ABSTRACT
Lipoic acid is an organosulfur cofactor essential for several key enzyme complexes in oxidative and one-carbon metabolism. It is covalently bound to the lipoyl domain of the E2 subunit in some 2-oxoacid dehydrogenase complexes and the H-protein in the glycine cleavage system. Lipoate-protein ligase (Lpl) is involved in the salvage of exogenous lipoate and attaches free lipoate to the E2 subunit or the H-protein in an ATP-dependent manner. In the hyperthermophilic archaeon Thermococcus kodakarensis, TK1234 and TK1908 are predicted to encode the N- and C-terminal regions of Lpl, respectively. TK1908 and TK1234 recombinant proteins form a heterodimer and together displayed significant ligase activity toward octanoate in addition to lipoate when a chemically synthesized octapeptide was used as the acceptor. The proteins also displayed activity toward other fatty acids, indicating broad fatty acid specificity. On the other hand, lipoyl synthase from T. kodakarensis only recognized octanoyl-peptide as a substrate. Examination of individual proteins indicated that the TK1908 protein alone was able to catalyze the ligase reaction although with a much lower activity. Gene disruption of TK1908 led to lipoate/serine auxotrophy, whereas TK1234 gene deletion did not. Acyl carrier protein homologs are not found on the archaeal genomes, and the TK1908/TK1234 protein complex did not utilize octanoyl-CoA, raising the possibility that the substrate of the ligase reaction is octanoic acid itself. Although Lpl has been considered as an enzyme involved in lipoate salvage, the results imply that in T. kodakarensis, the TK1908 and TK1234 proteins function in de novo lipoyl-protein biosynthesis.
IMPORTANCE Based on previous studies in bacteria and eukaryotes, lipoate-protein ligases (Lpls) have been considered to be involved exclusively in lipoate salvage. The genetic analyses in this study on the lipoate-protein ligase in T. kodakarensis, however, suggest otherwise and that the enzyme is additionally involved in de novo protein lipoylation. We also provide biochemical evidence that the lipoate-protein ligase displays broad substrate specificity and is capable of ligating acyl groups of various chain-lengths to the peptide substrate. We show that this apparent ambiguity in Lpl is resolved by the strict substrate specificity of the lipoyl synthase LipS in this organism, which only recognizes octanoyl-peptide. The results provide relevant physiological insight into archaeal protein lipoylation.
KEYWORDS: Archaea, lipoylation, lipoate-protein ligase, biosynthesis, metabolism, hyperthermophile
INTRODUCTION
Lipoic acid is an organosulfur compound containing two sulfur atoms at the C6 and C8 positions of octanoic acid to form a dithiolane ring. The compound is a cofactor that plays an essential role in the glycine cleavage system (GCS) and some enzymes involved in the oxidative decarboxylation of 2-oxoacids including pyruvate dehydrogenase (PDH), 2-oxoglutarate dehydrogenase (OGDH), branched-chain α-ketoacid dehydrogenase, and acetoin dehydrogenase (1–4). Lipoic acid is covalently bound to a lipoyl domain of the H-protein in GCS, as well as the E2 subunits of the lipoate-requiring 2-oxoacid dehydrogenases (1, 2). GCS is composed of four proteins and provides one-carbon (C1) units derived from glycine to C1 carriers such as tetrahydrofolate. During the transfer, the C1 unit from glycine is transiently attached to the lipoyl group on the H-protein (1, 5). The subsequent C1 donation from 5,10-methylenetetrahydrofolate to other compounds is vital in biosynthesis, such as the transfer of a hydroxymethyl group to glycine to form serine (5, 6).
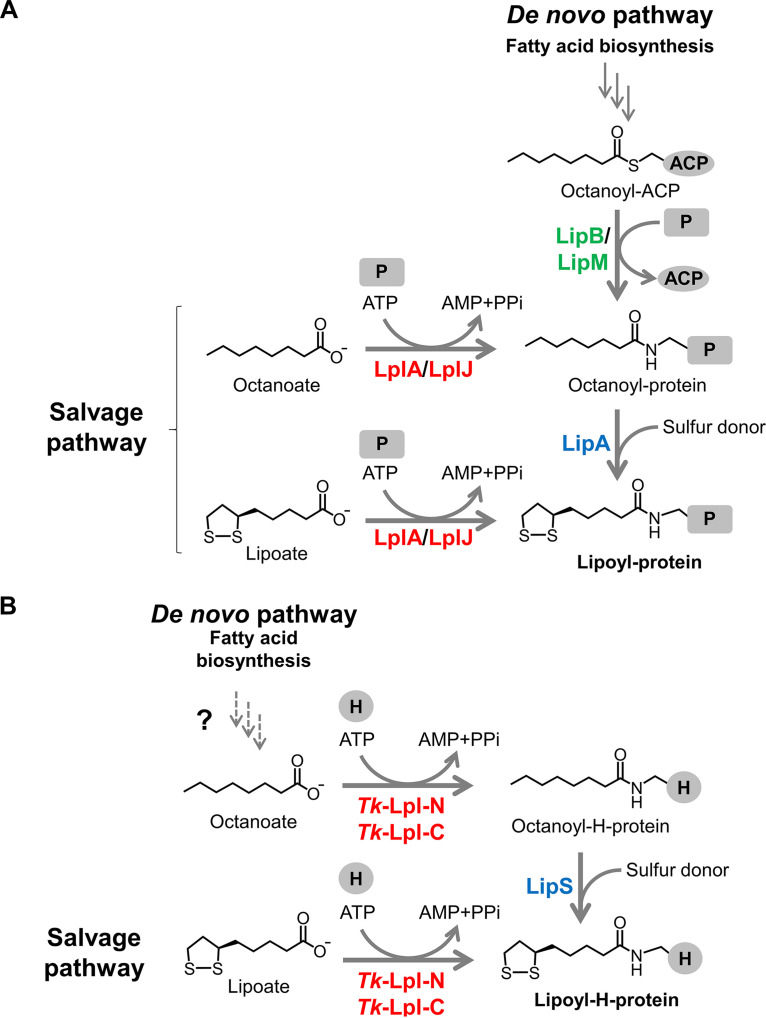
Lipoyl protein biosynthesis has been established in the Gram-negative bacterium Escherichia coli and the Gram-positive bacterium Bacillus subtilis (4). Both organisms utilize two pathways for protein lipoylation, the de novo biosynthesis pathway and the salvage pathway (Fig. 1A). In the de novo pathway in E. coli, octanoyl transferase Ec-LipB transfers an octanoyl moiety from octanoyl-acyl carrier protein (ACP), an intermediate in fatty acid biosynthesis, onto the lipoyl domain in the H-protein or the E2 subunit of 2-oxoacid dehydrogenases (7–10). Lipoyl synthase Ec-LipA then catalyzes the sulfur insertion to generate lipoyl-proteins (11–14). On the other hand, exogenous lipoate can be salvaged and attached to the H-protein or the E2 subunit by lipoate-protein ligase (Lpl), Ec-LplA (15–18). Octanoate is also recognized by the enzyme and is utilized for the generation of octanoyl-protein, which can be converted to lipoyl-protein by Ec-LipA (4, 15, 16). However, Ec-LplA displays relatively low specific activity toward octanoate (3.9 nmol min−1 mg−1) compared to that toward lipoate (25 nmol min−1 mg−1) (16). In B. subtilis, the octanoyl transferase Bs-LipM only recognizes the GCS H-protein as the octanoyl-acceptor and catalyzes the formation of octanoyl-H-protein (19, 20). Lipoyl synthase Bs-LipA then converts octanoyl-H-protein into lipoyl-H-protein. Upon the formation of lipoyl-H-protein, an amidotransferase Bs-LipL transfers the lipoyl moiety from lipoyl-H-protein to the E2 subunits of the dehydrogenases (20, 21). Concerning lipoate salvage, lipoate/octanoate-protein ligase in B. subtilis, Bs-LplJ, recognizes both the H-protein and the E2 subunit of OGDH as the acceptors of exogenous lipoate or octanoate (20, 22). B. subtilis possesses only one Lpl, but some Gram-positive bacteria, such as Listeria monocytogenes (23, 24) and Staphylococcus aureus (25), harbor multiple Lpls. To date, Lpls have also been studied in other bacteria and eukaryotes, including Chlamydia trachomatis serovar L2 (26), Streptomyces coelicolor (27), Mycoplasma hyopneumoniae (28, 29), Plasmodium falciparum (30), and rice (31).
FIG 1.
Classical and proposed pathways to synthesize lipoyl-protein. (A) De novo and salvage pathways for protein lipoylation in Escherichia coli and Bacillus subtilis. (B) The de novo lipoylation pathway in the hyperthermophilic archaeon Thermococcus kodakarensis proposed in this study and the salvage lipoylation pathway. Tk-Lpl-N and Tk-Lpl-C are predicted to be involved in both de novo and salvage lipoylation pathways in this organism. LplA, LplJ, and Tk-Lpl-N/Tk-Lpl-C are lipoate-protein ligases; LipB and LipM are octanoyl transferases; LipA and LipS are lipoyl synthases. ACP, acyl carrier protein; P, lipoate-dependent protein; H, H-protein of GCS; PPi, pyrophosphate.
Lpl is an ATP-dependent enzyme that catalyzes the attachment of lipoate to protein substrates through two-step reactions (see Fig. S1 in the supplemental material). Lipoate is initially activated by ATP to form the intermediate lipoyl-AMP (activation reaction), and then the lipoyl moiety is transferred to the lysine residue in the lipoyl domain of an apoprotein to generate the lipoyl-holoprotein (transfer reaction) (4, 17). In mammals, the activation reaction and transfer reaction are catalyzed by two separate enzymes, the lipoate-activating enzyme and lipoyl transferase (32–34). Although mammalian lipoyl transferase displays structural similarity with bacterial Lpls, it is only able to transfer the lipoyl moiety from lipoyl-AMP to the target proteins.
