ABSTRACT
Dental caries is a multifactorial disease driven by interactions between the highly complex microbial biofilm community and host factors like diet, oral hygiene habits, and age. The oral streptococci are one of the most dominant members of the plaque biofilm and are implicated in disease but also in maintaining oral health. Current methods used for studying the supragingival plaque community commonly sequence portions of the16S rRNA gene, which often cannot taxonomically resolve members of the streptococcal community past the genus level due to their sequence similarity. The goal of this study was to design and evaluate a more reliable and cost-effective method to identify oral streptococci at the species level by applying a new locus, the 30S-S11 rRNA gene, for high-throughput amplicon sequencing. The study results demonstrate that the newly developed single-copy 30S-S11 gene locus resolved multiple amplicon sequence variants (ASVs) within numerous species, providing much improved taxonomic resolution over 16S rRNA V4. Moreover, the results reveal that different ASVs within a species were found to change in abundance at different stages of caries progression. These findings suggest that strains of a single species may perform distinct roles along a biochemical spectrum associated with health and disease. The improved identification of oral streptococcal species will provide a better understanding of the different ecological roles of oral streptococci and inform the design of novel oral probiotic formulations for prevention and treatment of dental caries.
IMPORTANCE The microbiota associated with the initiation and progression of dental caries has yet to be fully characterized. Although much insight has been gained from 16S rRNA hypervariable region DNA sequencing, this approach has several limitations, including poor taxonomic resolution at the species level. This is particularly relevant for oral streptococci, which are abundant members of oral biofilm communities and major players in health and caries disease. Here, we develop a new method for taxonomic profiling of oral streptococci based on the 30S-S11 rRNA gene, which provides much improved resolution over 16S rRNA V4 (resolving 10 as opposed to 2 species). Importantly, 30S-S11 can resolve multiple amplicon sequence variants (ASVs) within species, providing an unprecedented insight into the ecological progression of caries. For example, our findings reveal multiple incidences of different ASVs within a species with contrasting associations with health or disease, a finding that has high relevance toward the informed design of prebiotic and probiotic therapy.
KEYWORDS: dental caries, 16S, streptococci, amplicon, microbiome, oral
INTRODUCTION
Next-generation sequencing has made possible significant advancements in our understanding of the complex community structure and diverse nature of the oral bacteriome without the need for culture-based methods. One of the most cost-effective, timely, and commonly used techniques is amplicon sequencing of hypervariable regions of the 16S rRNA gene (1, 2). The 16S rRNA gene is approximately 1,500 bp and is ubiquitous in bacteria (3, 4). It is composed of nine highly conserved regions and nine hypervariable regions (V1 to V9) (3). The highly conserved regions flank the hypervariable regions, providing an area where universal PCR primers can be designed and used to generate sequences capable of delineating between different bacterial taxa (5). Amplicon sequencing of 16S rRNA hypervariable regions is widely used in microbiome studies; however, there are several limitations associated with this approach. Importantly, hypervariable regions provide poor taxonomic resolution at the species level due to low sequence divergence (6). Additional problems include a wide range in copy number of the operon containing the 16S-5S-23S rRNA genes within a genome. For example, Hassler et al. (unpublished data) reported a range of 1 to 27 copies of the 16S rRNA gene among 165 species representing four divergent genera. This study also showed the hypervariable regions to have low phylogenetic performance. Both characteristics have potential to confound microbiome diversity metrics.
Although sequencing of the entire 16S rRNA gene can improve taxonomic resolution, it is more costly and requires long-read sequencing technology or shotgun sequencing (4). Notably, the 16S gene does not have good taxonomic resolution at the species level for many abundant oral genera, including Streptococcus, Neisseria, Pseudomonas, and Enterobacter (5). Oral streptococci are important members of the plaque biofilm. They are dominant species in the mouth and are known to be one of the first inhabitants of the oral cavity acquired directly after birth (7). These bacteria are mostly commensal and occupy all niches of the oral cavity; however, certain species of Streptococcus are pathogenic. The mutans streptococci, principally Streptococcus mutans, have been considered the most prominent cariogens in the oral cavity (7–9). S. mutans has many important biochemical properties that contribute to its enhanced virulence in caries development and progression, including its ability to metabolize a variety of different carbohydrate sources and produce acids, to tolerate acidic environments, and to produce extracellular polysaccharides (EPSs) that are key for biofilm formation and adherence to the tooth surface (8). In contrast, there are many oral streptococci (e.g., Streptococcus australis, Streptococcus sanguinis, Streptococcus gordonii, and Streptococcus parasanguinis) that are associated with maintenance of dental health by raising pH through ammonia production as a result of arginine catabolism via the arginine deaminase system (ADS) and that have shown antagonistic or inhibitory effects on the growth of S. mutans (7, 10–13).
With a deeper understanding of dental microbial community dynamics and the taxa that contribute to the development of caries, the concept of preventing and treating caries using prebiotics and probiotics has emerged (14). One of the major drivers believed to cause caries is the shift of the microbial community into a dysbiotic state dominated by microbes that can survive and thrive under acidic conditions. The goal of prebiotics and probiotics is to introduce either substrates or living microbes that can shift the dysbiotic microbiome back into symbiosis with its host (14, 15). However, to successfully use prebiotics and probiotics, knowledge of the exact species and strains of oral bacteria comprising the tooth biofilm is needed. Previous studies have shown a large spectrum of phenotypic differences and biochemical properties within strains, resulting in large variations in virulence (16–18). Because of this, detailed knowledge of which strains are present during health and throughout disease can allow for mechanistic and biochemical screening of these taxa. These tests can determine if they are promising candidates for the design of an oral probiotic formulation. Due to the high prevalence of Streptococcus species in the mouth and their importance in oral health and disease, a method to obtain a fast, reliable, and accurate taxonomic classification to the species level is needed for both diagnostic and research purposes. To accomplish this, we have developed PCR primers that target 378 bp of the 381-bp single-copy 30S-S11 gene in oral streptococci. The 30S-S11 ribosomal protein is an rRNA binding protein and a structural component of the ribosome (19). This protein forms a portion of the Shine-Dalgarno cleft of the 70S ribosomal subunit. The 30S-S11 gene is well conserved in bacteria and has been identified in multiple genera (19, 20).
A set of site-specific supragingival plaque samples (in which communities are categorized into six progressive stages of the disease) obtained from 33 children were amplified using the newly developed 30S-S11 primers and primers that target the V4 region of the 16S gene (806r and 515f) for comparison. The V4 region was targeted as it is one of the most used hypervariable regions in microbiome studies (21, 22). Comparison showed that our newly developed 30S-S11 primer set had higher taxonomic resolution. Specifically, the 30S-S11 amplicon data provided species-level identification for 10 oral streptococci, compared to only 2 species-level identifications when using V4 data. A key finding was that the 30S-S11 amplicon data could resolve multiple amplicon sequence variants (ASVs) per species. Importantly, our study found that within some species, different ASVs were correlated with health and others with disease. These results have important implications for oral probiotic design. A deeper understanding of the bacterial strains that are associated with dental health will allow for an accurate screening of oral probiotic candidates for caries prevention and treatment.
RESULTS
In silico taxonomic resolution of the 30S-S11 rRNA loci and mock community evaluation.
