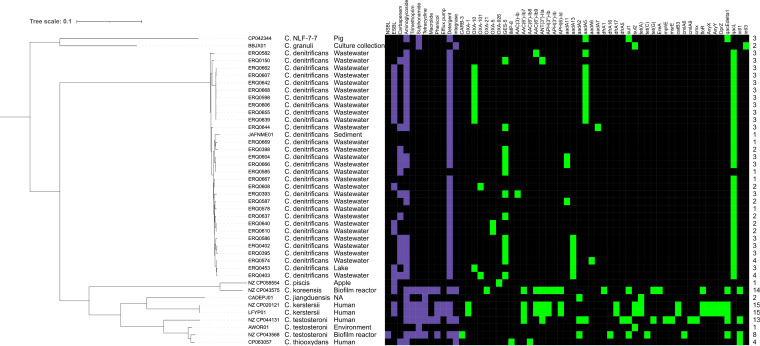
All 88 Comamonas spp. were screened for ARGs (Fig. 4). In the Australian collection, 30 of the 33 C. denitrificans isolates (29 from wastewater, 1 from a lake sample) carried at least one ARG or detergent resistance gene. Among these 33 Australian C. denitrificans isolates, 13 carried carbapenemase gene blaGES-5, 9 hosted blaOXA-10, 2 carried blaOXA-101, and 2 carried blaOXA-5-like alleles. The blaGES and blaOXA genes were only identified in C. dentrificans; therefore, we could not account for resistance to the carbapenem supplement in Oxoid Brilliance CRE Agar plates in 26 of 39 Comamonas spp. isolates.
FIG 4.
Antimicrobial resistance genes (ARGs) in Comamonas spp. Heat map of ARGs identified in all available Comamonas spp. genomes adjacent to a midpoint-rooted maximum-likelihood phylogenetic tree generated using Phylosift. Specific ARGs, detergent resistance genes, and integrases are shown in green. Resistance conferred by gene carriage is shown in purple. NSBL, narrow spectrum β-lactamase.
A total of 21 Australian C. denitrificans isolates carried at least 1 aminoglycoside resistance gene, with aadA5 being the most common (n = 9) followed by aadA13 (n = 6) (Fig. 4). C. denitrificans isolate ERQ0403 carried a combination of clinically important antimicrobial resistance genes (blaGES-5 and blaOXA-10) and aminoglycoside resistance (aadA13). All but one Australian C. denitrificans harbored detergent resistance gene qacL (Fig. 4).
A subset of Comamonas isolates sourced from GenBank (n = 11; 22%) also carried ARGs (Fig. 1 and 4), five of which, sourced from either humans or biofilm reactors, had multidrug resistance profiles (Fig. 4). However, outside the Australian C. denitrificans isolates, only one isolate, a C. thiooxydans sourced from a human sample, carried a carbapenem resistance gene (blaIMP-8).
Metal resistance genes were also identified using the MEGAres 2.0 database. All C. denitrificans isolates originating from Australia carried genes conferring resistance to copper (cop genes), and 11 (33%) additionally carried mercury resistance genes (mer genes). The singular Australian C. aquatica isolate also carried copper resistance genes, as did two C. aquatica isolates sourced from GenBank. The five Australian C. testosteroni isolates did not carry metal resistance genes; however, eight C. testosteroni isolates sourced from GenBank all carried mercury resistance genes, with one isolate carrying an additional gene conferring resistance to chromium. Virulence-associated genes were also screened for using the VFDB; however, none were identified in any isolate using a >80% nucleotide identity and >80% gene coverage cutoff.
(i) C. denitrificans chromosomally acquired blaGES-5 and blaOXA alleles.
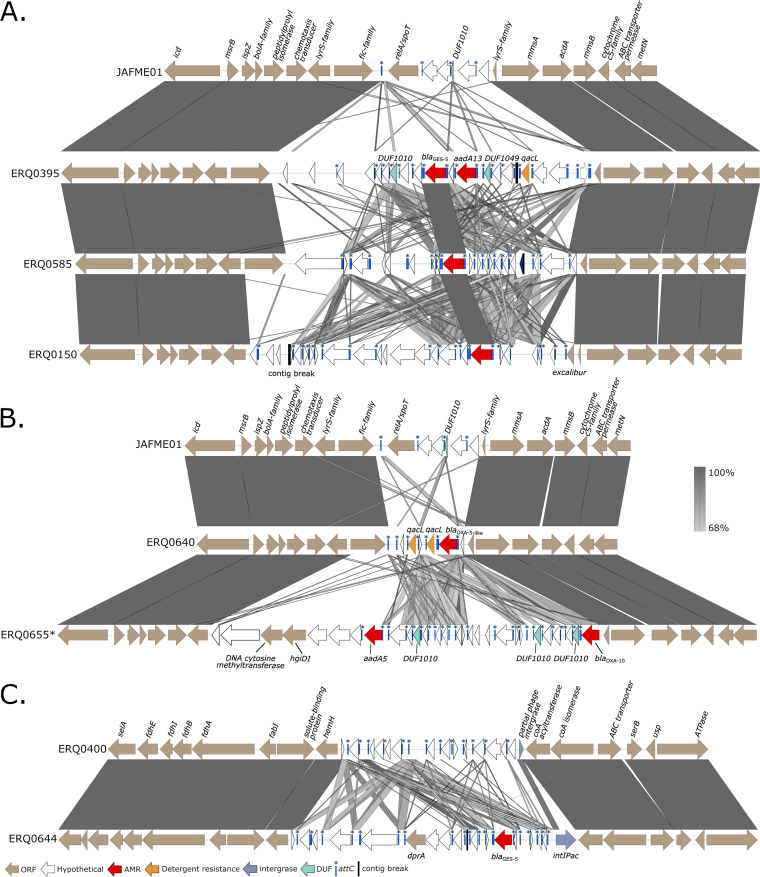
Regarding genetic context, blaGES-5 and blaOXA genes appeared on putative genomic islands (GIs). Upon comparing the genetic context around blaGES-5, it was determined that two chromosomal sites were hosting the gene, with related isolates ERQ0150, ERQ0395, and ERQ0585 encoding the gene in the same chromosomal loci. One isolate, ERQ0644, had acquired the gene at a separate chromosomal site. Two reference strains (JAFNME01 and ERQ0400) were used to resolve these chromosomal sites across contigs present in the isolates hosting AMR (Fig. 5A and C). In addition to blaGES-5, the GI present in ERQ0395 also harbored aadA13 (aminoglycoside resistance), qacL (quaternary ammonium compound resistance), and several DUF1010 genes. Although the GIs represented in Fig. 5A lacked any tRNA gene or known integrase, they were identified as GIs based on atypical codon usage by SIGI-HMM (55), and most ORFs within the GIs, including all AMR genes, had adjacent attC sites, indicating that these ORFs are mobile gene cassettes. The attC sites associated with blaGES-5 in ERQ0585 and ERQ0150 shared 98% sequence identity but only ~50% sequence identity with the ERQ0395 blaGES-5 attC site. However, the blaGES-5 gene and associated attC site in ERQ0395 was identical to that carried by an intI3-containing Tn7221 (also called TnPfu1; Tn402 family) in P. aeruginosa pIPM3H3-GES5. Unlike the three aforementioned blaGES-5-containing GIs, the GI carrying blaGES-5 in ERQ0644, located at a different chromosomal insertion site (Fig. 5C), contained a phage associated integrase intIPac, which was also partially present in ERQ0400 (Fig. 5C).
FIG 5.
Genomic islands (GIs) carrying AMR determinants. (A) Comparison of three GIs carrying blaGES-5 at the chromosomal insertion site. (B) Comparison of two GIs, one carrying blaOXA-10, the other blaOXA-5-like. GI in the same genetic context as panel A. (C) Comparison of GI carrying blaGES-5. The genetic context differs to panels A and B and is associated with integrase intIPac. ORF, open reading frame.
A comparison of two representative GIs from EQR0640 and EQR0655, encoding blaOXA-10 and blaOX-5-like, respectively, was also visualized (Fig. 5B). Alignments demonstrated that GIs hosting these AMR genes were inserted at the exact same chromosomal site as three GIs containing blaGES-5 (Fig. 5A). The shared chromosomal sequence at each structure’s termini included lysR (transcriptional regulator) and a fic family gene, often found in mobile GIs (56) at one end and a partial lysR and an mmsA (CoA-acylating methylmalonate-semialdehyde dehydrogenase) at the other. Like the blaGES-5 GIs, most ORFs, including blaOXA-5-like and two qacL genes in EQR0640 and blaOXA-10 and aadA5 in EQR0655, were associated with attC sites, although they lacked any known integrase or tRNA gene. All blaGES-5- and blaOXA-containing GIs appear to be unique to this Australian collection of C. denitrificans, with no significant homology to GIs previously deposited in GenBank, including a well characterized blaGES-5-containing GI residing in P. aeruginosa (57).