Knowledge on lipoyl-protein biosynthesis in archaea is still limited. Archaeal Lpl has been extensively studied in the thermoacidophilic archaeon Thermoplasma acidophilum (35–39). Lpls have a common overall structure: a large catalytic domain and a small accessory domain. Bacterial Lpls comprise a single polypeptide with two domains, whereas the Lpl from T. acidophilum (Ta-LplA) is composed of two separate proteins, Ta-LplA-N and Ta-LplA-C, whose three-dimensional structures are similar to the large domain and small domain of Ec-LplA, respectively (4, 37, 39). Both Ta-LplA-N and Ta-LplA-C proteins are required for the formation of lipoyl-AMP, while Ta-LplA-N alone is sufficient for the lipoyl transfer reaction from lipoyl-AMP to the E2 subunit of PDH (37–39). Concerning lipoyl synthase, a protein homologous to the bacterial LipAs from the thermoacidophilic archaeon Saccharolobus solfataricus (Ss-LipA) has been characterized (40, 41). However, Ss-LipA homologs are not widely distributed in Archaea. We have previously identified a structurally novel lipoyl synthase LipS in hyperthermophilic archaeon Thermococcus kodakarensis (42). Here, we examined a putative Lpl in T. kodakarensis (Tk-Lpl), which is encoded by two genes, TK1908 and TK1234. In contrast to Ta-LplA-N, TK1908 protein alone, the large domain of Tk-Lpl, is able to catalyze the complete lipoate attachment reaction including both the activation and transfer steps. Our biochemical and genetic analyses suggest that, unlike the previously studied Lpls that function in lipoate/octanoate salvage, Tk-Lpl is essential for the de novo biosynthesis of lipoyl-protein in T. kodakarensis.
RESULTS
Preparation of the purified recombinant TK1234 and TK1908 proteins.
In a previous study, the serine auxotrophy observed in lipoyl synthase gene disruption strains could be complemented by supplementing lipoate, suggesting the presence of a lipoate salvage pathway in T. kodakarensis (42). TK1908 and TK1234 genes are annotated as the N-terminal and C-terminal domains of lipoate-protein ligase in this organism. Their primary structures share 31% and 32% identities with those of Ta-LplA-N and Ta-LplA-C from T. acidophilum, respectively. To identify the reactions catalyzed by the TK1234 and TK1908 proteins, their recombinant proteins were prepared. The TK1234 and TK1908 genes were individually expressed in E. coli, and the recombinant proteins were purified by heat treatment, anion-exchange chromatography, and gel filtration chromatography. Judging from the results of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), the TK1234 (10.8 kDa) and TK1908 (28.3 kDa) recombinant proteins were purified to apparent homogeneity (Fig. S2A and B).
Interaction between the TK1234 and TK1908 proteins.
We examined whether the two proteins display physical interaction. When applied to gel filtration chromatography (Fig. S3A), the TK1908 protein alone eluted at 16.0 mL, corresponding to a molecular mass of 27.2 kDa (theoretical molecular mass of 28.3 kDa based on the primary structure). The TK1234 protein alone eluted at 17.2 mL, corresponding to a molecular mass of 16.0 kDa (theoretical molecular mass of 10.8 kDa). The major peak (P1) of an equimolar mixture of the TK1234 and TK1908 proteins eluted at 15.3 mL, corresponding to a molecular mass of 38.5 kDa. The result suggests that these two proteins form a heterodimer in the mixture (theoretical molecular mass of 39.1 kDa). A fraction including the P1 peak (from 15.0 to 15.5 mL) was analyzed by SDS-PAGE (Fig. S3B). Two bands whose positions corresponded to those of the TK1908 protein and TK1234 protein were observed, further supporting that the two proteins form a complex.
Detection of lipoate-protein ligase activity.
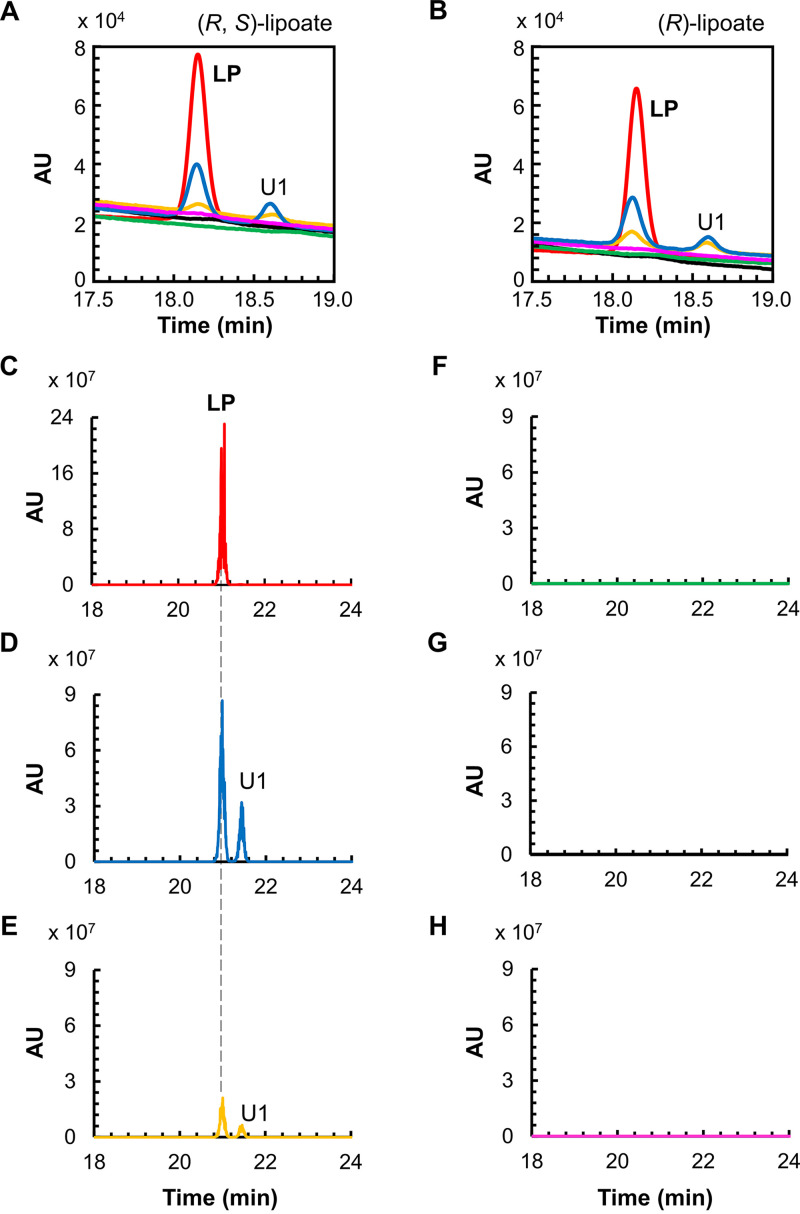
The lipoate-protein ligase activity of the two recombinant proteins toward (R, S)-lipoate was examined. A chemically synthesized octapeptide (NH2-ESVKAVSE-COOH), whose sequence corresponds to that of the lipoyl domain in the H-protein of T. kodakarensis (TK0150 protein), was used as the lipoyl moiety acceptor. The reaction products were analyzed by high-performance liquid chromatography (HPLC) (Fig. 2A). An octapeptide modified with a lipoyl group bound to the Lys residue of the octapeptide was also chemically synthesized and applied to HPLC as a standard. The lipoyl-peptide standard displayed a retention time of 18.1 min. When both TK1234 and TK1908 proteins were included in the reaction mixture, we observed a new peak after the reaction whose retention time corresponded to that of lipoyl-peptide. In the presence of only the TK1234 protein, production of lipoyl-peptide was not detected. In contrast, addition of the TK1908 protein alone led to the formation of the product lipoyl-peptide. However, the product yield was higher in the presence of both proteins than that with only the TK1908 protein. This was not entirely due to a dramatic decrease in thermostability of the TK1908 protein in the absence of TK1234 protein, as further product generation was observed when reaction times were prolonged up to approximately 2 h (Fig. S4). The results suggest that the TK1908 protein alone can catalyze the lipoate-protein ligase reaction and that the presence of the TK1234 protein enhances this activity. In addition to the product peak, another new peak (U1), with a retention time of 18.6 min, was observed after the reaction with both proteins or with the TK1908 protein alone. This peak was not observed when the peptide was omitted from the reaction, suggesting that the compound derives from the octapeptide. The lipoyl group of the lipoyl-peptide standard is the R-isomer, but the lipoate used in the reaction was a racemic mixture. To examine whether U1 is the S-isomer of lipoate linked to the peptide, we carried out the reaction using (R)-lipoate as the substrate. However, U1 was still observed (Fig. 2B), indicating that the generation of U1 is not due to the use of a racemic mixture of lipoate.
FIG 2.
Lipoate-protein ligase activity measurement with HPLC and LC-MS. HPLC analyses were carried out for the reaction products with the substrates (R, S)-lipoate (A), and (R)-lipoate (B). Red lines, standard peptides modified with fatty acids; blue lines, reaction mixtures with both TK1908 and TK1234 proteins; green lines, reaction mixtures with only TK1234 protein; yellow lines, reaction mixtures with only TK1908 protein; black lines, reaction mixtures without protein; pink lines, reaction mixtures without peptide. LC-MS analyses were carried out for standard product lipoyl-peptide (C), reaction products with both TK1908 and TK1234 proteins (D), with only TK1908 protein (E), with only TK1234 protein (F), without protein (G), and without peptide (H). The chromatograms of the compounds whose exact masses corresponded to that of lipoyl-peptide are shown. U1 indicates a by-product with the same exact mass to that of LP. LP, lipoyl-peptide; AU, arbitrary units; HPLC, high-performance liquid chromatography; LC-MS, liquid chromatography-mass spectrometry.