To identify a replacement locus for 16S rRNA hypervariable regions, we constructed global sequence alignments for 18 genes shared among the genomes of 1,406 clinical strains of oral streptococci (23 species) (Table 1; see Table S1 in the supplemental material). Following criteria outlined in Materials and Methods, which included locus length, sequence divergence, and conserved PCR primer sites, we found that 378 bp of the 30S-S11 rRNA gene (381 bp) to be the most suitable replacement. A global nucleotide sequence alignment showed the 1,406 strains to be represented by 92 unique sequence variants (6.7% sequence divergence), with many species represented by multiple variants. A maximum likelihood (ML) phylogeny containing these sequences was able to resolve the following 13 species: Streptococcus agalactiae, Streptococcus pyogenes, Streptococcus downei, Streptococcus sobrinus, S. parasanguinus, S. mutans, Streptococcus lactarius, Streptococcus salavarius, S. australis, S. sanguinis, Streptococcus cristatus, S. gordonii, and Streptococcus sinensis (Fig. 1A). Strains for the following 10 species did not show monophyletic branching patterns but fell into three complexes (clades): (i) Streptococcus mitis, Streptococcus oralis, Streptocccus pneumoniae, Streptococcus infantis, and Streptococcus peroris (Mitis complex); (ii) Streptococcus thermophilus and Streptococcus vestibularis (Vestibularis complex); and (iii) Streptococcus anginosus, Streptococcus intermedius, and Streptococcus constellatus (Anginosus complex). Using a gene clustering approach (see Materials and Methods), we evaluated different sequence identity clustering threshold values for use when constructing a custom taxonomy classifier database. The 13 species and three complexes were reliably recovered using a threshold of 98% sequence identity. Following this taxonomic delineation, the 92 30S-S11 haplotype sequences were used to build a custom reference database for QIIME2 (23). For comparative purposes, and using the same genomes, we extracted the 16S rRNA V3 and V4 regions and built an ML phylogeny of the combined V3-V4 regions (474 bp) and V4 region (297 bp). The V3-V4 regions recovered five species with 4.9% sequence divergence (Fig. 1B), and the V4 region recovered three species with 4.3% divergence (Fig. 1C).
TABLE 1.
Eighteen core oral streptococcal genes
| Representative locus tag | Gene product |
|---|---|
| WY5_RS110910 | 30S ribosomal protein S10 |
| WY5_RS111035 | 30S ribosomal protein S11 |
| D8785_RS04160 | 30S ribosomal protein S12 |
| WY5_RS110960 | 30S ribosomal protein S17 |
| WY5_RS118250 | 30S ribosomal protein S18 |
| WY5_RS110945 | 30S ribosomal protein S3 |
| D8880_RS00905 | 30S ribosomal protein S6 |
| WY5_RS118615 | 30S ribosomal protein S7 |
| WY5_RS117375 | 50S ribosomal protein L11 |
| SK137_RS00700 | 50S ribosomal protein L16 |
| DK43_RS01015 | 50S ribosomal protein L18 |
| WY5_RS116740 | 50S ribosomal protein L20 |
| WY5_RS110940 | 50S ribosomal protein L22 |
| WY5_RS110975 | 50S ribosomal protein L5 |
| CYK15_RS07680 | Elongation factor G |
| WY5_RS114220 | Elongation factor Tu |
| WY5_RS111950 | Fructose-1,6-bisphosphate aldolase, class II |
| SK642_RS05300 | Phosphopyruvate hydratase |
FIG 1.
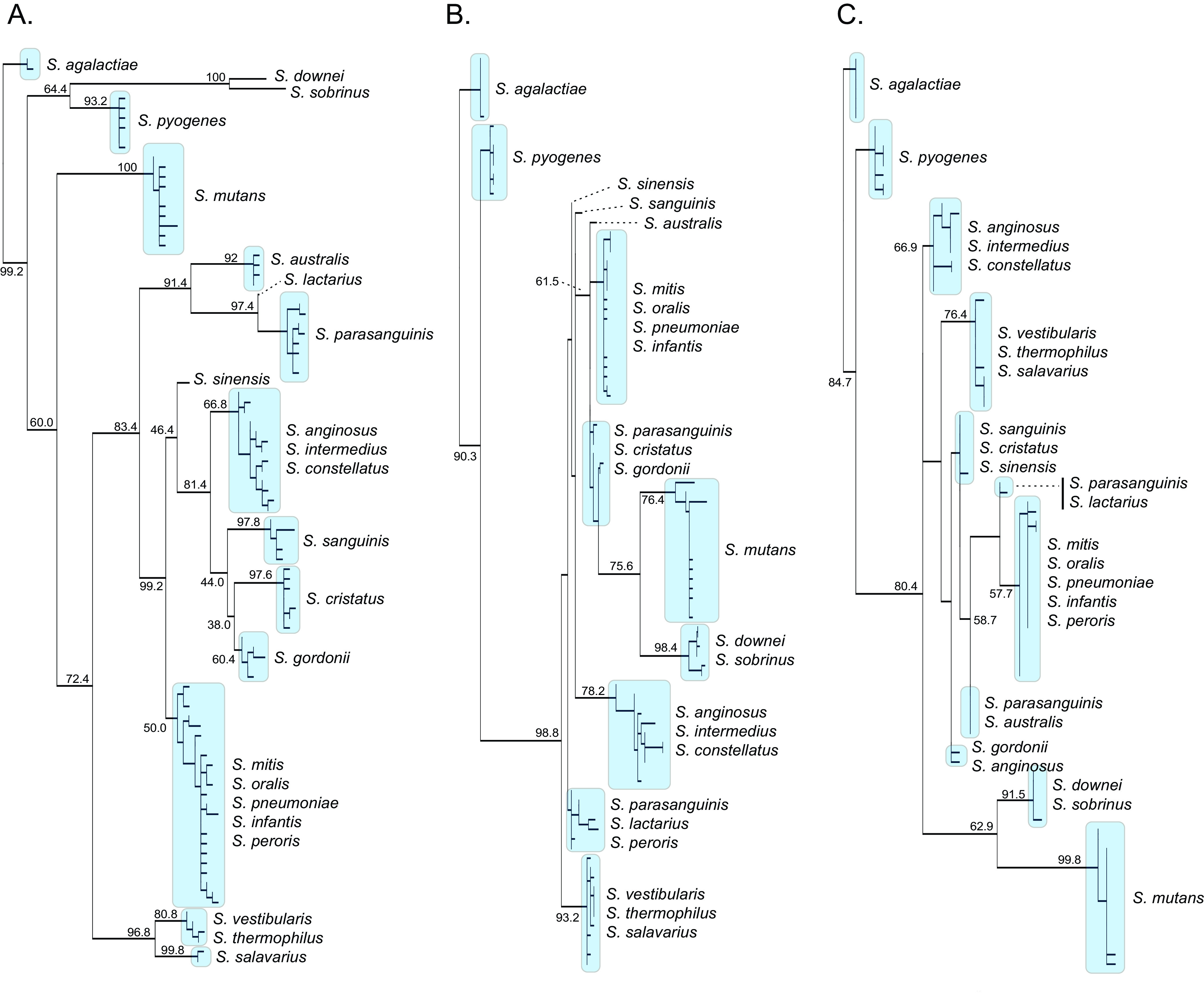
Maximum likelihood (ML) phylogenies comparing oral streptococcal taxonomic resolution for (A) the 30S-S11 rRNA gene, (B) the combined 16S rRNA V3 and V4 regions, and (C) the 16S rRNA V4 region. Bootstrap support values (500 replicates) are shown.
PCR primers designed to amplify 378 bp of the 30S-S11 gene were evaluated using DNA from two mock communities representing 10 common supragingival streptococcal species. For each community, amplicon sequence reads received taxonomic assignments that were concordant with the respective taxonomic composition of each community (Fig. S1). More specifically, DNA from all members of each community was successfully sequenced and taxonomically identified.
High-resolution evaluation of streptococcal community dynamics during early-childhood caries.