C. denitrificans IncP-1 plasmid.
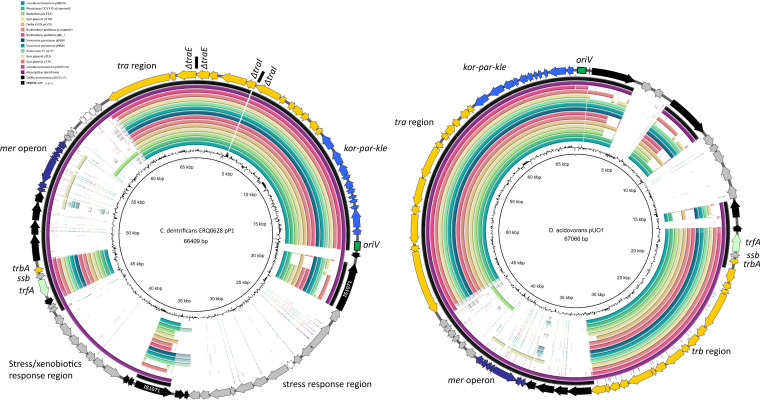
All 88 Comamonas genomes were screened for plasmid replicons, and three were identified: IncP-1 in C. denitrificans isolate ERQ0628 and a C. jiangduensis isolate (CADEPJ01), and rep21 in a C. kerstersii strain J29 (NZ_LFYP00000000.1). The IncP-1 replicon (trfA) in ERQ0628 was located on a 62,429-bp contig, beginning with a partial traI gene and ending with a partial traE gene. Another contig (3,980 bp) contained both partial traI and traE genes present at the contig start and end. Future long-read sequencing is needed to investigate any potential additional insertions into the tra region following traI or traE, although previously, insertions have only been reported following traM, between oriV and trfA, and between trb and tra regions (58). Nevertheless, a BLAST search of the NCBI database was used to identify high sequence identity IncP-1 plasmids, and the top 15 hits (ranging between 98.78 and 99.95% sequence identity and 40 and 79% sequence coverage) were used for content comparisons (Fig. 6).
FIG 6.
BRIG comparisons of IncP-1 plasmids. (Left) Plasmid map of putative IncP-1 plasmid from ERQ0628 and BLAST alignments to other IncP-1 plasmids. Contig breaks are noted in the tra operon. (Right) Plasmid map of D. acidovorans pUO1 and BLAST alignments to other IncP-1 plasmids.
All compared IncP-1 plasmids possessed an IncP-type conjugation tra region and the IncP kor-par-kle regulon region. The remaining regions present in ERQ628 IncP-1 (Fig. 6, left) were less common. An 11,709-bp inserted region containing a mercury resistance mer operon carried by Tn5031 (Tn402 family) was identified in two other similar plasmids, one carried by Alicycliphilus denitrificans pALIDE02 (CP002451) and the other by Delftia acidovorans pUO1 (AB063332) (59). These two Tn5031 sequences were nearly identical, differing by 1 SNP. A. denitrificans pALIDE02 was the only other plasmid to possess a 7,964-bp inserted region containing genes involved in stress response and xenobiotic degradation, including two genes encoding glutathione S-transferases and a short-chain dehydrogenase/reductase (SDR). The ERQ628 IncP-1 plasmid also possesses a unique 12,843-bp inserted region flanked by two IS1071 elements and contained genes involved in oxidative stress response, including msrA, msrB, and ydiU. In D. acidovorans pUO1 (Fig. 6, right), IS1071 elements flank genes responsible for haloacetate degradation (dehH1 and dehH2). Notably, the ERQ0628 IncP-1 plasmid only contained trbA, and the remaining trb region was missing (Fig. 6, right).
DISCUSSION
Wastewater has been flagged as a hot spot for interaction between diverse bacteria, genetic exchange, and AMR evolution and dissemination across ecosystems (60). However, despite the abundance of Comamonas species in wastewater, its ability to acquire AMR is poorly understood. Here, we added 39 draft Comamonas genomes, sourced from South Australian municipal wastewater, a lake, and a wetland, to make up a total pool of 88 Comamonas WGS currently available. Our analyses show that (i) carbapenem-resistant Comamonas was most prevalent in wastewater, (ii) C. denitrificans isolates carried genomic islands encoding carbapenemase blaGES-5 or extended-spectrum β-lactamase (ESBL) blaOXA alleles, (iii) Comamonas carry genes conferring resistance to copper and mercury, (iv) C. denitrificans, the most commonly isolated species, was genetically diverse, and (v) a C. denitrificans isolate carried an IncP-1 plasmid with genes involved in xenobiotic degradation and response to oxidative stress.
Carbapenem-resistant Comamonas is present in aquatic environments: implications for public health and wastewater treatments.
Carbapenem-resistant Gram-negative bacteria are an urgent threat to global health (61, 62) and have been detected in both wastewater and surface water worldwide (46, 63–65). Here, we isolated 39 carbapenem-resistant Comamonas isolates: 32 C. denitrificans and 5 C. testosteroni from wastewater, 1 C. denitrificans from a wetland, and 1 C. aquatica from a lake with public access. To the best of our knowledge, this is the first report of carbapenem resistance in C. denitrificans and C. aquatica, although there has been at least one report of a carbapenem-resistant C. testosteroni infection in Turkey in 2015 (66). Infections caused by carbapenem-resistant bacteria are associated with poor prognoses and increased morbidity and mortality rates (67). C. testosteroni has been isolated as the infectious agent in various sites, including blood, peritoneal fluid, cerebrospinal fluid, urine, and different tissues (68). C. aquatica reportedly has caused bacteremia and septic shock (69), and C. denitrificans has been detected in clinical samples (26). Therefore, the emergence of carbapenem resistance in these organisms warrants closer public health surveillance. To that end, it is important to note that Comamonas spp. grow poorly on routine clinical media (26), and biochemical tests often provide erroneous Pseudomonas spp. identifications (1). Similarly, as shown here and elsewhere, MALDI-TOF MS can misidentify Comamonas at the species level (70). Furthermore, C. denitrificans and Comamonas nitrativorans have highly similar 16S rRNA sequences and cannot be differentiated by phenotypic analysis (1). Conversely, WGS represents a high-resolution method that can accurately identify Comamonas species.
The presence of carbapenem-resistant Comamonas in wastewater also has implications for wastewater treatments. Wastewater treatment plants receive large volumes of sewerage enriched in inorganic constitutes, such as metals and chemicals, and organic constituents, such as feces and food waste (71, 72). Biological treatment of wastewater using microbes is effective and easy to implement at a low cost, particularly when using indigenous microbial communities (73). A recent meta-analysis of 20 diverse metagenomic wastewater samples across 11 countries found that Comamonas spp. was dominant in industrial, biological, and municipal wastewater (74). In wastewater, Comamonas spp. play important roles in the degradation of benzene, ethylbenzene, and toluene (75); dye (13); naphthalene-2-sulfonic acid (76); organics (14); pyridine (15); and quinoline and phenol (77) and in the denitrification (78) and bioremediation of petrochemicals (79). These capabilities, in addition to high biofilm-forming properties, make Comamonas good candidates for biofilm systems wastewater treatment (80, 81). However, a recent study demonstrated that spiking wastewater biofilm reactors with streptomycin and oxytetracycline caused the acquisition of class 1 integron integrases (intI1) and aadA genes (aminoglycoside resistance) in Comamonas strains (82). Given this recent observation, it is perhaps unsurprising that genes conferring resistance to aminoglycosides were identified in 30 C. denitrificans isolates (91%) in our collection, with aadA5 the most prevalent (n = 9), and while intI1 was not identified in this collection, the potential for capture should not be overlooked. On that note, bacteria in wastewater exist as heterogeneous communities, and the presence of Comamonas is positively correlated with the presence of Acinetobacter, Aeromonas, and Pseudomonas, suggesting that these microbes form a symbiotic relationship (74). Horizontal gene transfer occurs in proximal bacteria, and all three species are known producers of carbapenemases (46, 83). Therefore, a symbiotic relationship with these organisms may not only involve the cooperative degradation of pollutants but also the exchange of AMR determinants.
