ABSTRACT
Toluene o-xylene monooxygenase (ToMO) was found to oxidize chlorobenzene to form 2-chlorophenol (2-CP, 4%), 3-CP (12%), and 4-CP (84%) with a total product formation rate of 1.2 ± 0.17 nmol/min/mg protein. It was also discovered that ToMO forms 4-chlorocatechol (4-CC) from 3-CP and 4-CP with initial rates of 0.54 ± 0.10 and 0.40 ± 0.04 nmol/min/mg protein, respectively, and chlorohydroquinone (CHQ, 13%), 4-chlororesorcinol (4-CR, 3%), and 3-CC (84%) from 2-CP with an initial product formation rate of 1.1 ± 0.32 nmol/min/mg protein. To increase the oxidation rate and alter the oxidation regiospecificity of chloroaromatics, as well as to study the roles of active site residues L192 and A107 of the alpha hydroxylase fragment of ToMO (TouA), we used the saturation mutagenesis approach of protein engineering. Thirteen TouA variants were isolated, among which some of the best substitutions uncovered here have never been studied before. Specifically, TouA variant L192V was identified which had 1.8-, 1.4-, 2.4-, and 4.8-fold faster hydroxylation activity toward chlorobenzene, 2-CP, 3-CP, and 4-CP, respectively, compared to the native ToMO. The L192V variant also had the regiospecificity of chlorobenzene changed from 4% to 13% 2-CP and produced the novel product 3-CC (4%) from 3-CP. Most of the isolated variants were identified to change the regiospecificity of oxidation. For example, compared to the native ToMO, variants A107T, A107N, and A107M produced 6.3-, 7.0-, and 7.3-fold more 4-CR from 2-CP, respectively, and variants A107G and A107G/L192V produced 3-CC (33 and 39%, respectively) from 3-CP whereas native ToMO did not.
IMPORTANCE Chlorobenzene is a commonly used toxic solvent and listed as a priority environmental pollutant by the US Environmental Protection Agency. Here, we report that Escherichia coli TG1 cells expressing toluene o-xylene monooxygenase (ToMO) can successfully oxidize chlorobenzene to form dihydroxy chloroaromatics, which are valuable industrial compounds. ToMO performs this at room temperature in water using only molecular oxygen and a cofactor supplied by the cells. Using protein engineering techniques, we also isolated ToMO variants with enhanced oxidation activity as well as fine-tuned regiospecificities which make direct microbial oxygenations even more attractive. The significance of this work lies in the ability to degrade environmental pollutants while at the same time producing valuable chemicals using environmentally benign biological methods rather than expensive, complex chemical processes.
KEYWORDS: toluene o-xylene monooxygenase, chlorobenzene, microbial oxidation, protein engineering, saturation mutagenesis
INTRODUCTION
Chlorobenzene is a commonly used industrial solvent and is listed as a priority environmental pollutant by the US Environmental Protection Agency (EPA) (1, 2). Long-term exposure of humans to chlorobenzene affects the central nervous system and may damage the lungs, liver, and kidneys (3, 4). Because of its toxicity, it is also regulated under the Safe Drinking Water Act, which sets a maximum contaminant level of 0.1 ppm (3). Therefore, EPA regulations require companies and governmental agencies to detect its presence in effluents and remove them from contaminated sites.
Monooxygenases can hydroxylate chlorobenzene, but since monochlorophenols, the monohydroxylated products, are also common environmental pollutants and 2-chlorophenol (2-CP) is also listed as a priority pollutant (5), double hydroxylation of chlorobenzene to dihydroxy chloroaromatics is a more promising path toward bioremediation and green chemistry. Dihydroxy chloroaromatics are important industrial compounds. For example, 4-chlororesorcinol (4-CR) is used in hair dye formulations (6). It can also be used in the synthesis of coumarin-based compounds, carrying halogen and trifluoromethyl, which have been shown to have antifungal activities. Chlorohydroquinone (CHQ) is a component of polyethers based on organotin with potential applications for breast cancer treatment via growth inhibition (7). It is also used to make photographic developer, stabilizer, antioxidants, medicines, and other useful organic intermediates (2). Direct microbial oxidation of chlorobenzene or chlorophenols for the synthesis of dihydroxy chloroaromatics may provide a more cost-effective and environmentally benign approach compared to complex classical methods (8). In particular, the potential of regioselective catalysis makes direct microbial oxygenations attractive.