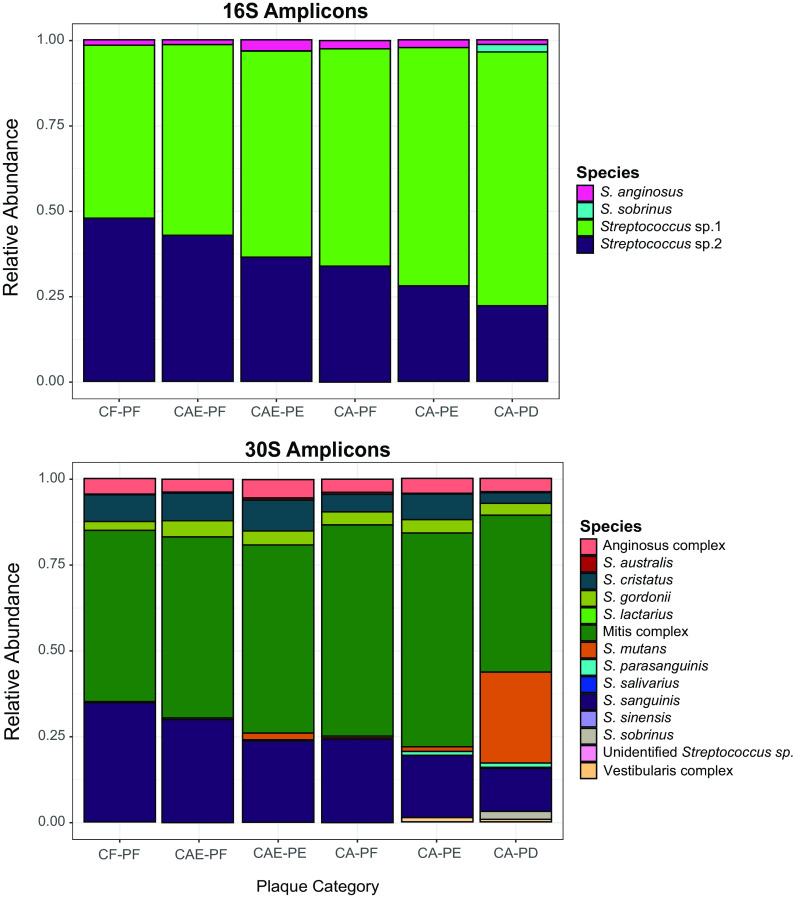
As described previously, variations in the supragingival environment can lead to changes in the bacterial community composition and subsequently drive microbial dysbiosis and lead to caries. To evaluate the 30S-S11 rRNA gene’s ability to taxonomically profile supragingival plaque communities during caries progression, we analyzed 82 supragingival plaque samples representing six progressive stages of the disease from 33 children ranging in age from 2 to 7 years and compared profiling using the 30S-S11 rRNA gene and the 16S rRNA V4 loci. Concordant with our phylogenetic analysis (and mock community evaluation) the 30S-S11 primers detected 10 species and three species complexes. This was a considerable improvement over the 16S V4 region, which was only able to resolve two species (S. anginosus and S. sobrinus) (Fig. 2). In addition to resolving more species than 16S V4, the 30S-S11 primers also detected multiple ASVs for each species (Fig. 3). In total, we detected 162 ASVs (80 in the species complexes) (Table 2). The number of ASVs ranged from 1 for S. sobrinus to 17 for S. australis. A total of 14 ASVs not belonging to a species complex showed significant differential abundance during stages of caries (Fig. 3; Table S2). These 14 ASVs represented six species: S. sanguinis, S. cristatus, S. gordonii, S. mutans, S. sobrinus, and S. parasanguinis. Importantly, S. sanguinis and S. cristatus each contained both health- and disease-associated ASVs (Fig. 3). For S. sanguinis, a total of nine ASVs were observed, with three showing statistical difference in their abundance (ASV3, ASV4, and ASV5). The distribution of ASV4 was skewed toward health and was found in highest abundance in plaque collected from caries-free tooth surfaces. In contrast, ASV3 and ASV5 were associated with plaque collected from carious teeth. ASV3 was enriched in the late stage of caries development and mostly absent in the earlier stages, whereas ASV5 occurred in all health categories but had a higher frequency in enamel caries. Although ASV1 did not show any significant differences, it did show a progressive decrease in abundance as caries severity increased. Of note, this ASV was the second most abundant in the entire data set, making up 17.7% of the community (Fig. 3; Table S3). Of the 11 S. cristatus ASVs, five were differentially abundant throughout caries progression. ASVs 3 and 6 were significantly enriched in plaque from dentinal caries lesions, whereas ASVs 4, 5, and 8 were enriched in health and early-stage enamel caries. Although not statistically significant, ASV 9 appeared enriched in dentin caries, ASV 7 appeared enriched in enamel caries, and ASVs 10, and 11 appeared enriched in caries-free teeth. S. gordonii, S. sobrinus, S. mutans, and S. parasanguinis all contained ASVs that were significantly enriched in caries. S. gordonii contained four ASVs, of which three (ASVs 2, 3, and 4) were significantly differentially abundant. ASV3 was enriched in dentin caries, whereas ASVs 2 and 4 were enriched in enamel caries. S. sobrinus contained only one ASV, which was found in increased abundance in late-stage dentin caries. A similar trend was observed for S. mutans (two ASVs), where ASV1 was significantly enriched in late-stage dentin caries. Although the second ASV of S. mutans, which occurred in much lower abundance, was not significant, it did show an increase in abundance as caries progressed. S. parasanguinis also contained only one significantly differentially abundant ASV (ASV1), which also increased in abundance as caries progressed. While not significant, ASVs 2, 3, 4, and 6 all showed increased abundance in plaque samples from active enamel and dentinal lesions and decreased abundance on plaque from healthy tooth surfaces.
FIG 2.
Stacked bar charts of the relative abundance (percentage) values of oral streptococcal species obtained after 16S rRNA V4 and 30S-S11 rRNA amplicon sequencing of site-specific supragingival plaque samples. See text for a full explanation of the six plaque categories (each representing progressive stages of caries). In brief, CF-PF is plaque from a caries-free child, whereas CA-PD is plaque from a dentin carious lesion (the most progressed stage).
FIG 3.
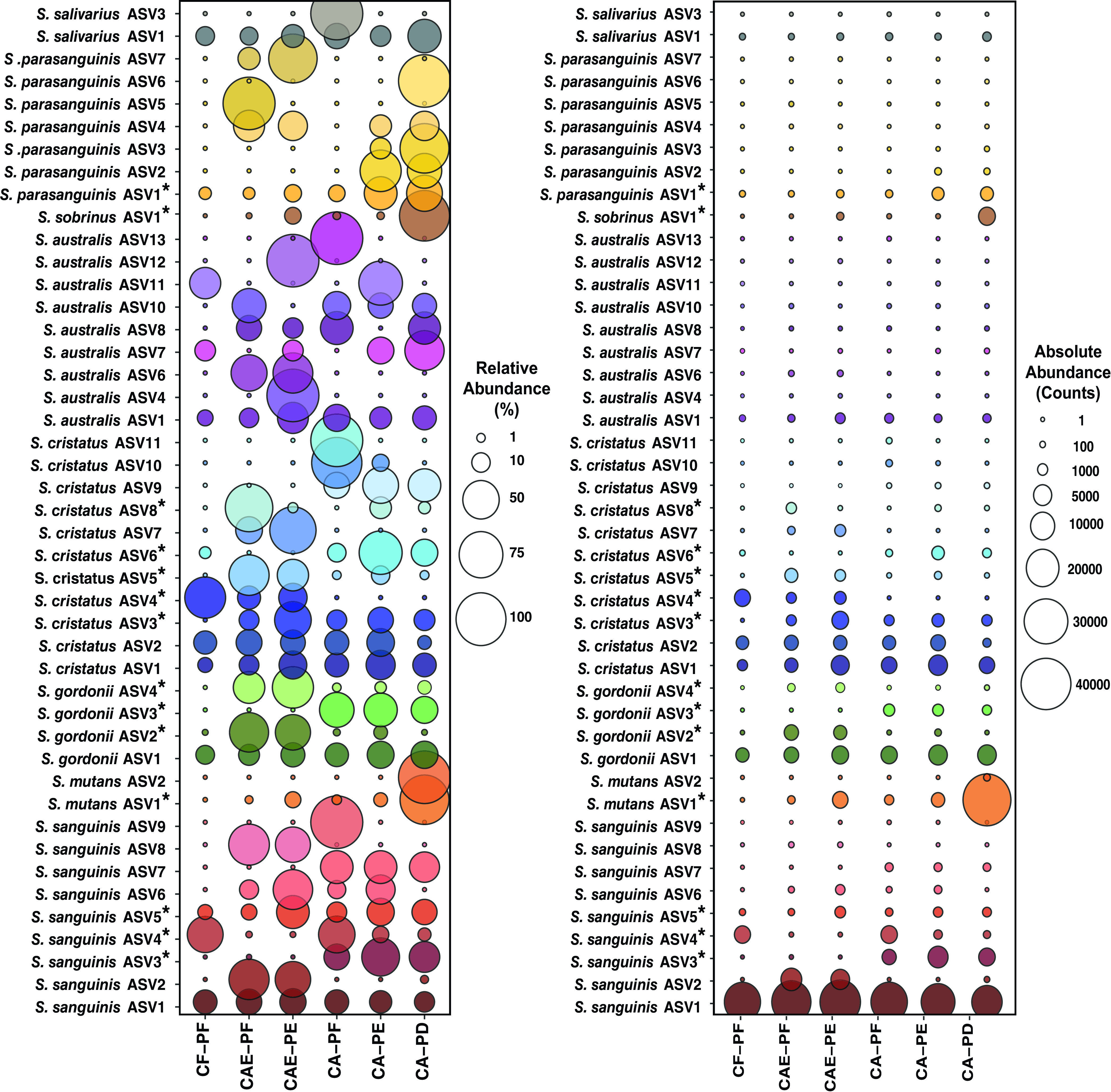
Bubble plot showing abundance of ASVs for six plaque categories (see Materials and Methods and Fig. 2 legend). ASVs showing a significant difference in abundance among categories are indicated with an asterisk. All species depicted occurred at least three times and had a relative abundance greater than 0.0001%. The left plot depicts relative abundance (percentage), and the right plot depicts absolute abundance (counts).