Identification of reaction products with liquid chromatography-mass spectrometry.
To identify the generated products, liquid chromatography-mass spectrometry (LC-MS) analysis was carried out (Fig. 2C to H). The standard lipoyl-peptide (Fig. 2C) eluted at 21.0 min under the conditions used in the LC-MS system. The compounds with the exact mass corresponding to lipoyl-peptide ([M-H]− = 1,034.5) were detected in the products from the reactions using either both TK1908 and TK1234 proteins or only TK1908 protein (Fig. 2D and E). As expected, lipoyl-peptide was not observed in the reaction mixture with only TK1234 protein (Fig. 2F), without protein (Fig. 2G), or without the peptide substrate (Fig. 2H). These results were consistent with those of the HPLC analysis and indicated that the TK1234 and TK1908 proteins display lipoate-protein ligase activity resulting in the generation of lipoyl-peptide. On the other hand, the LC-MS analysis also revealed that the unknown compound U1 possesses the same exact mass as that of lipoyl-peptide (U1 in Fig. 2D and E). This raised the possibility that compound U1 corresponds to the lipoyl group attached to the amino group of the N-terminal glutamic acid residue of the peptide.
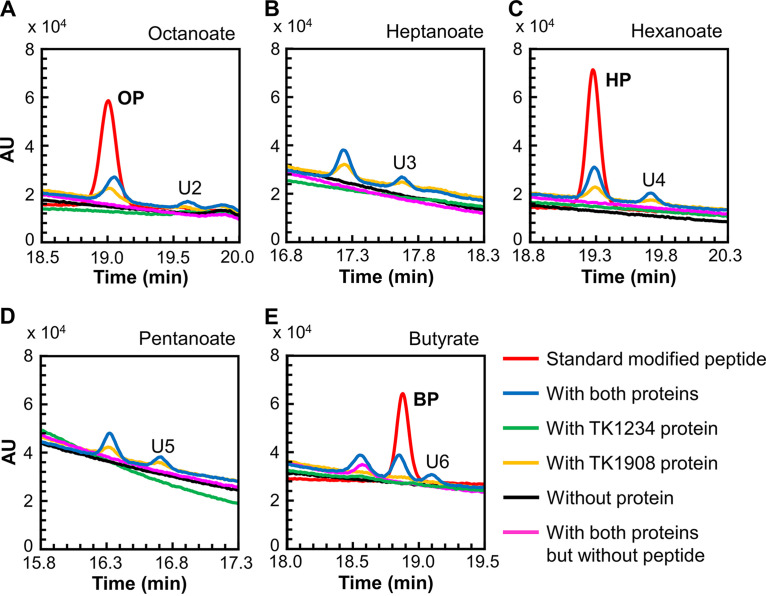
Ligase activity toward various fatty acids.
We examined the ligase activity of the two recombinant proteins toward octanoate (Fig. 3A). As in the case of the lipoyl-peptide standard, an octanoyl-peptide standard was chemically synthesized and applied to HPLC. This eluted at 19.0 min under the applied conditions. A peak with the same retention time was observed in the reaction mixture that included octanoate and the TK1234 and TK1908 proteins. The peak was also observed when only the TK1908 protein was added. The generated product was confirmed to be octanoyl-peptide by LC-MS (Fig. S5A). The results indicated that, in addition to lipoate, the TK1234 and TK1908 proteins could recognize octanoate and catalyze the formation of octanoyl-peptide. We further examined the substrate specificity of the proteins. Short-chain fatty acids, including heptanoate, hexanoate, pentanoate, and butyrate, were tested. HPLC analyses (Fig. 3B to E) and LC-MS analyses (Fig. S5B to E) showed that all fatty acids examined could be recognized by the complex, indicating that the proteins display a broad substrate specificity toward fatty acids. As in the case of the experiments with lipoate, by-products (U2–U6 in Fig. 3A to E and U1–U5 in Fig. S5) were also observed when these fatty acids were used as substrates. These compounds may also correspond to the acyl groups linked to the terminal amino group of the peptide.
FIG 3.
Fatty acid-protein ligase activity measurement with HPLC. HPLC analyses were carried out for the reaction products with the substrates octanoate (A), heptanoate (B), hexanoate (C), pentanoate (D), and butyrate (E). Red lines, standard peptides modified with fatty acids; blue lines, reaction mixtures with both TK1908 and TK1234 proteins; green lines, reaction mixtures with only TK1234 protein; yellow lines, reaction mixtures with only TK1908 protein; black lines, reaction mixtures without protein; pink lines, reaction mixtures without peptide. In experiments using heptanoate and pentanoate (B and D), standard modified peptides were not available. OP, octanoyl-peptide; HP, hexanoyl-peptide; BP, butyryl-peptide; U2–U6, unidentified compounds.
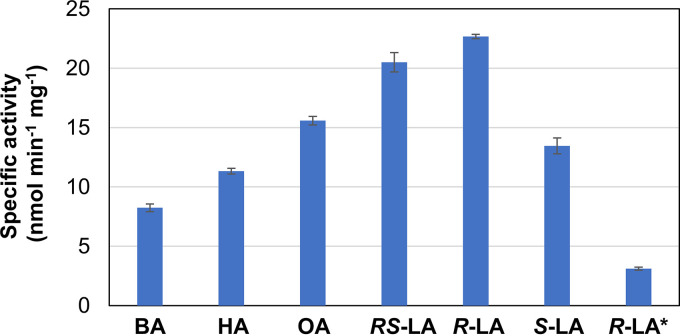
Specific activity toward various fatty acids.
Using the synthetic octapeptide, the specific activity of Tk-Lpl was measured toward six substrates; (R, S)-lipoate, (R)-lipoate, (S)-lipoate, octanoate, hexanoate, and butyrate (Fig. 4). In all cases, we were able to confirm product formation using chemically synthesized peptides modified with each substrate. Among the examined substrates, Tk-Lpl displayed the highest activity toward (R)-lipoate, which is the substrate of Ec-LplA in vivo (16, 17). The specific activity toward (R)-lipoate (22.7 nmol min−1 mg−1) was similar to that of Ec-LplA (25 nmol min−1 mg−1) (16). On the other hand, the specific activity toward octanoate (15.6 nmol min−1 mg−1) was 3-fold higher than that of Ec-LplA (3.9 nmol min−1 mg−1).
FIG 4.
Substrate specificity of TK1234 and TK1908 proteins. Specific activities toward a chemically synthesized octapeptide and various fatty acids were measured. BA, butyrate; HA, hexanoate; OA, octanoate; RS-LA, (R, S)-lipoate; R-LA, (R)-lipoate; S-LA, (S)-lipoate; R-LA*, ligase activity of the TK1908 protein alone toward (R)-lipoate.
Tk-Lpl also exhibited activities toward (R, S)-lipoate (20.5 nmol min−1 mg−1), (S)-lipoate (13.5 nmol min−1 mg−1), hexanoate (11.3 nmol min−1 mg−1), and butyrate (8.2 nmol min−1 mg−1), suggesting a broad fatty acid specificity. The TK1908 protein alone showed very low activity toward (R)-lipoate (3.1 nmol min−1 mg−1), corresponding to approximately 14% of that of Tk-Lpl, the TK1234-TK1908 protein complex. This supports the presumption that the low yield of product by the TK1908 protein alone observed in Fig. 2A was due to low activity.
Substrate specificity of lipoyl synthase in T. kodakarensis.
In a previous study (42), we identified a structurally novel lipoyl synthase LipS (TK2109 and TK2248 proteins) in T. kodakarensis, which recognizes a chemically synthesized octanoyl-peptide and catalyzes its conversion to lipoyl-peptide (Fig. S6A). As the TK1234 and TK1908 proteins could catalyze the attachment of various fatty acids onto the peptide, this poses a problem if we assume that the chain length of the acyl group, or the group after sulfur insertion, is important for the activity of GCS. We thus examined whether LipS exhibits the sulfur insertion activity toward hexanoyl-peptide and butyryl-peptide. Although several small new peaks (U1–U4 in Fig. S6B and C) were observed in the reaction mixture with LipS proteins, we could not detect the predicted products or intermediates through LC-MS analysis (data not shown). The lipoyl synthase activity toward free octanoate (or free octanoate mixed with peptide) was also examined. However, the expected product, lipoate or reduced lipoate, was not generated (Fig. S6D). The results suggested that the substrate specificity of LipS is strict and the enzyme recognizes only octanoyl-peptide. Although we cannot rule out the possibility that the native LipS recognizes a wider range of substrates than those we observed with the reconstituted enzyme, the present results suggest that the structural integrity of the lipoyl group on the GCS H-protein in T. kodakarensis is mainly accomplished through the specificity of LipS.
Construction of TK1234 and TK1908 gene disruption strains.
To identify the physiological function of these two genes, we constructed the gene disruption strains ΔTK1234 and ΔTK1908 (Fig. S7A). The genotypes of the selected transformants were examined by PCR (Fig. S7B to E), which indicated that the target genes were successfully deleted. In both transformants, the absence of unintended mutations in the homologous regions was also confirmed by DNA sequencing analysis.
Growth properties of TK1234 and TK1908 gene disruption strains.