Due to its wide substrate range and malleable catalytic activity, toluene o-xylene monooxygenase (ToMO) of Pseudomonas sp. OX1 (9–17) was investigated as a chloroaromatic oxidizer for the first time. ToMO is a soluble, non-heme, O2- and NADH-dependent monooxygenase which hydroxylates toluene and o-xylene as natural substrates. The six Pseudomonas genes encoding ToMO are touABE (three-component hydroxylase with two catalytic oxygen-bridged dinuclear centers, A2B2E2), touC (ferredoxin), touD (regulatory protein), and touF (NADH-oxidoreductase). TouA (499 amino acids) contains the active site, where dioxygen activation and substrate hydroxylation occur. The active site is connected to the exterior of the protein through a large access channel, a probable route for aromatic substrate access and/or product egress (9–20).
Residues TouA I100, E103, A107, Q141, F176, M180, L192, F196, T201, Q204, and F205 form the active site substrate binding pocket (21) and are mostly conserved among toluene monooxygenase (TMO) family members (9, 16, 22, 23). Most of these TouA residues have been the subject of several protein engineering studies and have been shown to influence the catalytic activity and/or regiospecificity of ToMO (9–11, 13, 15, 17, 24–27). Of these residues, L192 and A107 are especially noteworthy since these positions have not been comprehensively studied through saturation mutagenesis in ToMO. All TMOs have leucine at the position analogous to the L192 in ToMO, except for toluene ortho-monooxygenase (TOM) of Burkholderia cepacia G4 (28), which has valine; TouA position A107 is conserved in all of the TMOs. Residue A107 of ToMO has been studied previously using the rational design approach of site-directed mutagenesis by generating variants A107V and A107I (24). It was found that insertion of valine or isoleucine at position 107 resulted in an enhanced para-hydroxylation preference compared to the native ToMO. Through random mutagenesis/directed evolution, we also previously isolated a TouA double variant, E214A/A107T, which also acted like a para enzyme (13). Studies with other TMOs have also reported the important role of position 107 on regioselective aromatic hydroxylations (29–32). In this study, our goals were to evaluate the ability of native ToMO to oxidize chlorobenzene and chlorophenols; to use the semirational approach of saturation mutagenesis at TouA positions A107 and L192 to investigate their roles in catalysis; to enhance the oxidation rates of chlorobenzene and chlorophenols, hence enhancing the synthesis rates of dihydroxy chloroaromatics; and to alter the regiospecific hydroxylation of chloroaromatic compounds by ToMO. It was discovered that native ToMO forms 2-CP, 3-CP, and 4-CP from chlorobenzene and dihydroxy chloroaromatics from CPs. Overall, four new reactions were found for native ToMO, and protein engineering was used to construct 13 variants, of which 10 were novel in ToMO and 9 among the related family enzymes.