TABLE 2.
Total number of ASVs identified for each streptococcal species and their total abundance across the data set
| Taxonomy | No. of ASVs identified | % of abundance in data set |
|---|---|---|
| Mitis complex | 62 | 54.5992 |
| Streptococcus sanguinis | 11 | 22.3498 |
| Streptococcus cristatus | 11 | 6.571 |
| Streptococcus mutans | 7 | 6.2707 |
| Anginosus complex | 13 | 4.2874 |
| Streptococcus gordonii | 6 | 3.807 |
| Streptococcus parasanguinis | 10 | 0.601 |
| Streptococcus sobrinus | 1 | 0.5156 |
| Vestibularis complex | 5 | 0.4072 |
| Streptococcus australis | 17 | 0.4016 |
| Streptococcus salivarius | 5 | 0.1822 |
| Streptococcus sp. | 9 | 0.0063 |
| S. sinensis | 3 | 0.0005 |
| S. lactarius | 2 | 0.0006 |
It is of note that there were one or two predominant ASVs for some species that remained consistent in their abundance throughout caries progression. Specifically, these ASVs were S. sanguinis ASV1, S. gordonii ASV1, S. cristatus ASV1 and ASV2, S. australis ASV1, and S. salivarius ASV1. Intriguingly, these were the most abundant ASVs for their respective species. The Mitis complex (which can capture five species) contained 62 ASVs, the Anginosus complex (which can capture three species) contained 13 ASVs, and the Vestibularis complex (which can capture two species) contained 5 ASVs. The most abundant ASV was within the S. mitis complex (ASV1). This ASV made up 35.1% of all reads in the data set, and its abundance was not statistically different among the stages of caries progression (Table 2). Similarly, ASVs for S. australis and S. salivarius did not show any significant difference in their abundance at any stage of caries progression.
DISCUSSION
Dental caries is a multifaceted disease characterized by microbial interactions operating across many taxonomic levels within the tooth biofilm. The complexity of these interactions is considerable, and until recently, the technology available to study the composition of the dental plaque microbiome was limited (24). Much of the research on caries has relied on culture-based studies or short-read 16S rRNA amplicon sequencing, both of which have limitations. The largest of these are the uncultivability of some organisms, loss of species-level resolution for important health and disease-associated taxa, and low sequence divergence among members of the same genus (25). In the present study, we developed a new locus for high-throughput amplicon sequencing, the 30S-S11 rRNA gene, which provides a cost-effective method for better taxonomic resolution of species within the genus Streptococcus. The new locus can resolve multiple ASVs within numerous species, providing much improved taxonomic resolution over the 16S rRNA V4 region. Importantly, the ability to resolve multiple ASVs within a species provides unprecedented insights into the ecological progression of caries. This study revealed multiple incidences of different ASVs within a species with contrasting associations with health and disease, a finding that has high relevance toward the informed design of pre- and probiotic therapies.
Oral commensal bacteria are frequently found in high abundance on healthy teeth and have capabilities to outcompete and antagonize pathogens (7, 12). One method that commensal organisms employ against pathogens is their ability to colonize the tooth early. This prevents pathogenic bacteria from binding to the tooth, forming a biofilm, and producing acids in excess, which drives down biofilm pH and encourages caries formation (13). Another mechanism commensal oral bacteria utilize to promote dental health is the production of alkaline compounds via the metabolism of urea and arginine. Urea is metabolized by bacterial ureases, which yield ammonia. Arginine is catabolized via the arginine deaminase system (ADS), which also produces ammonia (7, 12, 26). The production of ammonia results in increased biofilm pH at the tooth surface, reducing the potential for enamel demineralization and restricting growth of aciduric/acidogenic pathogens such as S. mutans (7, 27). Many oral bacteria can also synthesize bacteriocin proteins as forms of defense against other microbes in the biofilm. Often bacteriocins work as antimicrobials by disrupting the cell membrane or preventing synthesis of the cell wall (28).
Streptococcus sanguinis is considered a commensal member of the plaque community and is an early colonizer of plaque biofilms (29). In previous studies, S. sanguinis has been classified as a health-associated taxon because it is often found in increased abundance in plaque samples obtained from caries-free teeth (11, 29–31). S. sanguinis can produce hydrogen peroxide (H2O2), which can limit the growth and physiological fitness of S. mutans cells via oxidation of their cell walls (10, 27, 32). Moreover, S. sanguinis metabolizes arginine via ADS. Many of the studies associating S. sanguinis with dental health utilized 16S sequencing or lab-grown single- and dual-species biofilm assays. In both instances, the lack of strain-level identification causes a broad-level assumption that all members within a species contribute the same beneficial characteristics to the community. Our results suggest that not all strains of the same species are equivalent in this regard. For example, S. sanguinis ASVs 1 and 4 most likely represent commensal strains in the oral cavity. ASV1 was present ubiquitously throughout all stages of caries, but was more abundant in tooth health, while ASV4 was significantly more abundant in tooth health. Importantly, the remaining ASVs were present in increased abundance throughout different stages of disease. ASVs 3 and 5 were significantly increased in dentinal lesions and in enamel lesions, respectively. Interestingly, recent studies using different strains of S. sanguinis showed phenotypic variation in exopolysaccharide (glucan) and extracellular production, H2O2 production, and ADS pathway activity (18, 33, 34). In one of these studies, a laboratory strain of S. sanguinis (SK150) was shown to produce less EPS in the presence of sucrose than a clinical isolate (BCC04) that could produce a robust biofilm characterized by considerable glucan production in the presence of sucrose (27). Glucans are synthesized from dietary sucrose via glucosyltransferase enzymes that are found on the tooth pellicle. Glucan EPS production is essential to biofilm formation and acts as a scaffold for other microbes to adhere (29, 35). A robust EPS layer leaves ample space for cariogens to bind and possibly outcompete oral commensals, leading to caries initiation (29). Similarly, in another study, ADS-positive strains of S. sanguinis were shown to have different antagonistic capabilities on S. mutans (10, 18).