C. denitrificans carry genomic islands encoding either blaGES-5 or blaOXA alleles.
Genes encoding carbapenemases confer resistance to carbapenems, penicillins, cephalosporins, and monobactams, thus severely limiting treatment options (67). While all 39 Comamonas isolates described here had a carbapenem-resistant phenotype, only 13 isolates, all C. denitrificans, carried a known carbapenemase gene, blaGES-5, suggesting novel AMR mechanisms for the remaining isolates. The blaGES-5 gene cassette has previously been detected in the genera of Citrobacter, Enterobacter, Escherichia, Leclercia, and Lelliottia found in wastewater (46, 63, 84); in Citrobacter and Klebsiella in surface water (44, 85) and was commonly associated with class 1 and 3 integrons. The gene has also been detected in clinical P. aeruginosa strains carried by plasmids and genomic islands (57, 86, 87).
Genomic islands are fragments of DNA inserted into the chromosome via horizontal gene transfer and play an important role in the evolution and adaptation of bacteria through the dissemination of ARGs and virulence genes and the formation of catabolic pathways (88). We were able to resolve four examples in which a blaGES-5 gene cassette was located on predicted genomic islands, and in one instance, the blaGES-5 gene cassette was identical to that carried by intI3-containing Tn7221 structure found on a P. aeruginosa plasmid. However, genomic islands are generally characterized by atypical G+C content and codon usage and the presence of integrons and are typically inserted at tRNA loci (89). Of the four putative genomic islands, only one carried an integrase, IntIPac, previously reported promoting AMR in Acidithiobacillus ferrooxidans (90). Apart from atypical G+C content and codon usage, the remaining three genomic islands did not contain any typical genomic island characteristics and appeared to consist of a collection of gene cassettes located in the same chromosomal loci. Curiously, we also resolved two predicted genomic islands, one carrying ESBL blaOXA-10, commonly found in Acinetobacter baumannii (91), and the other a novel blaOXA-5-like gene, both located at the same chromosomal loci as the three blaGES-5 containing genomic islands. Although rare, gene cassettes have been found outside an integron context (92); however, the mechanisms behind their insertion have not been determined.
Comamonas spp. carry heavy metal and biocide resistance genes.
In addition to residual antibiotics, it has been postulated that the high AMR levels in water matrices are also due to the presence of metals and biocides, which can exert selective pressure for ARGs through coselection (93). Additionally, previous studies have observed a correlation between metal resistance and β-lactamase production in bacteria, particularly in regard to mercury resistance (94). Although we did not find any apparent relationship between ARG, heavy metal, and biocide carriage, we did identify 11 C. denitrificans isolates (33%) that carried mercury resistance genes (mer genes), and genes conferring resistance to copper (cop genes) were present in all C. denitrificans and the singular C. aquatica isolate in this collection. Additionally, 29 C. denitrificans isolates (89%) carried detergent resistance gene qacL. Interestingly, a study on the association of metal tolerance with antibiotic susceptibility in C. acidovorans found that mercury-resistant phenotypes were resistant to nitrofuran, β-lactams, aminoglycosides, glycopeptides, and tetracycline, while copper-resistant phenotypes were susceptible to all tested antibiotics (95). However, ARG carriage was not determined.
IncP-1 plasmid present in C. denitrificans.
Sensing stressful conditions and adjusting cellular metabolism accordingly are essential for bacteria to survive in variable environments such as wastewater (96). IncP-1 plasmids are promiscuous self-transmissible plasmids with broad host ranges and an ability to swiftly acquire and transfer genes involved in the degradation of introduced xenobiotics (97). IncP-1 plasmids have been previously identified in Comamonas spp. isolated from wastewater that were involved in the degradation of dyes (98). However, only one IncP-1 replicon was identified in Comamonas from Australian aquatic environments: a C. denitrificans isolate from wastewater. The IncP-1 replicon was situated on a putative 66,409-bp plasmid containing three inserted regions. One inserted region contained a functional mer operon, a well characterized metal resistance system capable of degrading highly toxic mercury into volatile, nontoxic forms (9). Another inserted region carried genes involved in oxidative stress response and was flanked by IS1071 elements. IS1071 is known to flank many catabolic genes in diverse Gram-negative and Gram-positive bacteria (99) and has been shown to transpose at high frequencies in C. testosteroni (100). The last inserted region carried two glutathione S-transferases, known to degrade a wide range of toxic chemicals, including carcinogens, environmental pollutants, and oxidative stress products (101), and a short-chain dehydrogenase/reductase, involved in the metabolism of aromatic hydrocarbons, including steroids and sugars (99). Unlike most, if not all, IncP-1 plasmids, the majority of trb genes, which form the sex pili, were missing in the C. denitrificans IncP-1 plasmid, meaning that in all probability, it is nonconjugative.
Concluding remark.
At the time of this study, only one C. denitrificans genome was publicly available (JAFNME01). Thus, this study has contributed significantly to our understanding of the genetic diversity of this species. Our phylogenetic and pangenome analyses identified at least 25 distinct C. denitrificans lineages, and C. denitrificans JAFNME01 isolated from sediment in Denmark in 2019 was approximately 6000 SNPs from its closest neighbor from this collection. Acquisition of the critically important antibiotic resistance genes blaGES-5 and blaOXA on genomic islands and an IncP-1 plasmid carrying metal resistance is a serious cause for concern and should be monitored given the important role C. denitrificans plays in bioremediation and wastewater treatment. This may require: (i) improvements in growth medium to ensure reliable culture; (ii) more reliable diagnostic assays, including improvements to MALDI-TOFF species identification databases; and (iii) introduction of long-read metagenomic sequencing strategies to monitor wastewater. Finally, we could not identify the genetic basis for carbapenem resistance in a significant subset of the Comamonas isolates described here. Further studies are required to rectify this knowledge gap.
MATERIALS AND METHODS
Sample collection and bacterial isolation.
Within a period of 1 year, from July 2018 to July 2019, water samples (~10 L) were collected in triplicate, monthly, from three sources: influent wastewater, a lake, and a wetland in South Australia.
Influent wastewater was collected from four municipal wastewater treatment plants (here referred to as WWTPs A, B, C, and D). WWTP A serves ≈150,000 inhabitants and treats approximately 33 ML/day of primarily domestic sewage. WWTP B serves ≈198,000 inhabitants and treats around 55 ML/day, primarily from households and commercial establishments with minor industrial inputs. WWTP C serves ≈700,000 inhabitants and treats around 175 ML/day with a large industrial/commercial component, as well as residential and hospital sources. WWTP D is a rural wastewater treatment plant that serves 5,000 inhabitants and treats around 1.2 ML/day primarily from households and seasonal meat-processing facilities. Raw wastewater to all WWTPs is classified as low-to-medium organic strength (102). All samples were stored on ice directly after collection and processed within 2 to 3 h.
The lake is a shallow artificial lake fed by recycled water, which is composed of a mix of treated wastewater and cleansed storm water. The lake is sporadically fringed by aquatic reads and vegetation and attracts silver gulls (Larus novaehollandiae), pigeons (Columba livia), Eurasian coots (Fulica atra), and indigenous and migratory birds.
The inland wetland covers 42 ha and is recharged by seasonal rainfall/runoff inflows. The annual mean rainfall is 438 mm (11.4 mm in February; 59.6 mm in July) and mean annual temperature ranging between 21.6°C (28.2°C in January) and 11.5°C (7.1°C in July) (bom.gov.au). The wetland is habitat to over 160 species of indigenous and migratory birds, including purple swamp hem, stilt, and herons, as well as L. novaehollandiae, F. atra, C. livia, and Haliaeetus leucogaster. The water of this inland wetland is harvested and pumped underground into natural sandy limestone aquifers.
Isolation of carbapenem-resistant Comamonas species and MALDI-TOF MS species identification.
All of the samples were plated, in triplicate, on Oxoid Brilliance CRE agar plates (Thermo Fisher Scientific Australia, Adelaide, SA, Australia) after 10-fold serial dilutions, using 500 μL from 2 to 3 consecutive dilutions. Oxoid Brilliance CRE agar plates are chromogenic screening plates selective for carbapenem-resistant Enterobacteriaceae (CRE). The formulation contained a modified carbapenem at a level recommended by both the European Committee on Antimicrobial Susceptibility Testing (EUCAST) and the Clinical and Laboratory Standards Institute (CLSI). The two-chromogen system differentiated Escherichia coli (pale pink) from the KESC group organisms (Klebsiella, Enterobacter, Serratia, and Citrobacter) (blue). Other carbapenem-resistant organisms not from the Enterobacteriaceae family (e.g., Acinetobacter) produced white or naturally pigmented colonies. All cultures were incubated at 25, 37, and 44°C for 24 h. Single colonies growing on CRE agar were picked up and steaked on a plate counting agar (PCA; Thermo Fisher Scientific). Then, PCA cultures were incubated at 37°C for 18 to 24 h to have sufficient bacterial growth. A total of 40 Comamonas isolates were sampled, with their identity resolved initially by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). MALDI-TOF MS was performed using Bruker Daltonics, operated in linear positive mode under MALDI Biotyper 3.0 real-time classification (version 3.1, Bruker Daltonics) (103). All isolates were preserved in glycerol (40% v/v) at −80°C. Clinical Klebsiella pneumoniae and C. testosteroni carbapenem resistance strains were used as controls to ensure the isolation conditions and that carbapenem plates were efficient and reproducible. The control strains were identified by MALDI-TOF, blaKPC, blaGES, and blaOXA qPCR assays (104) and WGS.