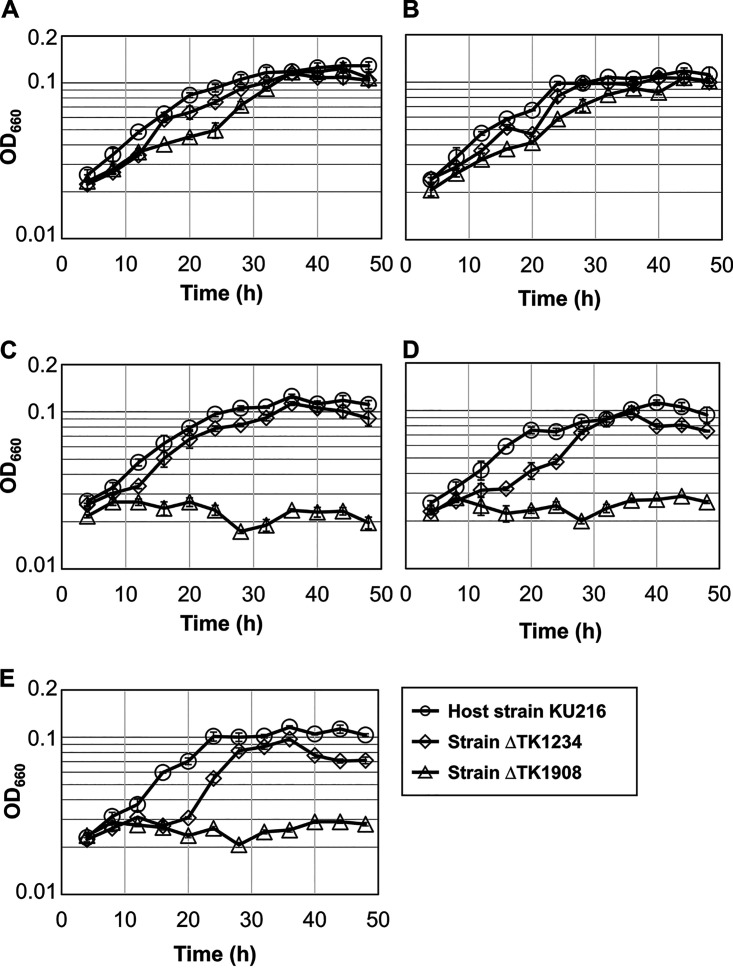
The growth properties of the host strain and the gene disruption strains (ΔTK1234 and ΔTK1908) were examined in a synthetic medium with or without Ser and/or lipoate (Fig. 5). Previous studies indicated that serine is synthesized from glycine by glycine/serine hydroxymethyltransferase (GlyA, TK0528) when T. kodakarensis is cultured in the synthetic medium (43). The C1 group for serine biosynthesis is most likely provided by GCS, whose H-protein requires lipoate modification. Therefore, a mutant with defects in de novo lipoyl-H-protein biosynthesis capabilities would display serine auxotrophy in the absence of lipoate in the synthetic medium. On the other hand, a mutant impaired in a salvage pathway would grow in the absence of lipoate and serine, as the de novo pathway can synthesize lipoyl-protein, resulting in serine production. As our results indicated that TK1234 and TK1908 encode an Lpl, which until now has been shown to be involved in lipoate salvage, the TK1234 and TK1908 gene disruption strains were predicted not to exhibit serine/lipoate auxotrophy. In a medium containing serine regardless of the presence or absence of lipoate, the ΔTK1234 and ΔTK1908 strains displayed growth (Fig. 5A and B). The ΔTK1234 strain could also grow in a serine-free medium, but a delay was observed when cultivated in a medium without lipoate or only supplemented with octanoate (Fig. 5C, D, and E). In contrast to the host strain KU216 and the ΔTK1234 strain, the ΔTK1908 strain displayed serine auxotrophy regardless of the presence or absence of lipoate (Fig. 5C and D). This result and the in vitro analyses described above suggest that the TK1908 gene is involved not only in the salvage of lipoate but also in the de novo biosynthesis of lipoyl-H-protein, most likely by ligating octanoate onto the H-protein to form octanoyl-H-protein, which is subsequently converted to lipoyl-H-protein by lipoyl synthase LipS (Fig. 1B). In vitro and in vivo analyses indicate a supporting role of the TK1234 protein in protein lipoylation.
FIG 5.
Growth properties of the host strain KU216 and the gene disruption strains ΔTK1234 and ΔTK1908. Growth properties of the host KU216 strain (circles), ΔTK1234 (diamonds), and ΔTK1908 (triangles) were examined in the following five synthetic media based on ASW-AA-Ser−-LA−-S0: supplemented with both 0.71 mM serine and 1 mM lipoate (Ser[+]-LA[+]) (A); only supplemented with serine (Ser[+]-LA[−]) (B); only supplemented with 1 mM lipoate (Ser[−]-LA[+]) (C); without any supplement (Ser[−]-LA[−]) (D); and only supplemented with 1 mM octanoate (Ser[−]-OA[+]) (E). Error bars indicate the standard deviations of three independent culture experiments. The vertical axis is represented in logarithmic scale. OD660, optical density at 660 nm.
DISCUSSION
In this study, we demonstrated that the TK1234 and TK1908 genes encode an Lpl in T. kodakarensis. Similar to the archaeal Lpl from T. acidophilum Ta-LplA (37–39), Tk-Lpl consists of two proteins corresponding to the large and small domains of previously characterized Lpls and forms a heterodimer. Tk-Lpl and Ta-LplA share 32% identity, but several significant differences were found between the two Lpls. The large domain of Ta-LplA (Ta-LplA-N) alone cannot catalyze protein lipoylation (37–39), but Tk-Lpl-N was able to catalyze the complete ligase reaction, as in the case of the C-terminal-truncated mutant of the Lpl from M. hyopneumoniae (Mhp-Lpl) (28). Both Ta-LplA and Tk-Lpl recognize octanoate, but Ta-LplA displays relatively low levels of activity toward octanoate (20–30%) compared to lipoate, whereas Tk-Lpl displays about 70% activity toward octanoate. The lower activity of Ta-LplA toward octanoate may reflect the fact that lipoyl synthase homologs (LipA or LipS) are not found on the T. acidophilum genome (37, 39, 42). The presence of a LipS in T. kodakarensis, which allows the conversion of octanoyl-H-protein to lipoyl-H-protein, brings about physiological relevance to the octanoate ligase activity.
We aligned the amino acid sequences of Tk-Lpl, Ta-LplA, and some bacterial Lpls (Fig. S8 and Fig. S9). The amino acid sequences of previously characterized Lpl proteins display low identity to each other, especially in the small domain (Fig. S9). However, the key motifs RRX(T/S)GGG, KX2GXA, and KX3KX3SX3RV, which play important roles in lipoylation (17, 18, 28, 35, 39), are conserved in the large domains of these Lpl proteins (Fig. S8). These motifs are also present in Tk-Lpl-N. Ta-LplA-N harbors an extended sequence (residues 124–137; Fig. S8), which forms a loop structure (highlighted in yellow in Fig. S10A) supposed to repress the catalytic activity of Ta-LplA-N in the absence of Ta-LplA-C by partially capping the ATP binding pocket (39). A portion of the capping loop is reorganized to a β-strand structure upon interaction between Ta-LplA-N and Ta-LplA-C, resulting in removal of the capping structure from the active site (highlighted in yellow in Fig. S10B) (39). On the other hand, most of the sequences corresponding to the capping structure are not found in Tk-Lpl-N and other Lpls (Fig. S8) and might explain why Tk-Lpl-N exhibits activity alone. To examine this possibility, the structure of Tk-Lpl-N was predicted with AlphaFold2, and superimposed with those of Ta-LplA-N (Protein Data Bank [PDB] entry 2ARS; Fig. S10A) and Ta-LplA (Ta-LplA-N and Ta-LplA-C complex, PDB entry 3R07; Fig. S10B). As expected from the sequence, a capping loop structure was absent in Tk-Lpl-N and the corresponding region constituted a short loop, resembling the structure of the loop in Ta-LplA-N when bound to Ta-LplA-C (Fig. S10B). In addition, the structures of the large domains of the active single subunit enzymes Mhp-Lpl (PDB entry 6JOM) and Ec-LplA (PDB entry 1X2G) (with root mean square deviation [RMSD] values 2.2 Å and 3.3 Å, respectively) (Fig. S10C and D) are similar to the entire structure of the Tk-Lpl-N protein and also form short loops. This structural property also supports our prediction that Tk-Lpl-N alone can display activity probably due to the absence of the capping loop.
Our results suggest that Tk-Lpl is involved in the de novo biosynthesis of lipoyl-protein in addition to the salvage pathway in T. kodakarensis. As Tk-LipS recognizes octanoyl-peptide but not free octanoate, we can presume that synthesis of octanoyl-H-protein by Tk-Lpl precedes sulfur insertion in de novo protein lipoylation. However, as the source of the octanoyl moiety has yet to be identified in archaea, the in vivo substrate of Tk-Lpl in de novo protein lipoylation cannot be firmly concluded. We cannot rule out the possibilities that the octanoyl moieties are provided by acyl carriers such as the ACPs in bacteria and eukaryotes or thiol compounds such as coenzyme A (CoA). It at least seems unlikely that ACPs are the octanoyl carriers in archaea, as ACP homologs are not found in any of the archaeal genomes. CoA has been proposed as a candidate for the acyl carrier in archaeal fatty acid biosynthesis (44, 45). We thus examined whether Tk-Lpl recognizes octanoyl-CoA as a substrate. We found however that neither Tk-Lpl (Tk-Lpl-N and Tk-Lpl-C) nor the Tk-Lpl-N protein alone recognized octanoyl-CoA as a substrate (Fig. S11). Based on (i) the activity of Tk-Lpl toward free octanoate, (ii) the lack of Tk-Lpl activity toward octanoyl-CoA, (iii) the specificity of LipS activity toward octanoyl-peptide, and (iv) the absence of ACP homologs on archaeal genomes, we currently propose that free octanoate is the substrate for de novo synthesis of lipoyl-protein (Fig. 1B). We cannot rule out the possibility that an unidentified octanoyl carrier other than ACP or CoA is utilized, and further studies will be necessary to clarify this matter.