RESULTS
The pathways for the oxidation of chlorobenzene to CPs, and dihydroxy chlorobenzene derivatives with native ToMO, are shown in Tables 1 and 2. To our knowledge, there are no previous reports about the hydroxylation of CB or CPs by ToMO. Here, we discovered that Escherichia coli TG1 cells expressing native ToMO oxidize chlorobenzene to form 2-CP (4%), 3-CP (12%), and 4-CP (84%) with a total product formation rate of 1.2 ± 0.17 nmol/min/mg protein (Table 1). ToMO oxidized 2-CP to form CHQ (13%), 4-CR (3%), and 3-chlorocatechol (3-CC, 84%) with a total product formation rate of 1.1 ± 0.32 nmol/min/mg protein (Table 2). Both 3-CP and 4-CP were oxidized to form 4-CC (100%), with initial formation rates of 0.54 ± 0.10 and 0.40 ± 0.04 nmol/min/mg protein, respectively. As expected, we also discovered that native TOM (28) oxidized chlorobenzene primarily at the ortho position and formed 93% of 2-CP, 3% of 3-CP, and 4% of 4-CP; whereas native toluene-4-monooxygenase of Bradyrhizobium sp. BTAi1 (T4MO·BTAi1) (16) oxidized chlorobenzene at the para position and formed only 4-CP (100%) (Table 1). None of these products were detected when E. coli cells with an empty vector was used.
TABLE 1.
Oxidation of chlorobenzene by E. coli TG1 expressing native ToMO, TOM (28), and T4MO·BTAi (16); saturation mutagenesis of TouA A107 and L192 variants, single variants Q204H (15) and F176H (17), and double variants A107G/L192V and A107T/L192Va
| Enzymeb | Regiospecificity (%)c |
Total rated | ||
|---|---|---|---|---|
| 2-CP | 3-CP | 4-CP | ||
| ToMO | 4 | 12 | 84 | 1.2 ± 0.17 |
| L192V | 13 | 12 | 75 | 2.2 ± 0.11 |
| L192A | 8 | 20 | 72 | 1.3 ± 0.45 |
| L192C | 14 | 17 | 69 | 0.27 |
| L192K | 12 | 13 | 75 | 0.18 |
| L192P | – | – | – | NA |
| A107G | 68 | 15 | 17 | 0.97 ± 0.20 |
| A107T | 0 | 2 | 98 | 0.76 ± 0.39 |
| A107M | 0 | 0 | 100 | 0.06 |
| A107N | 2 | 3 | 95 | 0.04 |
| A107I | 0 | 2 | 98 | 0.02 |
| A107V | 3 | 4 | 93 | 0.09 |
| L192V/A107G | 60 | 18 | 22 | 1.7 |
| L192V/A107T | 0 | 5 | 95 | 0.94 |
| Q204He | 2 | 11 | 87 | 0.47 |
| F176He | 2 | 7 | 91 | NM |
| TOM | 93 | 3 | 4 | 0.95 ± 0.10 |
| T4MO·BTAi1 | 0 | 0 | 100 | 0.1 |
ToMO, toluene o-xylene monooxygenase; TouA, alpha hydroxylase fragment of ToMO; CP, chlorophenol; NA, no activity; NM, not measured; TOM, toluene ortho-monooxygenase; T4MO·BTAi1, toluene-4-monooxygenase of Bradyrhizobium sp. BTAi1.
The accession numbers of the enzymes used are AY621080 for native ToMO, AF349675 for native ToM, and CP000494.1 for native T4MO·BTAi1. TouA variants were generated from AY621080.
Standard deviation values were <1%.
Initial concentration was 5 mM chlorobenzene. Initial total product formation rates are given in nmol/min/mg protein.
TABLE 2.