DNA extraction and WGS.
DNA from MALDI-TOF MS identified isolates (scores ≥ 2‐3) were extracted using the DNeasy blood and tissue kit (Qiagen) according to the manufacturer’s instructions. Nucleic acid quality was measured with Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific). DNA concentrations for all samples were measured by fluorometric quantitation using a Qubit instrument and high sensitivity double-stranded DNA HS assay kit (Thermo Fisher Scientific), and purified DNA extracts were stored at −80°C until sequencing.
WGS was performed on an Illumina Nextseq500 instrument according to the manufacturer’s instructions. The data were uploaded to Basespace (www.basespace.illumina.com), which converted the raw data to eight FASTQ files for each sample. Genome assembly was performed using Shovill version 1.0.4 (https://github.com/tseemann/shovill) using the SPAdes option. A single genome was cut due to high scaffolding, with the remaining 39 sequences passing basic quality control using assembly stats (github.com/sanger-pathogens/assembly-stats).
Phylogenetic analysis.
A phylogenetic analysis was performed on Comamonas isolates from multiple species, including: 39 draft genomes from aquatic environments in South Australia (this study) alongside 49 Comamonas genomes (22 complete and 27 draft) sourced from GenBank (downloaded on 5 March 2021) (105) (ncbi.nlm.nih.gov/GenBank/) including five clinical Comamonas isolates. GenBank sourced genomes are referred to by their accession numbers throughout this article. A maximum-likelihood phylogenetic tree of all species were constructed using PhyloSift (106), and SNP-based phylogenetic analyses were generated using Parsnp (107) (github.com/marbl/parsnp). All trees were resolved using FastTree2 (108) and visualized using Interactive Tree Of Life (iTOL) software version 4 (109) (itol.embl.de/). The C. denitrificans pangenome was calculated using Roary version 3.11.2 (110) (github.com/sanger-pathogens/Roary) and visualized using Phandango version 1.3.0 (111) (jameshadfield.github.io/phandango/#/main). A pangenome-wide gene association study on C. denitrificans isolates was performed using Scoary (112) (github.com/AdmiralenOla/Scoary).
Genotyping.
In addition to MALDI-TOF MS, in silico species identification was also performed using Kraken2 (113) (ccb.jhu.edu/software/kraken2/). To confirm the identification of C. denitrificans and C. testosteroni, pairwise genome comparisons were performed against C. denitrificans JAFNME01 and C. testosteroni NZ_CP067086.1 using both the ANIb and ANIm algorithms available on the JSpecies web server (jspecies.ribohost.com/jspeciesws) using a 95% cutoff value for species delimitation (114). Predicted DDH results were ascertained using the Genome-to-Genome Distance Calculator (GGDC) tool (115) (ggdc.dsmz.de) with a 70% cutoff value for species delimitation using the recommended Formula 2. Virulence-associated genes, antimicrobial resistance (AMR) genes, metal resistance genes, and plasmid replicons were screened for using ABRicate (github.com/tseemann/abricate) in conjunction with the following databases: VFDB (116) (mgc.ac.cn/VFs), CARD (117) (card.mcmaster.ca), MEGARes 2.0 (118) (megares.meglab.org), and PlasmidFinder (119) (cge.cbs.dtu.dk/services/PlasmidFinder), respectively.
Genome annotation.
All draft environmental genomes in this study were annotated using prokka (github.com/tseemann/prokka) and managed using SnapGene version 4.1.9 (snapgene.com). The RAST annotation pipeline (120) (rast.nmpdr.org/rast.cgi) was also utilized on genomes representative of each clade to cross-check annotations. Transposons were identified using TnCentral (tncentral.proteininformationresource.org). Putative genomic islands (GIs) were identified by Islandviewer 4 (121) (pathogenomics.sfu.ca/islandviewer/) using reference genome Comamonas sp. strain NLF 7-7 (accession NZ_CP042344.1) and confirmed in detail using progressive Mauve (122). Mobile gene cassette-associated recombination (attC) sites were screened for using HattCI (github.com/maribuon/HattCI) (123). Only attC sites with V scores greater than 7.5 were considered. GIs were visualized using EasyFig (124) (mjsull.github.io/Easyfig/). BLASTn was utilized to determine whether putative GIs and AMR regions identified in this study had been previously deposited into NCBI. The Aliview software version 3.0 (GPLv3) (125) (github.com/AliView), utilizing MUSCLE and FastTree2 was used to align gene alleles and perform maximum-likelihood phylogenetic analyses. Plasmid comparisons were performed using the BLAST Ring Image Generator (BRIG) (126) (http://brig.sourceforge.net/).
ACKNOWLEDGMENTS
We respectfully acknowledge the Peramangk, Kaurna, and Ngarrindjeri people as the traditional owners and custodians of the waters and lands where the environmental samples used in this research were collected.
Also acknowledge Gianluca Brunetti for helping with the water sample collection and processing and Robin Lockington for bacterial culturing and isolation.
We declare no conflict of interest.
S.H.: writing – original draft, formal analysis, investigation, writing – review & editing; E.R.W.: investigation, methodology, validation, conceptualization, supervision, writing – review & editing; B.D.: investigation, conceptualization, validation, formal analysis, data curation, writing – review & editing; E.D.: conceptualization, funding acquisition, supervision, writing – review & editing; I.G.C.: investigation, resources; D.J.B.: resources, methodology; V.M.J.: writing – original draft, formal analysis, investigation, supervision, conceptualization, writing – review & editing; S.P.D.: conceptualization, resources, supervision, funding acquisition, project administration, writing – review & editing.
This research was partly supported by Australian Government Medical Research Future Fund Project MRFF75873 and the Australian Centre for Genomic Epidemiological Microbiology (Ausgem), a collaborative research initiative between the New South Wales Department of Primary Industries and the University of Technology Sydney; and by Quadram Institute Bioscience Biotechnology and Biological Sciences Research Council (BBSRC)-funded Core Capability Grant Project BB/CCG1860/1 and BBSRC Institute Strategic Program Microbes in the Food Chain Project BB/R012504/1.
Footnotes
Supplemental material is available online only.
Contributor Information
Veronica M. Jarocki, Email: veronica.jarocki@uts.edu.au.
Steven P. Djordjevic, Email: steven.djordjevic@uts.edu.au.
Hideaki Nojiri, University of Tokyo.