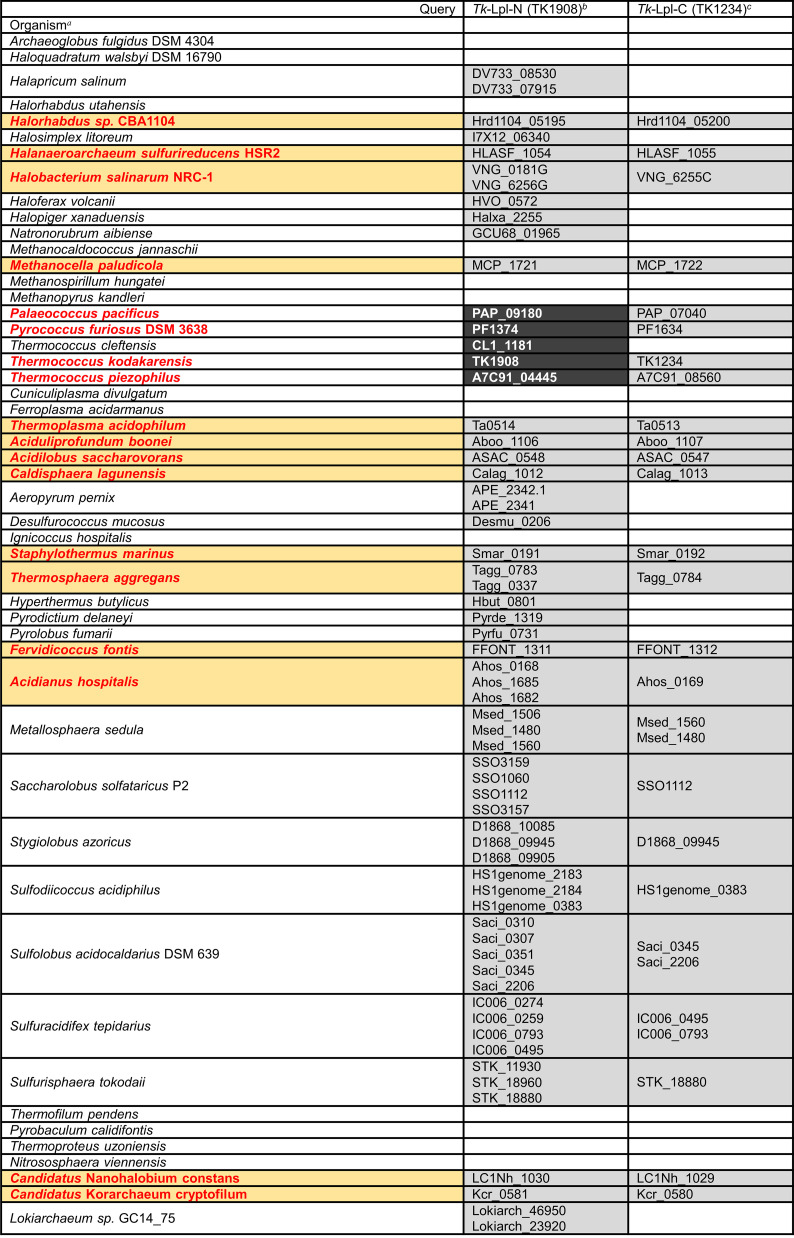
The distributions of homologs of genes encoding Tk-Lpl-N and Tk-Lpl-C proteins were examined in Archaea (Table 1 and expanded in Table S1). Tk-Lpl-N homolog (E-value lower than or equal to 9e-4) is widely distributed in archaea, including almost all haloarchaea and many (hyper)thermophilic archaea. On the other hand, the distribution of Tk-Lpl-C homolog is relatively limited. Intriguingly, Tk-Lpl-C homologs are absent on the genomes of Pyrococcus horikoshii, Pyrococcus sp. ST04, Pyrococcus yayanosii, Thermococcus chitonophagus, and Thermococcus cleftensis, which are phylogenetically close to T. kodakarensis. As Tk-Lpl-N alone displayed lipoate/octanoate-protein ligase activity, there is a possibility that these species utilize their Tk-Lpl-N homologs alone as their Lpl. Only members of Sulfolobales possess the genes encoding single polypeptides containing both the Lpl large and small domains. On the other hand, among the organisms that utilize two separate genes encoding Tk-Lpl-N and Tk-Lpl-C homologs (indicated in bold and red in Table 1 and Table S1), the genes are located next to each other on the genomes of all members of 10 orders (highlighted in orange background in Table 1 and Table S1), with members of Thermococcales the only exception. We also found that Lpl gene homologs are located near the lipoyl synthase homologs (LipA or LipS) on the genomes of a number of archaea (32.7% [56/171] of the organisms with both Lpl and lipoyl synthase gene homologs; Table S1), including all members of Methanocellales (LipA) and Sulfolobales, which possess multiple Lpl homologs (LipA and/or LipS), and some members from Thermococcales (LipS) and Desulfurococcales (LipA or LipS). Since these lipoyl synthases catalyze the sulfur insertion to octanoyl-H-protein, this gene arrangement raises the possibility that the main function of the archaeal lipoate-protein ligases whose genes form an operon with lipoyl synthase genes is the transfer of octanoate, rather than lipoate, to H-protein.
TABLE 1.
Distribution of Lpl homologs in selected species of Archaea
Species whose lipoate-protein ligases (Lpls) are separately encoded by two genes are indicated in bold and red. Orange boxes indicate the organisms in which the two genes are present next to each other.
The Tk-Lpl-N homologs whose E-values are lower than or equal to 1e-125 are shown with white letters on a black background, while those whose E-values are between 3e-69 and 9e-4 are shown with black letters on a gray background.
The Tk-Lpl-C homologs whose E-values are lower than or equal to 8e-4 are shown with black letters on a gray background.
MATERIALS AND METHODS
Chemicals, strains, media, and culture conditions.
Unless mentioned otherwise, chemical reagents were purchased from Nacalai Tesque (Kyoto, Japan) or Fujifilm Wako Pure Chemicals (Osaka, Japan). T. kodakarensis KOD1 (wild type), KU216 (ΔpyrF) (46), and KU216 derivative strains were cultured under anaerobic conditions at 85°C in a nutrient-rich medium (ASW-YT-S0) or synthetic media (ASW-AA-S0, ASW-AA-m1-S0, and ASW-AA-Ser−-LA−-S0). ASW-YT-S0 medium was composed of 0.8 × artificial seawater (ASW) (47), 5.0 g L−1 of yeast extract, 5.0 g L−1 of tryptone, and 2.0 g L−1 of elemental sulfur. ASW-AA-S0 medium consisted of 0.8 × ASW, a mixture of 20 amino acids, modified Wolfe’s trace minerals, a vitamin mixture, and 2.0 g L−1 of elemental sulfur (48). ASW-AA-m1-S0 medium and ASW-AA-Ser−-LA−-S0 medium are modified versions of ASW-AA-S0 medium. In ASW-AA-m1-S0 medium, concentrations of l-arginine hydrochloride (250 mg L−1 from 125 mg L−1) and l-valine (200 mg L−1 from 50 mg L−1) were increased and 20 μM KI, 20 μM H3BO3, 10 μM NiCl2·6H2O, and 10 μM tungsten were supplemented. ASW-AA-Ser−-LA−-S0 medium is ASW-AA-m1-S0 medium without l-serine and lipoate. In all media, 0.8 mg L−1 of resazurin sodium salt was supplemented to detect dissolved oxygen and Na2S·9H2O was added until the media became colorless. For solid medium, elemental sulfur and Na2S·9H2O were replaced with 2 mL L−1 of a polysulfide solution (10 g of Na2S·9H2O and 3 g of sulfur flowers in 15 mL of H2O) and 10 g L−1 of Gelrite was added to solidify the medium. E. coli strain DH5α (TaKaRa, Ohtsu, Japan) used for plasmid construction and E. coli strain BL21-CodonPlus(DE3)-RIL (Agilent Technologies, Santa Clara, CA) used for gene expression were cultivated at 37°C in lysogeny broth medium containing ampicillin (100 mg L−1).
Gene expression and purification of TK1234 and TK1908 recombinant proteins.
Plasmids for the expression of TK1234 and TK1908 were constructed as follows. Coding regions of TK1234 and TK1908 with NdeI and BamHI restriction sites were amplified by PCR with the primer sets eTK1234-F/-R and eTK1908-F/-R, respectively, and inserted into plasmid pET21a(+) at the NdeI-BamHI sites utilizing In-Fusion HD cloning kit (TaKaRa Bio, Shiga, Japan). The resulting plasmids were named pET1234 and pET1908, respectively. After confirming the absence of unintended mutations by DNA sequencing, pET1234 and pET1908 were individually introduced into E. coli strain BL21-CodonPlus(DE3)-RIL. Transformants were cultivated at 37°C until optical density at 660 nm (OD660) reached 0.4–0.6, and gene expression was induced by the addition of isopropyl β-d-1-thiogalactopyranoside at a final concentration of 0.1 mM. Cells were further cultivated at 37°C for 7 h, harvested, resuspended in 50 mM Tris-HCl buffer (pH 7.5), and disrupted by sonication. After centrifugation (4°C, 5,000 × g, 10 min), the soluble cell extracts were incubated at 75°C for 15 min to remove thermolabile proteins from E. coli. After centrifugation (4°C, 5,000 × g, 20 min), the supernatants including the TK1234 and TK1908 recombinant proteins were individually applied to an anion-exchange column, ResourceQ (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom), and proteins were eluted with a linear gradient of NaCl (0-1.0 M) in 50 mM Tris-HCl (pH 7.5) at a flow rate of 2.0 mL min−1. After exchanging buffer of the relevant fractions using Amicon Ultra centrifugal filter unit (molecular weight cutoff [MWCO] 3 K for TK1234 protein and MWCO 10 K for TK1908 protein) (EMD Millipore, Billerica, MA), the proteins were separated by a gel filtration column, Superdex 200 Increase 10/300 GL (GE Healthcare), with a mobile phase of 50 mM HEPES (pH 7.5), 0.15 M NaCl at a flow rate of 0.5 mL min−1. Relevant fractions were concentrated using Amicon Ultra centrifugal filter unit (MWCO 3 K for TK1234 protein and MWCO 10 K for TK1908 protein). Protein concentrations were determined with the Protein Assay System (Bio-Rad, Hercules, CA) using bovine serum albumin (Thermo Fisher Scientific, Waltham, MA) as a standard. The purities of TK1234 and TK1908 recombinant proteins were analyzed by SDS-PAGE.