Another health-associated commensal organism in the supragingival plaque biofilm is Streptococcus gordonii. As with S. sanguinis, S. gordonii is an arginolytic species and can produce H2O2 as a competitive mechanism against acidogenic and aciduric pathogens like S. mutans (10, 27, 36, 37). Previous research has found that S. gordonii can block a peptide signaling pathway important for bacteriocin production in S. mutans, reducing its competitiveness against commensals in the plaque (38). Our results showed that the most abundant S. gordonii ASV (ASV1) was present in high abundance across all stages of health and disease. This suggests that S. gordonii ASV (ASV1) is likely an indigenous strain in the supragingival plaque biofilm. Given this distribution, more research is needed to determine the ecological role of S. gordonii ASV1 in health and disease. As observed with S. sanguinis, we found that different ASVs of S. gordonii were present at different stages of disease. Four ASVs of S. gordonii showed increased abundance in early-stage enamel caries, with one ASV (ASV3) significantly increased in abundance during late-stage caries. These results suggest that different strains may have distinct roles in the community at different stages of disease. Our results are supported by previous studies that have found increased prevalence of S. gordonii in active carious lesions and evidence that S. gordonii will produce acidic end products when in a mixed community (39, 40).
Although we found ASVs for S. cristatus in health and in disease, the role that S. cristatus plays in the dental plaque biofilm is less clear. Our results found three ASVs (ASVs 4, 5, and 8) significantly enriched in health and early-stage enamel caries lesions and two ASVs (3 and 6) significantly enriched in disease. These results are supported by the contradicting data in previous studies examining the role of S. cristatus in caries progression (27, 38). Some strains of S. cristatus have shown ADS activity under certain conditions and have also shown capabilities of producing H2O2. S. cristatus has been found to be more prevalent in plaque biofilms from caries-free tooth surfaces (24). Conversely, S. cristatus has also been associated with both enamel and dentin caries (41–43). Intriguingly, S. cristatus has been found in severe carious lesions, even in the absence of high concentrations of S. mutans (43). It is possible that in the absence of S. mutans, opportunistic and pathogenic strains of S. cristatus may thrive in the biofilm community.
Streptococcus mutans is a known caries pathogen due to its aciduric and acidogenic capabilities (7). S. mutans can also produce bacteriocins, known as mutacins, that provide a competitive advantage in the plaque biofilm (27, 44). However, previous studies have shown that not all strains are equivalent in their virulence capabilities (7, 45, 46). For example, Valdez et al. examined different genotypes of S. mutans strains isolated from caries-free children and children with severe early-childhood caries (ECC) and found that strains of S. mutans isolated from children with ECC had better capacity to form biofilms and increased acid tolerance (8). Other studies have found similar results supporting strain-level differences in virulence factors, including susceptibility to host salivary antimicrobial peptides, ability to metabolize different sugars, and ability to cope with oxidative stress (7, 17, 27). Our findings showed that the most abundant S. mutans ASV was enriched in the late stage of caries progression (dentinal caries lesions, which may indicate high cariogenic potential of this strain, while another ASV was present during earlier stages of caries progression (enamel lesions).
The presence of multiple ASVs within a species throughout different stages of caries progression may also be attributed, in part, to host factors such as diet and dental hygiene habits. A major driver of bacterial acid production is the availability of easily fermentable carbohydrates (47). Diets that include frequent or high consumption of sugars or carbohydrates provide ample substrate for microbes to produce acids via fermentation. The oral streptococci have a large diversity of acidogenic and aciduric capabilities, with these capabilities differing between strains (47, 48). S. mutans and S. sobrinus generally produce the most acids at a pH range from 7.0 to 5.0; however, some strains of S. mitis and S. oralis have been found to exceed the acid production levels of S. mutans and S. sobrinus at pH values ranging from 5.5 to 7.0 (48). It is possible that when exposed to high-sugar diets or frequent sugar consumption, acidogenic and aciduric strains may proliferate, which could lead to the abundance shifts observed in the different ASVs throughout caries progression.
In the current study, we developed an alternative method to 16S rRNA amplicon sequencing that provided accurate classification of most supragingival oral streptococci to the species level. Furthermore, multiple ASVs were detected for most species, with an average of 75 ASVs among the different categories of dental plaque. Our global alignment of 30S-S11 gene sequences frequently showed strains within the same species to possess identical sequence. Specifically, 1,406 sequences were collapsed into 92 distinct sequence variants, suggesting that actual strain diversity is at least an order of magnitude higher than ASV diversity, which provides new insight into the level of bacterial diversity within supragingival plaque communities and emphasizes the need for future studies that utilize whole-genome metagenomics. Importantly, the results of our study indicate that different ASVs of a species (and strains represented by these ASVs) may perform distinct roles along a biochemical spectrum associated with health and disease. Specifically, while some lineages of non-mutans streptococci may contribute to the maintenance of health, others (in addition to species such as S. mutans) may contribute to the initiation and/or progression of caries. These findings are supported by previous biochemical studies showing the cariogenic potential for strains of non-mutans species (49–52) and are crucial to the informed design of an oral probiotic.
MATERIALS AND METHODS
Primer design.
To generate an alternative to 16S, we required a locus shared among all oral streptococci that possessed conserved nucleotide regions suitable for universal oral streptococcal PCR primer annealing that (i) flanked a region of approximately 200 to 300 nucleotides and (ii) possessed sufficient variation to resolve different Streptococcus species. The size range of 200 to 300 nucleotides is required for Illumina MiSeq v2 sequencing chemistry, which produces 250-bp paired-end (PE) reads. To evaluate these criteria, we needed to construct global nucleotide alignments that contained sufficient nucleotide diversity (taxa) to adequately represent the oral streptococci. To accomplish this, we utilized genome sequence data from RefSeq and SRA at NCBI. A total of 1,220 genomes representing 22 oral Streptococcus species were obtained from RefSeq (Table 3). Of the RefSeq genomes, 91 were also present in the Human Oral Microbiome Database (HOMD) (see Table S1 in the supplemental material) and, with the exception of Streptococcus agalactiae, all 22 oral Streptococcus species were represented. Due to the large number of genomes for Streptococcus pyogenes (2,203), Streptococcus pneumoniae (8,863), and Streptococcus agalactiae (1,509) at RefSeq (28 September 2021), we randomly subsampled 100 genomes for each of these species. Sequence reads for an additional 255 strains of Streptococcus mutans and the type strain of Streptococcus lactarius (MV1) were obtained from SRA. To this we added 168 genomes from RefSeq that were labeled as Streptococcus sp. Reads were assembled using Spades v3.1.1 (53) and annotated using Prokka (54). To determine genes shared among all strains, we performed a gene clustering analysis using the software vsearch (55). Most sequence data used for this study (91%) were from whole-genome shotgun sequencing. To account for gene sequence truncation or absence due to the high number of genomes and nature of the sequence data, we targeted gene sequences in the clustering analysis that occurred as a single copy in at least 90% of genomes. Using these criteria, we identified 18 core genes that were mostly ribosomal (eight 30S and six 50S). The corresponding gene clusters, which each contained 1,406 sequences (representing this many strains) were aligned and visually inspected to find a gene that satisfied the criteria outlined. Global alignments were performed using MAFFT (56) as implemented in Geneious v7.1 (57). To evaluate taxonomic resolution (see the Results section), maximum likelihood (ML) phylogenies were constructed using PhyML (GTR+G+I) (58). Branch support was obtained via 500 bootstrap replicates. The 30S-S11 ribosomal gene best fit the criteria. The gene was 381 bp, with highly conserved regions at the 5′ and 3′ ends, which facilitated the design of PCR primers. The forward primer started three nucleotides downstream of the 5′ end of the gene. The reverse primer started one nucleotide upstream of the 3′ end of the gene. The exact primer DNA sequence is shown below. Using vsearch, we evaluated different sequence identity clustering threshold values for use when constructing the custom taxonomy classifier database for use in the microbiome pipeline QIIME2 (59).