Oxidation of 2-CP, 3-CP, and 4-CP by E. coli TG1/pBS(Kan)ToMO expressing native ToMO and saturation mutagenesis TouA A107 and L192 variants, single variants Q204H (15) and F176H (17), and double variants A107G/L192V and A107T/L192Va
| Enzyme | 2-CPb oxidation |
3-CPb oxidation |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Regiospecificity (%)c |
Total rated | Regiospecificity (%)c |
4-CPb oxidation |
||||||
| CHQ | 4-CR | 3-CC | 4-CC | 3-CC | CHQ | Total rated | 4-CC formation rated | ||
| ToMO | 13 | 3 | 84 | 1.1 ± 0.32 | 100 | 0 | 0 | 0.54 ± 0.10 | 0.40 ± 0.04 |
| L192V | 15 | 3 | 82 | 1.5 ± 0.26 | 96 | 4 | 0 | 1.3 ± 0.34 | 1.9 ± 0.10 |
| L192A | 13 | 0 | 87 | 1.3 ± 0.46 | 100 | 0 | 0 | 0.59 ± 0.05 | 0.73 ± 0.19 |
| L192C | 14 | 2 | 84 | 0.32 | 100 | 0 | 0 | 0.09e | 0.35e |
| L192K | 11 | 3 | 86 | 0.25 | 98 | 2 | 0 | 0.21 | 0.41e |
| L192P | – | – | – | NA | – | – | – | NA | NA |
| A107G | 48 | 0 | 52 | 0.37 ± 0.05 | 57 | 33 | 10 | 0.60 ± 0.25 | 0.27 ± 0.05 |
| A107T | 28 | 19 | 53 | 0.87 ± 0.33 | 100 | 0 | 0 | 0.56 ± 0.12 | 0.39 ± 0.13 |
| A107M | 36 | 22 | 42 | 0.14e | 100 | 0 | 0 | 0.01 | 0.003 ± 0.001 |
| A107N | 8 | 21 | 71 | 0.018 | 100 | 0 | 0 | 0.025 ± 0.008 | 0.026 ± 0.012 |
| A107I | 11 | 2 | 87 | 0.003 | 100 | 0 | 0 | 0.004 | 0.003 |
| A107V | 13 | 1 | 86 | 0.032 ± 0.01 | 100 | 0 | 0 | 0.045 ± 0.007 | 0.019 ± 0.008 |
| L192V/A107T | 18 | 5 | 77 | 0.90 | 100 | 0 | 0 | 0.78 | 0.71 |
| L192V/A107G | 33 | 0 | 67 | NM | 50 | 39 | 11 | NM | NM |
| Q204Hf | 19 | 4 | 77 | 0.31 | 100 | 0 | 0 | 0.10 | 0.14 |
| F176Hf | 81 | 1 | 18 | NM | 95 | 0 | 5 | NM | NM |
ToMO, toluene o-xylene monooxygenase; TouA, alpha hydroxylase fragment of ToMO; NM, not measured; NA, no activity; CP, chlorophenol; CHQ, chlorohydroquinone; CR, chlororesorcinol; CC, chlorocatechol.
Initial concentrations were 500 μM 2-CP, 500 μM 3-CP, and 500 μM 4-CP.
Standard deviation values were <1%.
Initial total product formation rate in nmol/min/mg protein.
Induced with IPTG (isopropyl-β-d-thiogalactopyranoside).
Saturation mutagenesis was performed independently at amino acid positions TouA A107 and L192 (Fig. 1) in attempt to improve the ToMO catalytic activity and regiospecificity for chlorobenzene, 2-CP, 3-CP, and 4-CP oxidation. Libraries of around 1,000 colonies each were generated from both positions. Overall, 500 colonies from both libraries were screened with chlorobenzene and 2-CP as the substrates to ensure that all of the possible saturation variants were sampled, based on the statistical analysis reported by Rui et al. (28). Twelve possible variants from the A107 library were selected after three rounds of screening, and sequencing revealed that six different enzymes were obtained (Tables 1 and 2). TouA variants A107G and A107T were selected three and two times, respectively. Colonies expressing TouA variant A107G were also brown, similar to the color of native TOM (28) (E. coli TG1 cells expressing native ToMO are blue [Fig. S1 in Supplemental File S1]). Five different enzymes were obtained from the L192 library; variant L192V was selected three times (Tables 1 and 2). Variants L192P and A107I were originally chosen for their inability to produce blue-colored indigo on Luria-Bertani (LB) (100 μg/mL kanamycin) medium (Fig. S1).
FIG 1.