Protein interaction analysis by gel filtration chromatography.
The interaction analysis between TK1908 and TK1234 proteins was performed utilizing gel filtration chromatography. Molecular masses of TK1908, TK1234, and proteins in their mixture were determined using a Superdex 200 Increase 10/300 GL column. The column was equilibrated with a mobile phase of 50 mM HEPES (pH 7.5) including 0.15 M NaCl at a constant flow rate of 0.5 mL min−1. The absorbance derived from protein was monitored at 280 nm. The Blue Dextran 2000 and standard proteins (aprotinin [6.5 kDa], RNase A [13.7 kDa], carbonic anhydrase [29 kDa], ovalbumin [43 kDa], conalbumin [75 kDa], and aldolase [158 kDa]) in Gel Filtration Calibration Kits (GE Healthcare) were used to measure the void volume of the column and to prepare a standard curve (Fig. S12), respectively. TK1234 protein (100 μM) alone, TK1908 protein (100 μM) alone, or a mixture of TK1234 protein (100 μM) and TK1908 protein (100 μM) after heat treatment (85°C for 15 min) was applied to the gel filtration column. Fractions containing proteins in the mixture were analyzed by SDS-PAGE.
Fatty acid-protein ligase activity measurement.
A chemically synthesized peptide (Eurofins Genomics K.K., Tokyo, Japan), whose amino acid sequence is ESVKAVSE, was utilized as a substrate for the detection of protein ligase activity. The peptide is a portion of the H-protein from T. kodakarensis (TK0150), which is predicted to be the lipoyl domain based on the lipoyl domain sequences of the H-proteins from E. coli (18) and M. tuberculosis (49). The ligase reaction mixture (100 μL) contained 50 mM HEPES buffer (pH 7.5 at 85°C), 2 mM MgCl2, 10 mM lipoate [(R, S)-lipoate or (R)-lipoate] or fatty acid (octanoate, heptanoate, hexanoate, pentanoate, or butyrate), 100 μM peptide, 5 mM ATP, 10 μg of TK1234 protein, and/or 10 μg of TK1908 protein. The reaction mixture without proteins was set as a blank control. After preincubation at 85°C for 3 min, reactions were initiated by adding 5 mM ATP. After incubation at 85°C for 30 min, the reaction was stopped by rapid cooling on ice for 5 min and removal of enzyme by ultrafiltration using an Amicon Ultra centrifugal filter unit (MWCO 10 K). When monitoring product formation by TK1908 protein alone for longer periods of time, 70 μg of TK1908 protein was included in the reaction mixture and incubation at 85°C was prolonged up to 180 min. Reaction products were analyzed by HPLC (see below).
For measuring specific activity, the reaction mixture (100 μL) was composed of 50 mM HEPES buffer (pH 7.5 at 85°C), 2 mM MgCl2, 10 mM lipoate [(R, S)-lipoate, (R)-lipoate or (S)-lipoate] or fatty acid (octanoate, hexanoate or butyrate), 600 μM peptide, 5 mM ATP, 1.6 μg of TK1234 protein (1.5 μM), and 4.2 μg of TK1908 protein (1.5 μM). The reactions were performed at 85°C for 3, 5, and 10 min [(R, S)-lipoate and (R)-lipoate], 5, 10, and 20 min [(S)-lipoate], 5, 10, and 15 min (octanoate and hexanoate), or 10, 15, and 20 min (butyrate). The specific activity of TK1908 protein alone toward (R)-lipoate was also measured.
Examination of substrate specificity of lipoyl synthase.
The substrate specificity of LipS (TK2109 and TK2248 proteins) was examined using octanoyl-peptide, hexanoyl-peptide, butyryl-peptide, and free octanoate as the substrates. The iron-sulfur clusters of recombinant LipS proteins were reconstituted as previously described (42). The reaction mixture (50 μL) contained 50 mM HEPES buffer (pH 8.0 at 75°C), 50 mM NaCl, 10% (vol/vol) glycerol, 5 mM dithiothreitol, 2 mM S-adenosyl-l-methionine, 600 μM modified peptide (octanoyl-peptide, hexanoyl-peptide, or butyryl-peptide) or 10 mM octanoate (with or without 600 μM peptide), 10 mM sodium dithionite, 300 μM reconstituted TK2109 protein and 300 μM reconstituted TK2248 protein. The reaction mixture without proteins was set as a blank control. The reactions were carried out at 75°C for 2 h. The reaction products were analyzed by HPLC and LC-MS.
Measurement and identification of reaction products by HPLC and LC-MS.
Four kinds of the modified peptides (lipoyl-peptide, octanoyl-peptide, hexanoyl-peptide, and butyryl-peptide) (Eurofins Genomics K.K.), in which lipoyl, octanoyl, hexanoyl, and butyryl groups are connected with the Lys residue by amide bond, were used as the standards of reaction products. The enzyme reaction products were analyzed by HPLC with a COSMOSIL 5C18-PAQ column (4.6 mm × 250 mm, 5-μm particle size) using a Nexera X2 system equipped with an SPD-M20A Prominence Photodiode Array Detector (Shimadzu, Kyoto, Japan). The mobile phase consisted of solvent A (MilliQ water with 0.1% formic acid) and solvent B (acetonitrile with 0.1% formic acid). The elution programs were as follows: (i) for the ligase reaction products of lipoate and heptanoate, a gradient of 15–40% solvent B was applied from 0 to 25 min, followed by a 10-min maintenance; then the proportion returned back to 15% solvent B, and the column was reequilibrated for 15 min; (ii) for the ligase reaction products of octanoate, a gradient of 18–40% solvent B was applied from 0 to 30 min, followed by a 5-min maintenance; then the proportion returned back to 18% solvent B, and the column was reequilibrated for 15 min; (iii) For the ligase reaction products of hexanoate and pentanoate, a gradient of 10–40% solvent B was applied from 0 to 30 min, followed by a 5-min maintenance; then the proportion returned back to 10% solvent B, and the column was reequilibrated for 15 min; (iv) For the ligase reaction products of butyrate, a gradient of 5–40% solvent B was applied from 0 to 35 min; then the proportion returned back to 5% solvent B, and the column was reequilibrated for 15 min; (v) for the lipoyl synthase reaction products of modified peptides, a gradient of 5–40% solvent B was applied from 0 to 35 min; then the proportion returned back to 5% solvent B, and the column was reequilibrated for 15 min; and (vi) For the lipoyl synthase reaction products of octanoate, a gradient of 30–40% solvent B was applied from 0 to 10 min, followed by a 25-min maintenance; then the proportion returned back to 30% solvent B, and the column was reequilibrated for 5 min. A flow rate of 0.7 mL min−1 was maintained throughout all the elution programs. Absorbance at 200 nm was monitored by a UV detector.
The reaction products were also analyzed by LC-MS. The compounds in reaction products were separated by COSMOSIL 5C18-PAQ column with the same conditions of HPLC. Detection of analytes was performed using UltiMate 3000 (Thermo Fisher Scientific) and Exactive Plus Fourier-transform (orbitrap) mass spectrometer (Thermo Fisher Scientific). Ionization was performed by heated-electrospray ionization in negative ion mode and parameters were set as follows: Aux gas heater temperature at 300°C, a capillary temperature at 350°C, a spray voltage at 3.5 kV, and a mass range between m/z 300 and 1,500.
Construction of gene disruption strains of T. kodakarensis.
To construct TK1234 and TK1908 gene disruption vectors, the respective genes with 1-kbp of their 5′- and 3′-flanking regions were amplified by PCR using genomic DNA from the host strain KU216 as a template and the primer sets dTK1234-F/-R and dTK1908-F/-R, respectively. Primers used in this study are listed in Table 2. The amplified fragments were inserted into the HincII site of plasmid pUD3 (50), which harbors a selectable pyrF marker gene. The coding regions of each gene were removed by inverse PCR with the primer sets invdTK1234-F/-R and invdTK1908-F/-R and the amplified linear DNA fragments were self-ligated. The 3′-terminal 4 bp of the TK1908 gene were left intact since the regions are shared by the TK1907 gene. The resulting plasmids were named pUDTK1234 and pUDTK1908, respectively.
TABLE 2.