TABLE 3.
Number of strains for each species used to delineate core genes
| Species | No. of strains |
|---|---|
| Streptococcus agalactiae | 88 |
| Streptococcus anginosus | 42 |
| Streptococcus australis | 5 |
| Streptococcus constellatus | 11 |
| Streptococcus cristatus | 25 |
| Streptococcus downei | 2 |
| Streptococcus gordonii | 43 |
| Streptococcus infantis | 6 |
| Streptococcus intermedius | 39 |
| Streptococcus lactarius | 1 |
| Streptococcus mitis | 76 |
| Streptococcus mutans | 436 |
| Streptococcus oralis | 94 |
| Streptococcus parasanguinis | 36 |
| Streptococcus peroris | 1 |
| Streptococcus pneumoniae | 97 |
| Streptococcus pyogenes | 98 |
| Streptococcus salivarius | 52 |
| Streptococcus sanguinis | 58 |
| Streptococcus sinensis | 1 |
| Streptococcus sobrinus | 23 |
| Streptococcus sp. | 116 |
| Streptococcus thermophilus | 49 |
| Streptococcus vestibularis | 7 |
Sample collection of site-specific supragingival plaque.
A total of 82 site-specific supragingival plaque samples were collected from 33 children ranging in age from 2 to 7 years. The sampling process and patient demographics of the study population are described elsewhere (60, 61). Informed consent was obtained from the parents or legal guardians of each child, and approval for the study was granted by the Institutional Review Board of the University of Florida Health Science Center. Briefly, caries lesions were detected and diagnosed by a single examiner (M.M.N.) using the International Caries Detection and Assessment System (ICDAS-II) visual criteria (62). Lesion activity was determined by clinical appearance, plaque stagnation, and tactile sensation. Children were grouped by caries status as follows: caries free (CF), caries active with active enamel caries lesions only (CAE), and caries active with at least two active and unrestored dentin carious lesions (CA). The ICDAS scores (ranging from 0 to 6) as a function of caries status group were as follows: CF (no activity), ICDAS score = 0; CAE (active enamel lesions), ICDAS score = 0 to 3; and CA (active dentin lesions), ICDAS score = 0 to 6. The threshold for the CA group was the presence of at least two ICDAS scores of 5 or 6 (cavitated dentin lesions). Supragingival plaque samples were collected from separate tooth surfaces and classified as follows: PF for samples collected from healthy tooth surfaces (ICDAS score = 0), PE for samples collected from enamel carious lesions (ICDAS score = 1 to 3), and PD for samples collected from dentinal carious lesions (ICDAS score = 4 to 6). Multiple plaque samples were collected from children presenting more than one plaque category. Combining children caries status with plaque caries status produced the following six different study groups: CF-PF (n = 8), CAE-PF (n = 12), CAE-PE (n = 16), CA-PF (n = 13), CA-PE (n = 16), and CA-PD (n = 17) (total = 82) (Table S4).
DNA extraction and 16S V4 rRNA and 30S rRNA gene library construction.
Genomic DNA was extracted using the Qiagen DNeasy PowerBiofilm kit (Qiagen, USA) according to the manufacturer’s protocol. An extraction blank was included in each set of extractions to monitor for external contamination. The V4 region of the 16S rRNA gene was amplified using PCR primers 515F and 806R (63). The oral streptococcal 30S-S11 gene region was amplified using PCR. Nucleotide sequences for the primers were as follows: 30S-S11-OS-F1, 5′-GGCTAAACCMACRCGYAAACGTC-3′; and 30S-S11-OS-R1, 5′-ACACGRCGACGTTTTGGAGGA-3′. The primers produced a 378-bp amplicon. Custom barcoded primers were designed using the both the 16S (64) and the 30S-S11 (61) primers as outlined previously. Positive- and negative-control samples were run with each PCR. The positive control used was S. mutans genomic DNA, and the negative control used was ultrapure water. After PCR amplification, samples were run on 1.5% agarose TAE (Tris-actetate-EDTA) gels. The concentration of the samples was then measured using Qubit 3.0 (ThermoFisher, USA). Samples were prepared for sequencing following Illumina’s “NextSeq Denature and Dilute Libraries Guide Protocol A: Standard Normalization Method” for libraries at a concentration of 2 nM and the Illumina 16S “Metagenomic Sequencing Library Preparation Guide.” Custom read 1, read 2, and index primers were spiked into the Illumina reagent cartridge as outlined previously (64).
Mock community construction.
Mock community DNA was used to confirm that the newly designed primers could successfully amplify the 30S-S11 gene from oral streptococci and that the resulting amplicon sequences received the correct taxonomic assignment. Focusing on those oral streptococci that are common supragingival commensal species and those that are typically associated with caries, two communities were built (Table S5). To assess cross-contamination between the two communities, five separate species were restricted to only occur in one community (community 1, S. sanguinis, Streptococcus salivarius, and Streptococcus intermedius; community 2, Streptococcus sobrinus and S. australis). Recovery of these strains in the opposing community would indicate contamination. With the exception of S. mutans and S. sobrinus, DNA for all strains was extracted from supragingival plaque isolates obtained during a previous study (18). The S. mutans type strain (Clarke) (isolated from a carious lesion) was obtained from ATCC 25175 and extracted from pure culture cells. The S. sobrinus strain, 6715, was obtained from the Burne lab at the University of Florida (brain heart infusion [BHI] agar stabs). Stabs were incubated at 37°C with a 5% CO2 concentration for 48 h. Growth from the agar stabs was streaked onto BHI agar plates and incubated at 37°C with a 5% CO2 concentration for 48 h. DNA was extracted from pure culture cells. Genomic DNA was quantified using Qubit 3.0, and a total of 10 ng of genomic DNA (gDNA) from each strain was pooled for each community, respectively.
Amplicon sequencing, quality control, and community analyses.
The 30S-S11 rRNA and 16S rRNA V4 amplicons were sequenced on Illumina’s MiSeq platform (Illumina, San Diego, CA) using a V2 500-cycle kit (250-bp paired-end reads). Read quality control and clustering were completed using the DADA2 pipeline incorporated into QIIME2 (22). Specifically, DADA2 was used to filter and trim reads, remove chimeras, assign reads to amplicon sequence variants (ASVs), and generate a read count table (65). The minimum read depth across all samples was 9,556, and all samples were rarefied to this depth using QIIME2. Species relative abundance plots were generated using Phyloseq (v1.26.1) (66) and ggplots2 (67). Differential abundance testing was completed for all six health categories in a pairwise manner using DESeq2 (v1.22.2) (68) within Phyloseq. The nonrarefied read table generated in QIIME2 after DADA2 quality control was used for the differential abundance testing. The data were log transformed using variance stabilizing transformation (VSD) to generate ASV frequencies. P values were corrected for multiple comparisons using the false-discovery rate (FDR) by the method of Benjamini and Hochberg (69).
Ethics approval and consent to participate.
Informed consent was obtained from the parents or legal guardians for each child in the study, and the protocol was approved by the Institutional Review Board of the University of Florida Health Science Center (UF IRB no. 201600154).
Data availability.
Sequence data have been deposited in the Sequence Read Archive database under BioProject accession no. PRJNA810871.
ACKNOWLEDGMENTS
L.M.O. and V.P.R. conceptualized this project and its experimental design. M.M.N. developed and completed sample collection and sample collection methods. L.M.O., T.B., and A.S. completed sample processing. We thank Brinta Chakraborty for providing the streptococcal genomic DNA of the strains used for mock community construction. V.P.R. and L.M.O. completed phylogenies and primer design. L.M.O. completed sequencing and data analyses. L.M.O., V.P.R., M.M.N., and R.A.B. contributed to manuscript revisions and improvements of the final manuscript. All authors have read and approved the final manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Vincent P. Richards, Email: vpricha@clemson.edu.