REFERENCES
- 1.Brady MT, Marcon MJ. 2008. Less commonly encountered nonenteric Gram-negative bacilli, p 828–831. In Long SS (ed), Principles and Practice of Pediatric Infectious Disease, 3rd ed. W.B. Saunders, Philadelphia, PA. 10.1016/B978-0-7020-3468-8.50157-7. [DOI] [Google Scholar]
- 2.Yu X-Y, Li Y-F, Zheng J-W, Li Y, Li L, He J, Li S-P. 2011. Comamonas zonglianii sp. nov., isolated from phenol-contaminated soil. Int J Syst Evol Microbiol 61:255–258. 10.1099/ijs.0.019612-0. [DOI] [PubMed] [Google Scholar]
- 3.Chipirom K, Tanasupawat S, Akaracharanya A, Leepepatpiboon N, Prange A, Kim K-W, Chul Lee K, Lee J-S. 2012. Comamonas terrae sp. nov., an arsenite-oxidizing bacterium isolated from agricultural soil in Thailand. J Gen Appl Microbiol 58:245–251. 10.2323/jgam.58.245. [DOI] [PubMed] [Google Scholar]
- 4.Sun L-N, Zhang J, Chen Q, He J, Li Q-F, Li S-P. 2013. Comamonas jiangduensis sp. nov., a biosurfactant-producing bacterium isolated from agricultural soil. Int J Syst Evol Microbiol 63:2168–2173. 10.1099/ijs.0.045716-0. [DOI] [PubMed] [Google Scholar]
- 5.Patureau D, Zumstein E, Delgenes JP, Moletta R. 2000. Aerobic denitrifiers isolated from diverse natural and managed ecosystems. Microb Ecol 39:145–152. 10.1007/s002480000009. [DOI] [PubMed] [Google Scholar]
- 6.Chang Y-H, Han J, Chun J, Lee KC, Rhee M-S, Kim Y-B, Bae KS. 2002. Comamonas koreensis sp. nov., a non-motile species from wetland in Woopo, Korea. Int J Syst Evol Microbiol 52:377–381. 10.1099/00207713-52-2-377. [DOI] [PubMed] [Google Scholar]
- 7.Guo Y, Gong H, Guo X. 2015. Rhizosphere bacterial community of Typha angustifolia L. and water quality in a river wetland supplied with reclaimed water. Appl Microbiol Biotechnol 99:2883–2893. 10.1007/s00253-014-6182-9. [DOI] [PubMed] [Google Scholar]
- 8.Kim K-H, Ten LN, Liu Q-M, Im W-T, Lee S-T. 2008. Comamonas granuli sp. nov., isolated from granules used in a wastewater treatment plant. J Microbiol 46:390–395. 10.1007/s12275-008-0019-0. [DOI] [PubMed] [Google Scholar]
- 9.Palanisamy V, Gajendiran V, Mani K. 2021. Meta-analysis to identify the core microbiome in diverse wastewater. Int J Environ Sci Technol 10.1007/s13762-021-03349-4. [DOI] [Google Scholar]
- 10.Boon N, Goris J, De Vos P, Verstraete W, Top EM. 2000. Bioaugmentation of activated sludge by an indigenous 3-chloroaniline-degrading Comamonas testosteroni strain, I2gfp. Appl Environ Microbiol 66:2906–2913. 10.1128/AEM.66.7.2906-2913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu J, Jiang C, Wang B, Ma Y, Liu Z, Liu S. 2006. Novel partial reductive pathway for 4-chloronitrobenzene and nitrobenzene degradation in Comamonas sp. strain CNB-1. Appl Environ Microbiol 72:1759–1765. 10.1128/AEM.72.3.1759-1765.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang Q, Lu T, Liu S-J. 2018. Developing a synthetic biology toolkit for Comamonas testosteroni, an emerging cellular chassis for bioremediation. ACS Synth Biol 7:1753–1762. 10.1021/acssynbio.7b00430. [DOI] [PubMed] [Google Scholar]
- 13.Geng Z, Yu Y, Zhu S, Yu H, Liu J, Bian D, Yang X, Huo H, Huo M. 2017. Comparing polyethersulfone and polyurethane-immobilized cells of Comamonas testosteroni QYY in treatment of an accidental dye wastewater. Chem Res Chin Univ 33:36–43. 10.1007/s40242-017-6356-y. [DOI] [Google Scholar]
- 14.Yuan K, Li S, Zhong F. 2020. Characterization of a newly isolated strain Comamonas sp. ZF-3 involved in typical organics degradation in coking wastewater. Bioresour Technol 304:123035. 10.1016/j.biortech.2020.123035. [DOI] [PubMed] [Google Scholar]
- 15.Zhu G, Zhang Y, Chen S, Wang L, Zhang Z, Rittmann BE. 2021. How bioaugmentation with Comamonas testosteroni accelerates pyridine mono-oxygenation and mineralization. Environ Res 193:110553. 10.1016/j.envres.2020.110553. [DOI] [PubMed] [Google Scholar]
- 16.Gumaelius L, Magnusson G, Pettersson B, Dalhammar G. 2001. Comamonas denitrificans sp. nov., an efficient denitrifying bacterium isolated from activated sludge. Int J Syst Evol Microbiol 51:999–1006. 10.1099/00207713-51-3-999. [DOI] [PubMed] [Google Scholar]
- 17.Tago Y, Yokota A. 2004. Comamonas badia sp. nov., a floc-forming bacterium isolated from activated sludge. J Gen Appl Microbiol 50:243–248. 10.2323/jgam.50.243. [DOI] [PubMed] [Google Scholar]
- 18.Xing D, Cheng S, Logan BE, Regan JM. 2010. Isolation of the exoelectrogenic denitrifying bacterium Comamonas denitrificans based on dilution to extinction. Appl Microbiol Biotechnol 85:1575–1587. 10.1007/s00253-009-2240-0. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen AQ, Vu HP, Nguyen LN, Wang Q, Djordjevic SP, Donner E, Yin H, Nghiem LD. 2021. Monitoring antibiotic resistance genes in wastewater treatment: current strategies and future challenges. Sci Total Environ 783:146964. 10.1016/j.scitotenv.2021.146964. [DOI] [PubMed] [Google Scholar]
- 20.Andersson S, Kuttuva Rajarao G, Land CJ, Dalhammar G. 2008. Biofilm formation and interactions of bacterial strains found in wastewater treatment systems. FEMS Microbiol Lett 283:83–90. 10.1111/j.1574-6968.2008.01149.x. [DOI] [PubMed] [Google Scholar]
- 21.Farshad S, Norouzi F, Aminshahidi M, Heidari B, Alborzi A. 2012. Two cases of bacteremia due to an unusual pathogen, Comamonas testosteroni in Iran and a review literature. J Infect Dev Ctries 6:521–525. 10.3855/jidc.2215. [DOI] [PubMed] [Google Scholar]
- 22.Ojeda-Vargas M del M, Suárez-Alonso A, Pérez-Cervantes M de los A, Suárez-Gil E, Monzón-Moreno C. 1999. Urinary tract infection associated with Comamonas acidovorans. Clin Microbiol Infect 5:443–444. 10.1111/j.1469-0691.1999.tb00170.x. [DOI] [PubMed] [Google Scholar]
- 23.Young C-C, Chou J-H, Arun AB, Yen W-S, Sheu S-Y, Shen F-T, Lai W-A, Rekha PD, Chen W-M. 2008. Comamonas composti sp. nov., isolated from food waste compost. Int J Syst Evol Microbiol 58:251–256. 10.1099/ijs.0.65277-0. [DOI] [PubMed] [Google Scholar]
- 24.Latiefa B, Arnelia P, Vanessa J, Sehaam K, Wesaal K. 2011. Investigation into the microbial contamination in a spring water distribution system, Western Cape, South Africa. Afr J Microbiol Res 5:3200–3214. 10.5897/AJMR11.183. [DOI] [Google Scholar]
- 25.Bayhan Gİ, Tanır G, Karaman İ, Özkan Ş. 2013. Comamonas testosteroni: an unusual bacteria associated with acute appendicitis. Balkan Med J 30:447–448. 10.5152/balkanmedj.2013.9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abayasekara LM, Perera J, Chandrasekharan V, Gnanam VS, Udunuwara NA, Liyanage DS, Bulathsinhala NE, Adikary S, Aluthmuhandiram JVS, Thanaseelan CS, Tharmakulasingam DP, Karunakaran T, Ilango J. 2017. Detection of bacterial pathogens from clinical specimens using conventional microbial culture and 16S metagenomics: a comparative study. BMC Infect Dis 17:631. 10.1186/s12879-017-2727-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Y, Zaiden N, Cao B. 2018. The core- and pan-genomic analyses of the genus Comamonas: from environmental adaptation to potential virulence. Front Microbiol 9:3096. 10.3389/fmicb.2018.03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tiwari S, Nanda M. 2019. Bacteremia caused by Comamonas testosteroni, an unusual pathogen. J Lab Physicians 11:87–90. 10.4103/JLP.JLP_116_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsui T-L, Tsao S-M, Liu K-S, Chen T-Y, Wang Y-L, Teng Y-H, Lee Y-T. 2011. Comamonas testosteroni infection in Taiwan: reported two cases and literature review. J Microbiol Immunol Infect 44:67–71. 10.1016/j.jmii.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 30.Barbaro DJ, Mackowiak PA, Barth SS, Southern PM. 1987. Pseudomonas testosteroni infections: eighteen recent cases and a review of the literature. Rev Infect Dis 9:124–129. 10.1093/clinids/9.1.124. [DOI] [PubMed] [Google Scholar]