Primers used in this study
| Primer name | 5′-Sequence-3′ |
|---|---|
| dTK1234-F | TTGTACTTGCCCTCCGTCGCCCAGAAGAAG |
| dTK1234-R | ACATAAAGAGCATTCTTGAGGACGTCATAC |
| dTK1908-F | GAAAGCCAAAATCACAATCAGC |
| dTK1908-R | TTCCACGACCTCATTATCACTCTTTAGC |
| invdTK1234-F | GAAGGGTGAAACCCGTGGG |
| invdTK1234-R | GTAAGCTCGCGGGAGAAGTT |
| invdTK1908-F | GAAGTATAGAAATCCTAAGTGG |
| invdTK1908-R | CGTTAAAAACTCCGGAGGGC |
| idTK1234-F | TAAGGGGATGGGTATGAAGCAC |
| idTK1234-R | AGGGCCTTCTTCAACGCTATCTTG |
| idTK1908-F | GTGGTGGTTAAAATGACGATG |
| idTK1908-R | TCTTCTATCAGCTTTTCCGCT |
| odTK1234-F | CTTTCTTAAGTCCATCACGAAGG |
| odTK1234-R | ACCGAACCCATAAAGGCCAACTACTTCAAC |
| odTK1908-F | GCCCCACCACAACAGCCCTTTCTT |
| odTK1908-R | GCCATTCTTTACTTCAACAGGAC |
| eTK1234-F | AAGGAGATATACATATGAAGCACCACGTAGGTG |
| eTK1234-R | GCTCGAATTCGGATCCTCACCCTTCGAGGGCCTTC |
| eTK1908-F | AAGGAGATATACATATGCGCTTCATTCCGCTC |
| eTK1908-R | GCTCGAATTCGGATCCTAACGCATCTCGTTCCAC |
The TK1234 disruption strain and TK1908 disruption strain were prepared as follows. T. kodakarensis KU216 cells were grown in ASW-YT-S0 for 12 h (early stationary phase), harvested, resuspended in 200 μL of 0.8 × ASW, and kept on ice for 30 min. pUDTK1234 or pUDTK1908 (3 μg) was added to the cells, and the mixtures were kept on ice for over 1 h. After heat shock at 85°C for 45 s, the mixtures were kept on ice for 10 min. Cells were inoculated into ASW-AA-S0 liquid medium and incubated at 85°C for 2 days. Cells were cultured in the same medium again in order to enrich transformants harboring the pyrF gene via pop-in single crossover recombination. Cells were then grown at 85°C for 3–5 days on ASW-AA-S0 solid medium supplemented with 0.75% 5-fluoroorotic acid and 10 μg mL−1 uracil to select transformants in which target genes were removed along with the pyrF gene due to a pop-out recombination. Genotypes of the transformants were examined by PCR with primer sets that anneal within the target genes (idTK1234-F/-R or idTK1908-F/-R) and outside the homologous regions for homologous recombination (odTK1234-F/-R or odTK1908-F/-R). The resulting strains, in which TK1234 and TK1908 genes are disrupted, were named ΔTK1234 and ΔTK1908, respectively. The absence of mutations in the 5′- and 3′-flanking regions of TK1234 and TK1908 in the ΔTK1234 and ΔTK1908 strains, respectively, was confirmed by DNA sequencing.
Growth measurements of T. kodakarensis.
Growth properties of the host strain KU216 and the gene disruption strains were examined in the following five media based on the synthetic medium ASW-AA-Ser−-LA−-S0: supplemented with both serine and 1 mM lipoate (Ser[+]-LA[+]); only supplemented with serine (Ser[+]-LA[−]); only supplemented with 1 mM lipoate (Ser[−]-LA[+]); without any supplement (Ser[−]-LA[−]); only supplemented with 1 mM octanoate (Ser[−]-OA[+]). All media were supplemented with 10 μg mL−1 uracil. Cells were precultured in ASW-AA-m1-S0 for about 24 h until the stationary phase and then inoculated into the five media. Culture experiments were performed at 85°C in triplicate and their OD660 was measured.
Sequence alignment and structural homology modeling.
Amino acid sequence alignments were carried out by ClusterX (51) and colored by Web server ESPript 3.0 (52) (http://espript.ibcp.fr/ESPript/ESPript/). The three-dimensional structure model of Tk-Lpl-N, the large domain subunit of lipoate-protein ligase of T. kodakarensis (TK1908), was predicted by AlphaFold2 (53). The generated model of Tk-Lpl-N was validated by PROCHECK module of UCLA-DOE LAB-SAVES v6.0 (54) (https://saves.mbi.ucla.edu/) (Fig. S13). The RMSD values of the structural alignments were assessed by Pairwise structure comparison module of Dali server (55). The superimposition diagrams of the 3D-structures between Tk-Lpl-N and other Lpls were generated by PyMOL 1.8 (56).
Bioinformatic analysis of the homologs of Tk-Lpl-N and related proteins in archaea.
Gene distribution analysis was performed using the BLAST search tool in the KEGG database (57, 58) (https://www.genome.jp/tools/blast/). Amino acid sequences deduced from TK1908 and TK1234 were used as queries to search for the corresponding homologs in archaea. The N-terminal and C-terminal sections of the E2 subunit of pyruvate dehydrogenase complex (PDH E2) of S. solfataricus P2 (SSO1529 and SSO1530, respectively) and the H-protein of T. kodakarensis (TK0150) were used as queries to search for the homologs of archaeal PDH E2 and H-protein. The lipoyl synthases from S. solfataricus P2 (LipA [SSO3158]) and T. kodakarensis (LipS1 [TK2109] and LipS2 [TK2248]) were used as queries to search for the homologs of archaeal lipoyl synthase.
Data availability.
Accession numbers for TK1234 and TK1908 are BAD85423 and BAD86097, respectively. All relevant data are included in the manuscript and supplemental material.
ACKNOWLEDGMENTS
The authors are grateful to Karin Nishimura for LC-MS analysis.
J.-Q.J. acknowledges the scholarship provided by the China Scholarship Council (CSC) (No. 202008050047). This work was partially supported by SPIRITS 2021 of Kyoto University and a grant from the Japan Foundation for Applied Enzymology to T.S. and by the National Key Research and Development Project (No. 2017YFE0129400) and Japanese Society for the Promotion of Science (JSPS) KAKENHI Grant 18H03934 and JP19H05679 (Post-Koch Ecology) and JP19H05684 to H.A.
Footnotes
Supplemental material is available online only.
Contributor Information
Haruyuki Atomi, Email: atomi@sbchem.kyoto-u.ac.jp.
Jeremy D. Semrau, University of Michigan-Ann Arbor
REFERENCES
- 1.Perham RN. 2000. Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu Rev Biochem 69:961–1004. 10.1146/annurev.biochem.69.1.961. [DOI] [PubMed] [Google Scholar]
- 2.Spalding MD, Prigge ST. 2010. Lipoic acid metabolism in microbial pathogens. Microbiol Mol Biol Rev 74:200–228. 10.1128/MMBR.00008-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mayr JA, Feichtinger RG, Tort F, Ribes A, Sperl W. 2014. Lipoic acid biosynthesis defects. J Inherit Metab Dis 37:553–563. 10.1007/s10545-014-9705-8. [DOI] [PubMed] [Google Scholar]
- 4.Cronan JE. 2016. Assembly of lipoic acid on its cognate enzymes: an extraordinary and essential biosynthetic pathway. Microbiol Mol Biol Rev 80:429–450. 10.1128/MMBR.00073-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kikuchi G, Motokawa Y, Yoshida T, Hiraga K. 2008. Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc Jpn Acad, Ser B 84:246–263. 10.2183/pjab.84.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rao NA, Ambili M, Jala VR, Subramanya H, Savithri H. 2003. Structure-function relationship in serine hydroxymethyltransferase. Biochim Biophys Acta 1647:24–29. 10.1016/S1570-9639(03)00043-8. [DOI] [PubMed] [Google Scholar]
- 7.Jordan SW, Cronan JE. 1997. A new metabolic link. The acyl carrier protein of lipid synthesis donates lipoic acid to the pyruvate dehydrogenase complex in Escherichia coli and mitochondria. J Biol Chem 272:17903–17906. 10.1074/jbc.272.29.17903. [DOI] [PubMed] [Google Scholar]
- 8.Jordan SW, Cronan JE. Jr, 2003. The Escherichia coli lipB gene encodes lipoyl (octanoyl)-acyl carrier protein: protein transferase. J Bacteriol 185:1582–1589. 10.1128/JB.185.5.1582-1589.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao X, Miller JR, Cronan JE. 2005. The reaction of LipB, the octanoyl-[acyl carrier protein]: protein N-octanoyltransferase of lipoic acid synthesis, proceeds through an acyl-enzyme intermediate. Biochemistry 44:16737–16746. 10.1021/bi051865y. [DOI] [PubMed] [Google Scholar]
- 10.Nesbitt NM, Baleanu-Gogonea C, Cicchillo RM, Goodson K, Iwig DF, Broadwater JA, Haas JA, Fox BG, Booker SJ. 2005. Expression, purification, and physical characterization of Escherichia coli lipoyl (octanoyl) transferase. Protein Expr Purif 39:269–282. 10.1016/j.pep.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 11.Busby RW, Schelvis JP, Yu DS, Babcock GT, Marletta MA. 1999. Lipoic acid biosynthesis: LipA is an iron-sulfur protein. J Am Chem Soc 121:4706–4707. 10.1021/ja990134g. [DOI] [Google Scholar]
- 12.Ollagnier-de Choudens S, Fontecave M. 1999. The lipoate synthase from Escherichia coli is an iron-sulfur protein. FEBS Lett 453:25–28. 10.1016/s0014-5793(99)00694-8. [DOI] [PubMed] [Google Scholar]
- 13.Cicchillo RM, Iwig DF, Jones AD, Nesbitt NM, Baleanu-Gogonea C, Souder MG, Tu L, Booker SJ. 2004. Lipoyl synthase requires two equivalents of S-adenosyl-l-methionine to synthesize one equivalent of lipoic acid. Biochemistry 43:6378–6386. 10.1021/bi049528x. [DOI] [PubMed] [Google Scholar]
- 14.Cicchillo RM, Lee K-H, Baleanu-Gogonea C, Nesbitt NM, Krebs C, Booker SJ. 2004. Escherichia coli lipoyl synthase binds two distinct [4Fe-4S] clusters per polypeptide. Biochemistry 43:11770–11781. 10.1021/bi0488505. [DOI] [PubMed] [Google Scholar]
- 15.Morris TW, Reed KE, Cronan J. 1994. Identification of the gene encoding lipoate-protein ligase A of Escherichia coli. Molecular cloning and characterization of the lplA gene and gene product. J Biol Chem 269:16091–16100. 10.1016/S0021-9258(17)33977-7. [DOI] [PubMed] [Google Scholar]
- 16.Green D, Morris T, Green J, Cronan J, Jr, Guest J. 1995. Purification and properties of the lipoate protein ligase of Escherichia coli. Biochem J 309:853–862. 10.1042/bj3090853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujiwara K, Toma S, Okamura-Ikeda K, Motokawa Y, Nakagawa A, Taniguchi H. 2005. Crystal structure of lipoate-protein ligase A from Escherichia coli: determination of the lipoic acid-binding site. J Biol Chem 280:33645–33651. 10.1074/jbc.M505010200. [DOI] [PubMed] [Google Scholar]