Andrew J. McBain, University of Manchester
REFERENCES
- 1.Fuks G, Elgart M, Amir A, Zeisel A, Turnbaugh PJ, Soen Y, Shental N. 2018. Combining 16S rRNA gene variable regions enables high-resolution microbial community profiling. Microbiome 6:17. 10.1186/s40168-017-0396-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mukherjee C, Beall CJ, Griffen AL, Leys EJ. 2018. High-resolution ISR amplicon sequencing reveals personalized oral microbiome. Microbiome 6:153. 10.1186/s40168-018-0535-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boers SA, Jansen R, Hays JP. 2019. Understanding and overcoming the pitfalls and biases of next-generation sequencing (NGS) methods for use in the routine clinical microbiological diagnostic laboratory. Eur J Clin Microbiol Infect Dis 38:1059–1070. 10.1007/s10096-019-03520-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson JS, Spakowicz DJ, Hong B-Y, Petersen LM, Demkowicz P, Chen L, Leopold SR, Hanson BM, Agresta HO, Gerstein M, Sodergren E, Weinstock GM. 2019. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. 1. Nat Commun 10:5029. 10.1038/s41467-019-13036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janda JM, Abbott SL. 2007. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol 45:2761–2764. 10.1128/JCM.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeong J, Yun K, Mun S, Chung W-H, Choi S-Y, Nam Y, Lim MY, Hong CP, Park C, Ahn YJ, Han K. 2021. The effect of taxonomic classification by full-length 16S rRNA sequencing with a synthetic long-read technology. Sci Rep 11:1727. 10.1038/s41598-020-80826-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abranches J, Zeng L, Kajfasz JK, Palmer SR, Chakraborty B, Wen ZT, Richards VP, Brady LJ, Lemos JA. 2018. Biology of oral streptococci. Microbiol Spectr 6 10.1128/microbiolspec.GPP3-0042-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valdez RMA, Duque C, Caiaffa KS, Dos Santos VR, Loesch ML, de A, Colombo NH, Arthur RA, Negrini T, de C, Boriollo MFG, Delbem ACB. 2017. Genotypic diversity and phenotypic traits of Streptococcus mutans isolates and their relation to severity of early childhood caries. BMC Oral Health 17:115. 10.1186/s12903-017-0406-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banas JA, Drake DR. 2018. Are the mutans streptococci still considered relevant to understanding the microbial etiology of dental caries? BMC Oral Health 18:129. 10.1186/s12903-018-0595-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang X, Browngardt CM, Jiang M, Ahn S-J, Burne RA, Nascimento MM. 2018. Diversity in antagonistic interactions between commensal oral streptococci and Streptococcus mutans. Caries Res 52:88–101. 10.1159/000479091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richards VP, Alvarez AJ, Luce AR, Bedenbaugh M, Mitchell ML, Burne RA, Nascimento MM. 2017. Microbiomes of site-specific dental plaques from children with different caries status. Infect Immun 85:e00106-17. 10.1128/IAI.00106-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowen WH, Burne RA, Wu H, Koo H. 2018. Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol 26:229–242. 10.1016/j.tim.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker JL, Edlund A. 2018. Exploiting the oral microbiome to prevent tooth decay: has evolution already provided the best tools? Front Microbiol 9:3323. 10.3389/fmicb.2018.03323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaura E, Twetman S. 2019. Critical appraisal of oral pre- and probiotics for caries prevention and care. Caries Res 53:514–526. 10.1159/000499037. [DOI] [PubMed] [Google Scholar]
- 15.Twetman S. 2018. Prevention of dental caries as a non-communicable disease. Eur J Oral Sci 126(Suppl 1):19–25. 10.1111/eos.12528. [DOI] [PubMed] [Google Scholar]
- 16.Nascimento MM, Gordan VV, Garvan CW, Browngardt CM, Burne RA. 2009. Correlations of oral bacterial arginine and urea catabolism with caries experience. Oral Microbiol Immunol 24:89–95. 10.1111/j.1399-302X.2008.00477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmer SR, Miller JH, Abranches J, Zeng L, Lefebure T, Richards VP, Lemos JA, Stanhope MJ, Burne RA. 2013. Phenotypic heterogeneity of genomically-diverse isolates of Streptococcus mutans. PLoS One 8:e61358. 10.1371/journal.pone.0061358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Velsko IM, Chakraborty B, Nascimento MM, Burne RA, Richards VP. 2018. Species designations belie phenotypic and genotypic heterogeneity in oral streptococci. mSystems 3:e00158-18. 10.1128/mSystems.00158-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The UniProt Consortium. 2021. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res 49:D480–D489. 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaminishi T, Wilson DN, Takemoto C, Harms JM, Kawazoe M, Schluenzen F, Hanawa-Suetsugu K, Shirouzu M, Fucini P, Yokoyama S. 2007. A snapshot of the 30S ribosomal subunit capturing mRNA via the Shine-Dalgarno interaction. Structure 15:289–297. 10.1016/j.str.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 21.Ghyselinck J, Pfeiffer S, Heylen K, Sessitsch A, Vos PD. 2013. The effect of primer choice and short read sequences on the outcome of 16S rRNA gene based diversity studies. PLoS One 8:e71360. 10.1371/journal.pone.0071360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abellan-Schneyder I, Matchado MS, Reitmeier S, Sommer A, Sewald Z, Baumbach J, List M, Neuhaus K. 2021. Primer, pipelines, parameters: issues in 16S rRNA gene sequencing. mSphere 6:e01202-20. 10.1128/mSphere.01202-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fakhruddin KS, Ngo HC, Samaranayake LP. 2019. Cariogenic microbiome and microbiota of the early primary dentition: a contemporary overview. Oral Dis 25:982–995. 10.1111/odi.12932. [DOI] [PubMed] [Google Scholar]
- 25.Bennett JS, Watkins ER, Jolley KA, Harrison OB, Maiden MCJ. 2014. Identifying Neisseria species by use of the 50S ribosomal protein L6 (rplF) gene. J Clin Microbiol 52:1375–1381. 10.1128/JCM.03529-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borisy GG, Valm AM. 2021. Spatial scale in analysis of the dental plaque microbiome. Periodontol 2000 86:97–112. 10.1111/prd.12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L, Chakraborty B, Zou J, Burne RA, Zeng L. 2019. Amino sugars modify antagonistic interactions between commensal oral streptococci and Streptococcus mutans. Appl Environ Microbiol 85:e00370-19. 10.1128/AEM.00370-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawada-Matsuo M, Komatsuzawa H. 2017. Role of Streptococcus mutans two-component systems in antimicrobial peptide resistance in the oral cavity. Jpn Dent Sci Rev 53:86–94. 10.1016/j.jdsr.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu B, Macleod LC, Kitten T, Xu P. 2018. Streptococcus sanguinis biofilm formation & interaction with oral pathogens. Future Microbiol 13:915–932. 10.2217/fmb-2018-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valdebenito B, Tullume-Vergara PO, González W, Kreth J, Giacaman RA. 2018. In silico analysis of the competition between Streptococcus sanguinis and Streptococcus mutans in the dental biofilm. Mol Oral Microbiol 33:168–180. 10.1111/omi.12209. [DOI] [PubMed] [Google Scholar]
- 31.Díaz-Garrido N, Lozano CP, Kreth J, Giacaman RA. 2020. Competition and caries on enamel of a dual-species biofilm model with Streptococcus mutans and Streptococcus sanguinis. Appl Environ Microbiol 86:e01262-20. 10.1128/AEM.01262-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lozano CP, Díaz-Garrido N, Kreth J, Giacaman RA. 2019. Streptococcus mutans and Streptococcus sanguinis expression of competition-related genes, under sucrose. Caries Res 53:194–203. 10.1159/000490950. [DOI] [PubMed] [Google Scholar]