- 31.Cooper GR, Staples ED, Iczkowski KA, Clancy CJ. 2005. Comamonas (Pseudomonas) testosteroni endocarditis. Cardiovasc Pathol 14:145–149. 10.1016/j.carpath.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 32.Arda B, Aydemir S, Yamazhan T, Hassan A, Tünger A, Serter D. 2003. Comamonas testosteroni meningitis in a patient with recurrent cholesteatoma. APMIS 111:474–476. 10.1034/j.1600-0463.2003.1110404.x. [DOI] [PubMed] [Google Scholar]
- 33.Reddy AK, Murthy SI, Jalali S, Gopinathan U. 2009. Post-operative endophthalmitis due to an unusual pathogen, Comamonas testosteroni. J Med Microbiol 58:374–375. 10.1099/jmm.0.006072-0. [DOI] [PubMed] [Google Scholar]
- 34.Franzetti F, Cernuschi M, Esposito R, Moroni M. 1992. Pseudomonas infections in patients with AIDS and AIDS-related complex. J Intern Med 231:437–443. 10.1111/j.1365-2796.1992.tb00957.x. [DOI] [PubMed] [Google Scholar]
- 35.Ben-Tovim T, Eylan E, Romano A, Stein R. 1974. Gram-negative bacteria isolated from external eye infections. Infection 2:162–165. 10.1007/BF01642238. [DOI] [PubMed] [Google Scholar]
- 36.Zhou Y, Ma H, Dong Z, Shen M. 2018. Comamonas kerstersii bacteremia in a patient with acute perforated appendicitis: a rare case report. Medicine 97:e9296. 10.1097/MD.0000000000009296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Codjoe FS, Donkor ES. 2017. Carbapenem resistance: a review. Med Sci 6:1. 10.3390/medsci6010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsunaga N, Hayakawa K. 2018. Estimating the impact of antimicrobial resistance. Lancet Glob Health 6:e934–e935. 10.1016/S2214-109X(18)30325-5. [DOI] [PubMed] [Google Scholar]
- 39.Gandra S, Alvarez-Uria G, Turner P, Joshi J, Limmathurotsakul D, van DH. 2020. Antimicrobial resistance surveillance in low- and middle-income countries: progress and challenges in eight South Asian and Southeast Asian countries. Clin Microbiol Rev 33:e00048-19. 10.1128/CMR.00048-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larsson DGJ, Andremont A, Bengtsson-Palme J, Brandt KK, de Roda Husman AM, Fagerstedt P, Fick J, Flach C-F, Gaze WH, Kuroda M, Kvint K, Laxminarayan R, Manaia CM, Nielsen KM, Plant L, Ploy M-C, Segovia C, Simonet P, Smalla K, Snape J, Topp E, van Hengel AJ, Verner-Jeffreys DW, Virta MPJ, Wellington EM, Wernersson A-S. 2018. Critical knowledge gaps and research needs related to the environmental dimensions of antibiotic resistance. Environ Int 117:132–138. 10.1016/j.envint.2018.04.041. [DOI] [PubMed] [Google Scholar]
- 41.Djordjevic SP, Jarocki VM, Morgan B, Donner E. 2020. Genomic surveillance for one health antimicrobial resistance: understanding human, animal, and environmental reservoirs and transmission, p 71–100. In Manaia CM, Donner E, Vaz-Moreira I, Hong P (ed), Antibiotic Resistance in the Environment: A Worldwide Overview. Springer International Publishing, Cham, Switzerland. 10.1007/698_2020_626. [DOI] [Google Scholar]
- 42.Tarabai H, Wyrsch ER, Bitar I, Dolejska M, Djordjevic SP. 2021. Epidemic HI2 plasmids mobilising the carbapenemase gene blaIMP-4 in Australian clinical samples identified in multiple sublineages of Escherichia coli ST216 colonising silver gulls. Microorganisms 9:567. 10.3390/microorganisms9030567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piedra-Carrasco N, Fàbrega A, Calero-Cáceres W, Cornejo-Sánchez T, Brown-Jaque M, Mir-Cros A, Muniesa M, González-López JJ. 2017. Carbapenemase-producing Enterobacteriaceae recovered from a Spanish river ecosystem. PLoS One 12:e0175246. 10.1371/journal.pone.0175246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teixeira P, Tacão M, Pureza L, Gonçalves J, Silva A, Cruz-Schneider MP, Henriques I. 2020. Occurrence of carbapenemase-producing Enterobacteriaceae in a Portuguese river: blaNDM, blaKPC and blaGES among the detected genes. Environ Pollut 260:113913. 10.1016/j.envpol.2020.113913. [DOI] [PubMed] [Google Scholar]
- 45.Gomi R, Matsuda T, Yamamoto M, Chou P-H, Tanaka M, Ichiyama S, Yoneda M, Matsumura Y. 2018. Characteristics of carbapenemase-producing Enterobacteriaceae in wastewater revealed by genomic analysis. Antimicrob Agents Chemother 62:e02501-17. 10.1128/AAC.02501-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Araújo S, Sousa M, Tacão M, Baraúna RA, Silva A, Ramos R, Alves A, Manaia CM, Henriques I. 2021. Carbapenem-resistant bacteria over a wastewater treatment process: carbapenem-resistant Enterobacteriaceae in untreated wastewater and intrinsically-resistant bacteria in final effluent. Sci Total Environ 782:146892. 10.1016/j.scitotenv.2021.146892. [DOI] [Google Scholar]
- 47.Cooper AL, Carter C, McLeod H, Wright M, Sritharan P, Tamber S, Wong A, Carrillo CD, Blais BW. 2021. Detection of carbapenem-resistance genes in bacteria isolated from wastewater in Ontario. FACETS 6:569–591. 10.1139/facets-2020-0101. [DOI] [Google Scholar]
- 48.Nesporova K, Wyrsch ER, Valcek A, Bitar I, Chaw K, Harris P, Hrabak J, Literak I, Djordjevic SP, Dolejska M. 2020. Escherichia coli sequence type 457 is an emerging extended-spectrum-β-lactam-resistant lineage with reservoirs in wildlife and food-producing animals. Antimicrob Agents Chemother 65:e01118-20. 10.1128/AAC.01118-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jarocki VM, Heß S, Anantanawat K, Berendonk TU, Djordjevic SP. 2021. Multidrug-resistant lineage of enterotoxigenic Escherichia coli ST182 with serotype O169:H41 in airline waste. Front Microbiol 12:731050. 10.3389/fmicb.2021.731050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo X, Wang Q, Xu H, He X, Guo L, Liu S, Wen P, Gou J. 2021. Emergence of IMP-8-producing Comamonas thiooxydans causing urinary tract infection in China. Front Microbiol 12:585716. 10.3389/fmicb.2021.585716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nicolosi D, Nicolosi VM, Cappellani A, Nicoletti G, Blandino G. 2009. Antibiotic susceptibility profiles of uncommon bacterial species causing severe infections in Italy. J Chemother 21:253–260. 10.1179/joc.2009.21.3.253. [DOI] [PubMed] [Google Scholar]
- 52.Li B, Qiu Y, Glidle A, Cooper J, Shi H, Yin H. 2014. Single cell growth rate and morphological dynamics revealing an “opportunistic” persistence. Analyst 139:3305–3313. 10.1039/C4AN00170B. [DOI] [PubMed] [Google Scholar]
- 53.Dai W, Zhu Y, Wang X, Sakenova N, Yang Z, Wang H, Li G, He J, Huang D, Cai Y, Guo W, Wang Q, Feng T, Fan Q, Zheng T, Han A. 2016. Draft genome sequence of the bacterium Comamonas aquatica CJG. Genome Announc 4:e01186-16. 10.1128/genomeA.01186-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wauters G, De Baere T, Willems A, Falsen E, Vaneechoutte M. 2003. Description of Comamonas aquatica comb. nov. and Comamonas kerstersii sp. nov. for two subgroups of Comamonas terrigena and emended description of Comamonas terrigena. Int J Syst Evol Microbiol 53:859–862. 10.1099/ijs.0.02450-0. [DOI] [PubMed] [Google Scholar]
- 55.Waack S, Keller O, Asper R, Brodag T, Damm C, Fricke WF, Surovcik K, Meinicke P, Merkl R. 2006. Score-based prediction of genomic islands in prokaryotic genomes using hidden Markov models. BMC Bioinformatics 7:142. 10.1186/1471-2105-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Veyron S, Peyroche G, Cherfils J. 2018. FIC proteins: from bacteria to humans and back again. Pathog Dis 76:fty012. 10.1093/femspd/fty012. [DOI] [PubMed] [Google Scholar]
- 57.Roy Chowdhury P, Scott M, Worden P, Huntington P, Hudson B, Karagiannis T, Charles IG, Djordjevic SP. 2016. Genomic islands 1 and 2 play key roles in the evolution of extensively drug-resistant ST235 isolates of Pseudomonas aeruginosa. Open Biol 6:150175. 10.1098/rsob.150175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dennis JJ. 2005. The evolution of IncP catabolic plasmids. Curr Opin Biotechnol 16:291–298. 10.1016/j.copbio.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 59.Bilgin H, Sarmis A, Tigen E, Soyletir G, Mulazimoglu L. 2015. Delftia acidovorans: a rare pathogen in immunocompetent and immunocompromised patients. Can J Infect Dis Med Microbiol 26:277–279. 10.1155/2015/973284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Agrawal S, Orschler L, Sinn J, Lackner S. 2020. High-throughput profiling of antibiotic resistance genes in wastewater: comparison between a pond system in Namibia and an activated sludge treatment in Germany. J Water Health 18:867–878. 10.2166/wh.2020.018. [DOI] [PubMed] [Google Scholar]