- 18.Fujiwara K, Maita N, Hosaka H, Okamura-Ikeda K, Nakagawa A, Taniguchi H. 2010. Global conformational change associated with the two-step reaction catalyzed by Escherichia coli lipoate-protein ligase A. J Biol Chem 285:9971–9980. 10.1074/jbc.M109.078717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christensen QH, Cronan JE. 2010. Lipoic acid synthesis: a new family of octanoyltransferases generally annotated as lipoate protein ligases. Biochemistry 49:10024–10036. 10.1021/bi101215f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin N, Christensen QH, Mansilla MC, Cronan JE, de Mendoza D. 2011. A novel two-gene requirement for the octanoyltransfer reaction of Bacillus subtilis lipoic acid biosynthesis. Mol Microbiol 80:335–349. 10.1111/j.1365-2958.2011.07597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christensen QH, Martin N, Mansilla MC, de Mendoza D, Cronan JE. 2011. A novel amidotransferase required for lipoic acid cofactor assembly in Bacillus subtilis. Mol Microbiol 80:350–363. 10.1111/j.1365-2958.2011.07598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rasetto NB, Lavatelli A, Martin N, Mansilla MC. 2019. Unravelling the lipoyl-relay of exogenous lipoate utilization in Bacillus subtilis. Mol Microbiol 112:302–316. 10.1111/mmi.14271. [DOI] [PubMed] [Google Scholar]
- 23.Keeney KM, Stuckey JA, O’Riordan MX. 2007. LplA1-dependent utilization of host lipoyl peptides enables Listeria cytosolic growth and virulence. Mol Microbiol 66:758–770. 10.1111/j.1365-2958.2007.05956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christensen QH, Hagar JA, O’Riordan MX, Cronan JE. 2011. A complex lipoate utilization pathway in Listeria monocytogenes. J Biol Chem 286:31447–31456. 10.1074/jbc.M111.273607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zorzoli A, Grayczyk JP, Alonzo F, III.. 2016. Staphylococcus aureus tissue infection during sepsis is supported by differential use of bacterial or host-derived lipoic acid. PLoS Pathog 12:e1005933. 10.1371/journal.ppat.1005933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramaswamy AV, Maurelli AT. 2010. Chlamydia trachomatis serovar L2 can utilize exogenous lipoic acid through the action of the lipoic acid ligase LplA1. J Bacteriol 192:6172–6181. 10.1128/JB.00717-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao X, Cronan JE. 2015. The Streptomyces coelicolor lipoate-protein ligase is a circularly permuted version of the Escherichia coli enzyme composed of discrete interacting domains. J Biol Chem 290:7280–7290. 10.1074/jbc.M114.626879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu K, Chen H, Jin J, Wang N, Ma G, Huang J, Feng Y, Xin J, Zhang H, Liu H. 2020. Functional identification and structural analysis of a new lipoate protein ligase in Mycoplasma hyopneumoniae. Front Cell Infect Microbiol 10:156. 10.3389/fcimb.2020.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin J, Chen H, Wang N, Zhu K, Liu H, Shi D, Xin J, Liu H. 2020. A novel lipoate-protein ligase, Mhp-LplJ, is required for Lipoic acid metabolism in Mycoplasma hyopneumoniae. Front Microbiol 11:631433. 10.3389/fmicb.2020.631433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Afanador GA, Guerra AJ, Swift RP, Rodriguez RE, Bartee D, Matthews KA, Schön A, Freire E, Freel Meyers CL, Prigge ST. 2017. A novel lipoate attachment enzyme is shared by Plasmodium and Chlamydia species. Mol Microbiol 106:439–451. 10.1111/mmi.13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang SG, Jeong HK, Lee E, Natarajan S. 2007. Characterization of a lipoate-protein ligase A gene of rice (Oryza sativa L.). Gene 393:53–61. 10.1016/j.gene.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 32.Fujiwara K, Takeuchi S, Okamura-Ikeda K, Motokawa Y. 2001. Purification, characterization, and cDNA cloning of lipoate-activating enzyme from bovine liver. J Biol Chem 276:28819–28823. 10.1074/jbc.M101748200. [DOI] [PubMed] [Google Scholar]
- 33.Fujiwara K, Hosaka H, Matsuda M, Okamura-Ikeda K, Motokawa Y, Suzuki M, Nakagawa A, Taniguchi H. 2007. Crystal structure of bovine lipoyltransferase in complex with lipoyl-AMP. J Mol Biol 371:222–234. 10.1016/j.jmb.2007.05.059. [DOI] [PubMed] [Google Scholar]
- 34.Fujiwara K, Okamura-Ikeda K, Motokawa Y. 1994. Purification and characterization of lipoyl-AMP: Nϵ-lysine lipoyltransferase from bovine liver mitochondria. J Biol Chem 269:16605–16609. 10.1016/S0021-9258(19)89432-2. [DOI] [PubMed] [Google Scholar]
- 35.Kim DJ, Kim KH, Lee HH, Lee SJ, Ha JY, Yoon HJ, Suh SW. 2005. Crystal structure of lipoate-protein ligase A bound with the activated intermediate: insights into interaction with lipoyl domains. J Biol Chem 280:38081–38089. 10.1074/jbc.M507284200. [DOI] [PubMed] [Google Scholar]
- 36.McManus E, Luisi BF, Perham RN. 2006. Structure of a putative lipoate protein ligase from Thermoplasma acidophilum and the mechanism of target selection for post-translational modification. J Mol Biol 356:625–637. 10.1016/j.jmb.2005.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Christensen QH, Cronan JE. 2009. The Thermoplasma acidophilum LplA-LplB complex defines a new class of bipartite lipoate-protein ligases. J Biol Chem 284:21317–21326. 10.1074/jbc.M109.015016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Posner MG, Upadhyay A, Bagby S, Hough DW, Danson MJ. 2009. A unique lipoylation system in the Archaea: lipoylation in Thermoplasma acidophilum requires two proteins. FEBS J 276:4012–4022. 10.1111/j.1742-4658.2009.07110.x. [DOI] [PubMed] [Google Scholar]
- 39.Posner MG, Upadhyay A, Crennell SJ, Watson AJ, Dorus S, Danson MJ, Bagby S. 2013. Post-translational modification in the archaea: structural characterization of multi-enzyme complex lipoylation. Biochem J 449:415–425. 10.1042/BJ20121150. [DOI] [PubMed] [Google Scholar]
- 40.Bryant P, Kriek M, Wood RJ, Roach PL. 2006. The activity of a thermostable lipoyl synthase from Sulfolobus solfataricus with a synthetic octanoyl substrate. Anal Biochem 351:44–49. 10.1016/j.ab.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 41.Douglas P, Kriek M, Bryant P, Roach PL. 2006. Lipoyl synthase inserts sulfur atoms into an octanoyl substrate in a stepwise manner. Angew Chem Int Ed Engl 45:5197–5199. 10.1002/anie.200601910. [DOI] [PubMed] [Google Scholar]
- 42.Jin J-Q, Hachisuka S-I, Sato T, Fujiwara T, Atomi H. 2020. A structurally novel lipoyl synthase in the hyperthermophilic archaeon Thermococcus kodakarensis. Appl Environ Microbiol 86:e01359-20. 10.1128/AEM.01359-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Makino Y, Sato T, Kawamura H, Hachisuka S, Takeno R, Imanaka T, Atomi H. 2016. An archaeal ADP-dependent serine kinase involved in cysteine biosynthesis and serine metabolism. Nat Commun 7:13446. 10.1038/ncomms13446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lombard J, López-García P, Moreira D. 2012. An ACP-independent fatty acid synthesis pathway in archaea: implications for the origin of phospholipids. Mol Biol Evol 29:3261–3265. 10.1093/molbev/mss160. [DOI] [PubMed] [Google Scholar]
- 45.Dibrova DV, Galperin MY, Mulkidjanian AY. 2014. Phylogenomic reconstruction of archaeal fatty acid metabolism. Environ Microbiol 16:907–918. 10.1111/1462-2920.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sato T, Fukui T, Atomi H, Imanaka T. 2005. Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis. Appl Environ Microbiol 71:3889–3899. 10.1128/AEM.71.7.3889-3899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robb FT, Place AR. 1995. Media for thermophiles, p 167–168. In Robb FT, Place AR (ed), Archaea: a laboratory manual-thermophiles. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 48.Sato T, Fukui T, Atomi H, Imanaka T. 2003. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J Bacteriol 185:210–220. 10.1128/JB.185.1.210-220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lanz ND, Lee K-H, Horstmann AK, Pandelia M-E, Cicchillo RM, Krebs C, Booker SJ. 2016. Characterization of lipoyl synthase from Mycobacterium tuberculosis. Biochemistry 55:1372–1383. 10.1021/acs.biochem.5b01216. [DOI] [PubMed] [Google Scholar]
- 50.Yokooji Y, Tomita H, Atomi H, Imanaka T. 2009. Pantoate kinase and phosphopantothenate synthetase, two novel enzymes necessary for CoA biosynthesis in the Archaea. J Biol Chem 284:28137–28145. 10.1074/jbc.M109.009696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 52.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291. 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 55.Holm L. 2020. Using Dali for protein structure comparison, p 29–42. In Gáspári Z (ed), Structural bioinformatics: methods and protocols, vol 2112. Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 56.Schrodinger, LLC. 2015. The PyMOL Molecular Graphics System, Version 1.8.
- 57.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44:D457–D462. 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S13. Download aem.00644-22-s0001.pdf, PDF file, 6.2 MB (6.2MB, pdf)
Table S1. Download aem.00644-22-s0002.xlsx, XLSX file, 0.05 MB (49.7KB, xlsx)
Data Availability Statement
Accession numbers for TK1234 and TK1908 are BAD85423 and BAD86097, respectively. All relevant data are included in the manuscript and supplemental material.