- 33.Xu Y, Itzek A, Kreth J. 2014. Comparison of genes required for H2O2 resistance in Streptococcus gordonii and Streptococcus sanguinis. Microbiology (Reading) 160:2627–2638. 10.1099/mic.0.082156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Redanz S, Cheng X, Giacaman RA, Pfeifer CS, Merritt J, Kreth J. 2018. Live and let die: hydrogen peroxide production by the commensal flora and its role in maintaining a symbiotic microbiome. Mol Oral Microbiol 33:337–352. 10.1111/omi.12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koo H, Xiao J, Klein MI, Jeon JG. 2010. Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J Bacteriol 192:3024–3032. 10.1128/JB.01649-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miranda ML, Silva BNS, Salomão KB, de Oliveira AB, Gabbai-Armelin PR, Brighenti FL. 2020. Effect of arginine on microorganisms involved in dental caries: a systematic literature review of in vitro studies. Biofouling 36:696–709. 10.1080/08927014.2020.1802587. [DOI] [PubMed] [Google Scholar]
- 37.Chakraborty B, Burne RA. 2017. Effects of arginine on Streptococcus mutans growth, virulence gene expression, and stress tolerance. Appl Environ Microbiol 83:e00496-17. 10.1128/AEM.00496-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang X, Palmer SR, Ahn S-J, Richards VP, Williams ML, Nascimento MM, Burne RA. 2016. A highly arginolytic Streptococcus species that potently antagonizes Streptococcus mutans. Appl Environ Microbiol 82:2187–2201. 10.1128/AEM.03887-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neves BG, Stipp RN, da Silva Bezerra D, de Figueiredo Guedes SF, Rodrigues LKA. 2017. Molecular detection of bacteria associated to caries activity in dentinal lesions. Clin Oral Invest 21:2053–2061. 10.1007/s00784-016-1995-9. [DOI] [PubMed] [Google Scholar]
- 40.Hendrickson EL, Wang T, Dickinson BC, Whitmore SE, Wright CJ, Lamont RJ, Hackett M. 2012. Proteomics of Streptococcus gordonii within a model developing oral microbial community. BMC Microbiol 12:211. 10.1186/1471-2180-12-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kressirer CA, Chen T, Lake Harriman K, Frias-Lopez J, Dewhirst FE, Tavares MA, Tanner AC. 2018. Functional profiles of coronal and dentin caries in children. J Oral Microbiol 10:1495976. 10.1080/20002297.2018.1495976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanner ACR, Mathney JMJ, Kent RL, Chalmers NI, Hughes CV, Loo CY, Pradhan N, Kanasi E, Hwang J, Dahlan MA, Papadopolou E, Dewhirst FE. 2011. Cultivable anaerobic microbiota of severe early childhood caries. J Clin Microbiol 49:1464–1474. 10.1128/JCM.02427-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dzidic M, Collado MC, Abrahamsson T, Artacho A, Stensson M, Jenmalm MC, Mira A. 2018. Oral microbiome development during childhood: an ecological succession influenced by postnatal factors and associated with tooth decay. ISME J 12:2292–2306. 10.1038/s41396-018-0204-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hossain MS, Biswas I. 2011. Mutacins from Streptococcus mutans UA159 are active against multiple streptococcal species. Appl Environ Microbiol 77:2428–2434. 10.1128/AEM.02320-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cornejo OE, Lefébure T, Bitar PDP, Lang P, Richards VP, Eilertson K, Do T, Beighton D, Zeng L, Ahn S-J, Burne RA, Siepel A, Bustamante CD, Stanhope MJ. 2013. Evolutionary and population genomics of the cavity causing bacteria Streptococcus mutans. Mol Biol Evol 30:881–893. 10.1093/molbev/mss278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Argimón S, Konganti K, Chen H, Alekseyenko AV, Brown S, Caufield PW. 2014. Comparative genomics of oral isolates of Streptococcus mutans by in silico genome subtraction does not reveal accessory DNA associated with severe early childhood caries. Infect Genet Evol 21:269–278. 10.1016/j.meegid.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kilian M. 2018. The oral microbiome—friend or foe? Eur J Oral Sci 126:5–12. 10.1111/eos.12527. [DOI] [PubMed] [Google Scholar]
- 48.Bek-Thomsen M, Tettelin H, Hance I, Nelson KE, Kilian M. 2008. Population diversity and dynamics of Streptococcus mitis, Streptococcus oralis, and Streptococcus infantis in the upper respiratory tracts of adults, determined by a nonculture strategy. Infect Immun 76:1889–1896. 10.1128/IAI.01511-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Houte J, Sansone C, Joshipura K, Kent R. 1991. Mutans streptococci and non-mutans streptococci acidogenic at low pH, and in vitro acidogenic potential of dental plaque in two different areas of the human dentition. J Dent Res 70:1503–1507. 10.1177/00220345910700120601. [DOI] [PubMed] [Google Scholar]
- 50.Sansone C, Van Houte J, Joshipura K, Kent R, Margolis HC. 1993. The association of mutans streptococci and non-mutans streptococci capable of acidogenesis at a low pH with dental caries on enamel and root surfaces. J Dent Res 72:508–516. 10.1177/00220345930720020701. [DOI] [PubMed] [Google Scholar]
- 51.van Houte J, Lopman J, Kent R. 1996. The final pH of bacteria comprising the predominant flora on sound and carious human root and enamel surfaces. J Dent Res 75:1008–1014. 10.1177/00220345960750040201. [DOI] [PubMed] [Google Scholar]
- 52.van Ruyven FO, Lingström P, van Houte J, Kent R. 2000. Relationship among mutans streptococci, “low-pH” bacteria, and iodophilic polysaccharide-producing bacteria in dental plaque and early enamel caries in humans. J Dent Res 79:778–784. 10.1177/00220345000790021201. [DOI] [PubMed] [Google Scholar]
- 53.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 55.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Katoh K, Misawa K, Kuma K-i, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 59.Hall M, Beiko RG. 2018. 16S rRNA gene analysis with QIIME2. Methods Mol Biol 1849:113–129. 10.1007/978-1-4939-8728-3_8. [DOI] [PubMed] [Google Scholar]
- 60.Nascimento MM, Alvarez AJ, Huang X, Hanway S, Perry S, Luce A, Richards VP, Burne RA. 2019. Arginine metabolism in supragingival oral biofilms as a potential predictor of caries risk. JDR Clin Trans Res 4:262–270. 10.1177/2380084419834234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O’Connell LM, Santos R, Springer G, Burne RA, Nascimento MM, Richards VP. 2020. Site-specific profiling of the dental mycobiome reveals strong taxonomic shifts during progression of early-childhood caries. Appl Environ Microbiol 86:e02825-19. 10.1128/AEM.02825-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ekstrand KR, Martignon S, Ricketts DJN, Qvist V. 2007. Detection and activity assessment of primary coronal caries lesions: a methodologic study. Oper Dent 32:225–235. 10.2341/06-63. [DOI] [PubMed] [Google Scholar]
- 63.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 108(Suppl 1):4516–4522. 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wickham H. 2016. ggplot2: elegant graphics for data analysis, 2nd ed. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 68.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Methodol 57:289–300. 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download aem.00453-22-s0002.xlsx, XLSX file, 0.2 MB (189.2KB, xlsx)
Table S2. Download aem.00453-22-s0003.xlsx, XLSX file, 0.01 MB (12.8KB, xlsx)
Table S3. Download aem.00453-22-s0004.xlsx, XLSX file, 0.05 MB (56.5KB, xlsx)
Table S4. Download aem.00453-22-s0005.xlsx, XLSX file, 0.2 MB (239KB, xlsx)
Table S5. Download aem.00453-22-s0006.xlsx, XLSX file, 0.01 MB (9.4KB, xlsx)
Fig. S1. Download aem.00453-22-s0001.svg, SVG file, 0.03 MB (28.8KB, svg)
Data Availability Statement
Sequence data have been deposited in the Sequence Read Archive database under BioProject accession no. PRJNA810871.