- 61.Nordmann P, Dortet L, Poirel L. 2012. Carbapenem resistance in Enterobacteriaceae: here is the storm!. Trends Mol Med 18:263–272. 10.1016/j.molmed.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 62.Peri AM, Doi Y, Potoski BA, Harris PNA, Paterson DL, Righi E. 2019. Antimicrobial treatment challenges in the era of carbapenem resistance. Diagn Microbiol Infect Dis 94:413–425. 10.1016/j.diagmicrobio.2019.01.020. [DOI] [PubMed] [Google Scholar]
- 63.Runcharoen C, Moradigaravand D, Blane B, Paksanont S, Thammachote J, Anun S, Parkhill J, Chantratita N, Peacock SJ. 2017. Whole genome sequencing reveals high-resolution epidemiological links between clinical and environmental Klebsiella pneumoniae. Genome Med 9:6. 10.1186/s13073-017-0397-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yao Y, Lazaro-Perona F, Falgenhauer L, Valverde A, Imirzalioglu C, Dominguez L, Cantón R, Mingorance J, Chakraborty T. 2017. Insights into a novel blaKPC-2-encoding IncP-6 plasmid reveal carbapenem-resistance circulation in several Enterobacteriaceae species from wastewater and a hospital source in Spain. Front Microbiol 8:1143. 10.3389/fmicb.2017.01143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zarfel G, Lipp M, Gürtl E, Folli B, Baumert R, Kittinger C. 2017. Troubled water under the bridge: screening of River Mur water reveals dominance of CTX-M harboring Escherichia coli and for the first time an environmental VIM-1 producer in Austria. Sci Total Environ 593–594:399–405. 10.1016/j.scitotenv.2017.03.138. [DOI] [PubMed] [Google Scholar]
- 66.Duran A, Okur F, Sahin V, Uyar I, Abacilar A, Akpinar M, Alayunt E, Ates M. 2015. Comamonas testosteroni endocarditis in Turkey: a case report and review of the literature. Int Med J Sifa Univ 2:44. 10.4103/2148-7731.152117. [DOI] [Google Scholar]
- 67.Nordmann P, Poirel L. 2019. Epidemiology and diagnostics of carbapenem resistance in Gram-negative bacteria. Clin Infect Dis 69:S521–S528. 10.1093/cid/ciz824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yasayancan N, Koseoglu HI. 2017. The 20th Comamonas testosteroni bacteremia case in the literature from Turkey, mortal and polymicrobial: a case report and literature review. EJMO 1:168–171. 10.14744/ejmo.2017.16878. [DOI] [Google Scholar]
- 69.Kaeuffer C, Schramm F, Meyer A, Hansmann Y, Guffroy A, Argemi X. 2018. First case of Comamonas aquatica bacteremia complicated by septic shock. Med Mal Infect 48:540–542. 10.1016/j.medmal.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 70.Almuzara M, Barberis C, Traglia G, Famiglietti A, Soledad Ramirez M, Vay C. 2015. Evaluation of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry for species identification of nonfermenting Gram-negative bacilli. J Microbiol Methods 112:24–27. 10.1016/j.mimet.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 71.Lood R, Ertürk G, Mattiasson B. 2017. Revisiting antibiotic resistance spreading in wastewater treatment plants – Bacteriophages as a much neglected potential transmission vehicle. Front Microbiol 8:2298. 10.3389/fmicb.2017.02298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ng C, Tan B, Jiang X-T, Gu X, Chen H, Schmitz BW, Haller L, Charles FR, Zhang T, Gin K. 2019. Metagenomic and resistome analysis of a full-scale municipal wastewater treatment plant in Singapore containing membrane bioreactors. Front Microbiol 10:172. 10.3389/fmicb.2019.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aziz A, Basheer F, Sengar A, Irfanullah Khan SU, Farooqi IH. 2019. Biological wastewater treatment (anaerobic-aerobic) technologies for safe discharge of treated slaughterhouse and meat processing wastewater. Sci Total Environ 686:681–708. 10.1016/j.scitotenv.2019.05.295. [DOI] [PubMed] [Google Scholar]
- 74.Palanisamy V, Gajendiran V, Mani K. 2022. Meta-analysis to identify the core microbiome in diverse wastewater. Int J Environ Sci Technol 19:5079–5096. 10.1007/s13762-021-03349-4. [DOI] [Google Scholar]
- 75.Jiang B, Zhou Z, Dong Y, Tao W, Wang B, Jiang J, Guan X. 2015. Biodegradation of benzene, toluene, ethylbenzene, and o-, m-, and p-xylenes by the newly isolated bacterium Comamonas sp. JB. Appl Biochem Biotechnol 176:1700–1708. 10.1007/s12010-015-1671-6. [DOI] [PubMed] [Google Scholar]
- 76.Song Z, Edwards SR, Burns RG. 2005. Biodegradation of naphthalene-2-sulfonic acid present in tannery wastewater by bacterial isolates Arthrobacter sp. 2AC and Comamonas sp. 4BC. Biodegradation 16:237–252. 10.1007/s10532-004-0889-8. [DOI] [PubMed] [Google Scholar]
- 77.Liu J, Yu Y, Chang Y, Li B, Bian D, Yang W, Huo H, Huo M, Zhu S. 2016. Enhancing quinoline and phenol removal by adding Comamonas testosteroni bdq06 in treatment of an accidental dye wastewater. Int Biodeterior Biodegradation 115:74–82. 10.1016/j.ibiod.2016.07.016. [DOI] [Google Scholar]
- 78.Wang C-C, Lee C-M, Chen L-J. 2004. Removal of nitriles from synthetic wastewater by acrylonitrile utilizing bacteria. J Environ Sci Health A Tox Hazard Subst Environ Eng 39:1767–1779. 10.1081/ese-120037876. [DOI] [PubMed] [Google Scholar]
- 79.Jiang B, Zhou Z, Dong Y, Wang B, Jiang J, Guan X, Gao S, Yang A, Chen Z, Sun H. 2015. Bioremediation of petrochemical wastewater containing BTEX compounds by a new immobilized bacterium Comamonas sp. JB in magnetic gellan gum. Appl Biochem Biotechnol 176:572–581. 10.1007/s12010-015-1596-0. [DOI] [PubMed] [Google Scholar]
- 80.Andersson S, Dalhammar G, Land CJ, Rajarao GK. 2009. Characterization of extracellular polymeric substances from denitrifying organism Comamonas denitrificans. Appl Microbiol Biotechnol 82:535–543. 10.1007/s00253-008-1817-3. [DOI] [PubMed] [Google Scholar]
- 81.Wu Y, Shukal S, Mukherjee M, Cao B. 2015. Involvement in denitrification is beneficial to the biofilm lifestyle of Comamonas testosteroni: a mechanistic study and its environmental implications. Environ Sci Technol 49:11551–11559. 10.1021/acs.est.5b03381. [DOI] [PubMed] [Google Scholar]
- 82.Huyan J, Tian Z, Zhang Y, Zhang H, Shi Y, Gillings MR, Yang M. 2020. Dynamics of class 1 integrons in aerobic biofilm reactors spiked with antibiotics. Environ Int 140:105816. 10.1016/j.envint.2020.105816. [DOI] [PubMed] [Google Scholar]
- 83.Rosso F, Cedano JA, Parra-Lara LG, Sanz AM, Toala A, Velez JF, Hormaza MP, Moncada PA, Correa A. 2019. Emerging carbapenem-resistant Aeromonas spp. infections in Cali, Colombia. Braz J Infect Dis 23:336–342. 10.1016/j.bjid.2019.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.White L, Hopkins KL, Meunier D, Perry CL, Pike R, Wilkinson P, Pickup RW, Cheesbrough J, Woodford N. 2016. Carbapenemase-producing Enterobacteriaceae in hospital wastewater: a reservoir that may be unrelated to clinical isolates. J Hosp Infect 93:145–151. 10.1016/j.jhin.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 85.Manageiro V, Ferreira E, Caniça M, Manaia CM. 2014. GES-5 among the β-lactamases detected in ubiquitous bacteria isolated from aquatic environment samples. FEMS Microbiol Lett 351:64–69. 10.1111/1574-6968.12340. [DOI] [PubMed] [Google Scholar]
- 86.Hishinuma T, Tada T, Kuwahara-Arai K, Yamamoto N, Shimojima M, Kirikae T. 2018. Spread of GES-5 carbapenemase-producing Pseudomonas aeruginosa clinical isolates in Japan due to clonal expansion of ST235. PLoS One 13:e0207134. 10.1371/journal.pone.0207134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang J, Xu T, Ying J, Zhou W, Chen Q, Qian C, Zhu X, Shen K, Li P, Li K, Bao Q, Lu J. 2019. PAU-1, a novel plasmid-encoded ambler class A β-lactamase identified in a clinical Pseudomonas aeruginosa isolate. Infect Drug Resist 12:3827–3834. 10.2147/IDR.S225288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Juhas M. 2019. Genomic islands and the evolution of multidrug-resistant bacteria, p 143–153. In Villa TG, Viñas M (ed), Horizontal Gene Transfer: Breaking Borders between Living Kingdoms. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 89.Schmidt H, Hensel M. 2004. Pathogenicity islands in bacterial pathogenesis. Clin Microbiol Rev 17:14–56. 10.1128/CMR.17.1.14-56.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fariq A, Blazier JC, Yasmin A, Gentry TJ, Deng Y. 2019. Whole genome sequence analysis reveals high genetic variation of newly isolated Acidithiobacillus ferrooxidans IO-2C. Sci Rep 9:13049. 10.1038/s41598-019-49213-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pagano M, Martins AF, Barth AL. 2016. Mobile genetic elements related to carbapenem resistance in Acinetobacter baumannii. Braz J Microbiol 47:785–792. 10.1016/j.bjm.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Recchia GD, Hall RMY. 1995. Gene cassettes: a new class of mobile element. Microbiology 141:3015–3027. 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 93.Imran M, Das KR, Naik MM. 2019. Co-selection of multi-antibiotic resistance in bacterial pathogens in metal and microplastic contaminated environments: an emerging health threat. Chemosphere 215:846–857. 10.1016/j.chemosphere.2018.10.114. [DOI] [PubMed] [Google Scholar]
- 94.Deshpande LM, Kapadnis BP, Chopade BA. 1993. Metal resistance in Acinetobacter and its relation to β-lactamase production. Biometals 6:55–59. 10.1007/BF00154233. [DOI] [PubMed] [Google Scholar]
- 95.Rudakiya DM. 2018. Metal tolerance assisted antibiotic susceptibility profiling in Comamonas acidovorans. Biometals 31:1–5. 10.1007/s10534-017-0074-2. [DOI] [PubMed] [Google Scholar]
- 96.Yang Y, Yue Y, Song N, Li C, Yuan Z, Wang Y, Ma Y, Li H, Zhang F, Wang W, Jia H, Li P, Li X, Wang Q, Ding Z, Dong H, Gu L, Li B. 2020. The YdiU domain modulates bacterial stress signaling through Mn2+-dependent UMPylation. Cell Rep 32:108161. 10.1016/j.celrep.2020.108161. [DOI] [PubMed] [Google Scholar]
- 97.Dunon V, Bers K, Lavigne R, Top E, Springael D. 2018. Targeted metagenomics demonstrates the ecological role of IS1071 in bacterial community adaptation to pesticide degradation. Environ Microbiol 20:4091–4111. 10.1111/1462-2920.14404. [DOI] [PubMed] [Google Scholar]
- 98.Stolze Y, Eikmeyer F, Wibberg D, Brandis G, Karsten C, Krahn I, Schneiker-Bekel S, Viehöver P, Barsch A, Keck M, Top EM, Niehaus K, Schlüter A. 2012. IncP-1β plasmids of Comamonas sp. and Delftia sp. strains isolated from a wastewater treatment plant mediate resistance to and decolorization of the triphenylmethane dye crystal violet. Microbiology 158:2060–2072. 10.1099/mic.0.059220-0. [DOI] [PubMed] [Google Scholar]
- 99.Top EM, Springael D. 2003. The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr Opin Biotechnol 14:262–269. 10.1016/s0958-1669(03)00066-1. [DOI] [PubMed] [Google Scholar]
- 100.Sota M, Yano H, Nagata Y, Ohtsubo Y, Genka H, Anbutsu H, Kawasaki H, Tsuda M. 2006. Functional analysis of unique class II insertion sequence IS1071. Appl Environ Microbiol 72:291–297. 10.1128/AEM.72.1.291-297.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dasari S, Ganjayi MS, Yellanurkonda P, Basha S, Meriga B. 2018. Role of glutathione S-transferases in detoxification of a polycyclic aromatic hydrocarbon, methylcholanthrene. Chem Biol Interact 294:81–90. 10.1016/j.cbi.2018.08.023. [DOI] [PubMed] [Google Scholar]
- 102.Henze M, Loosdrecht M, Ekama G, Brdjanovic D. 2015. Wastewater treatment development. In Biological Wastewater Treatment: Principles, Modelling and Design. IWA Publishing, London, UK. [Google Scholar]
- 103.Schulthess B, Bloemberg GV, Zbinden A, Mouttet F, Zbinden R, Böttger EC, Hombach M. 2016. Evaluation of the Bruker MALDI biotyper for identification of fastidious Gram-negative rods. J Clin Microbiol 54:543–548. 10.1128/JCM.03107-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Drigo B, Brunetti G, Aleer SC, Bell JM, Short MD, Vasileiadis S, Turnidge J, Monis P, Cunliffe D, Donner E. 2021. Inactivation, removal, and regrowth potential of opportunistic pathogens and antimicrobial resistance genes in recycled water systems. Water Res 201:117324. 10.1016/j.watres.2021.117324. [DOI] [PubMed] [Google Scholar]
- 105.Leray M, Knowlton N, Ho S-L, Nguyen BN, Machida RJ. 2019. GenBank is a reliable resource for 21st century biodiversity research. Proc Natl Acad Sci USA 116:22651–22656. 10.1073/pnas.1911714116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Darling AE, Jospin G, Lowe E, Matsen FA, Bik HM, Eisen JA. 2014. PhyloSift: phylogenetic analysis of genomes and metagenomes. PeerJ 2:e243. 10.7717/peerj.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524. 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hadfield J, Croucher NJ, Goater RJ, Abudahab K, Aanensen DM, Harris SR. 2018. Phandango: an interactive viewer for bacterial population genomics. Bioinformatics 34:292–293. 10.1093/bioinformatics/btx610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Brynildsrud O, Bohlin J, Scheffer L, Eldholm V. 2016. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol 17:238. 10.1186/s13059-016-1108-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wood DE, Lu J, Langmead B. 2019. Improved metagenomic analysis with Kraken 2. Genome Biol 20:257. 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. 2016. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32:929–931. 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Meier-Kolthoff JP, Auch AF, Klenk H-P. 2013. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen L, Yang J, Yu J, Yao Z, Sun L, Shen Y, Jin Q. 2005. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res 33:D325–D328. 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen A-LV, Cheng AA, Liu S, Min SY, Miroshnichenko A, Tran H-K, Werfalli RE, Nasir JA, Oloni M, Speicher DJ, Florescu A, Singh B, Faltyn M, Hernandez-Koutoucheva A, Sharma AN, Bordeleau E, Pawlowski AC, Zubyk HL, Dooley D, Griffiths E, Maguire F, Winsor GL, Beiko RG, Brinkman FSL, Hsiao WWL, Domselaar GV, McArthur AG. 2020. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 48:D517–D525. 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Doster E, Lakin SM, Dean CJ, Wolfe C, Young JG, Boucher C, Belk KE, Noyes NR, Morley PS. 2020. MEGARes 2.0: a database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data. Nucleic Acids Res 48:D561–D569. 10.1093/nar/gkz1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Carattoli A, Zankari E, García-Fernández A, Larsen MV, Lund O, Villa L, Aarestrup FM, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA, Stevens R, Vonstein V, Wattam AR, Xia F. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365. 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bertelli C, Laird MR, Williams KP, Lau BY, Hoad G, Winsor GL, Brinkman FS, Simon Fraser University Research Computing Group . 2017. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res 45:W30–W35. 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pereira MB, Wallroth M, Kristiansson E, Axelson-Fisk M. 2016. HattCI: fast and accurate attC site identification using hidden Markov models. J Comput Biol 23:891–902. 10.1089/cmb.2016.0024. [DOI] [PubMed] [Google Scholar]
- 124.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Larsson A. 2014. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30:3276–3278. 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Set S1. Download aem.00646-22-s0001.xlsx, XLSX file, 0.01 MB (13.3KB, xlsx)
Data Set S2. Download aem.00646-22-s0002.xlsx, XLSX file, 0.02 MB (19.4KB, xlsx)
Data Set S3. Download aem.00646-22-s0003.xlsx, XLSX file, 0.01 MB (14.5KB, xlsx)