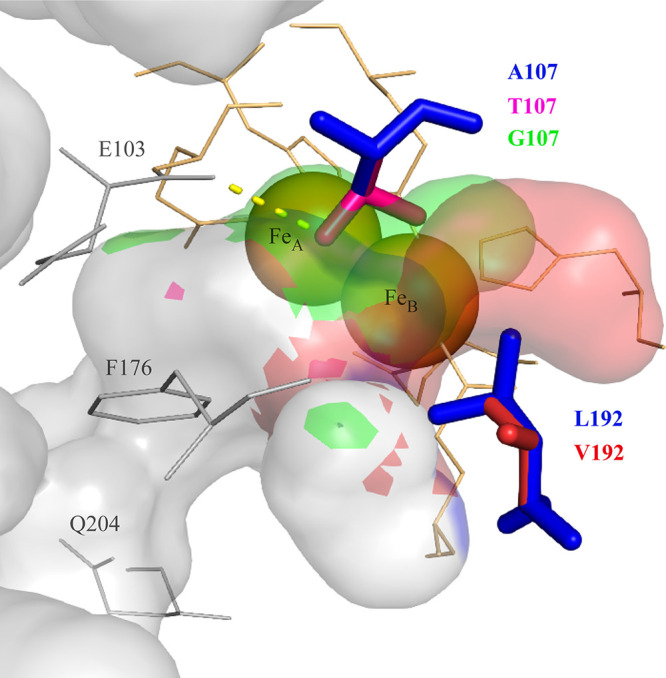
Three-dimensional structure of the ToMO alpha-subunit (TouA) (PDB code: 1T0Q [18]) showing catalytic residues A107 and L102 of the native ToMO (blue) along with the TouA variants A107T (pink), A107G (green), and L192V (red). For clarity, other variants generated are not shown. Other important positions E103 (active site residue), F176 (active site residue), and Q204 (channel residue) are shown in gray. Iron atoms are represented as orange spheres. Iron-binding residues (TouA E104, E134, H137, E197, E231, and H234) are depicted as orange lines. Surfaces of the hydrophobic cavities and channel detected by PyMOL are shown in gray for native ToMO, green for A107G, red for L192V, and pink for A107T. The side chain of T107 participates in the new hydrogen bond (yellow dashed lines) with the main chain of E103 (gray lines).
For chlorobenzene oxidation, there were 3.3-, 2.0-, 3.5-, and 3.0-fold changes in regiospecificity for 2-CP formation with variants L192V, L192A, L192C, and L192K, respectively (Table 1). No product was detected from chlorobenzene even after 24-h contact by variant L192P. Variant TouA A107G showed the most drastic change in regiospecificity among all the other variants by producing 68% 2-CP, which is 17-fold higher than the 2-CP percentage obtained with native ToMO. On the other hand, all of the other A107 variants showed a preference for C-4 hydroxylation and yielded more 4-CP compared to the native ToMO. Interestingly, previously isolated regiospecific variants TouA F176H and Q204H showed only small changes in chlorobenzene oxidation. In terms of activity, variant L192V was a better catalyst than native ToMO, exhibiting a 1.8-fold higher product formation rate (Table 1). Among the A107 and L192 variants, the total product formation rates for TouA L192A, A107G, and A107T were comparable to the rates for native ToMO, whereas TouA L192C, L192K, A107M, A107N, A107I, and A107V showed dramatically reduced rates (Table 1). In terms of regiospecificity, combinations of A107G/L192V or A107T/L192V were not additive for chlorobenzene oxidation; thus, double variants A107G/L192V and A107T/L192V behaved more like the single variants A107G and A107T, respectively.
For 2-CP oxidation, there were a 6.3-, 7.3-, and 7.0-fold changes in regiospecificity for 4-CR formation with variants A107T, A107M, and A107N, respectively (Table 2). Furthermore, TouA variants A107G, A107T, A107M, and the previously isolated regiospecific variant F176H produced 3.7-, 2.2-, 2.8-, and 6.2-fold more CHQ from 2-CP, respectively. On the other hand, TouA L192 variants, as well as the previously isolated regiospecific variant Q204H, showed only small changes. The combination of A107G/L192V might be additive for 2-CP regiospecific oxidation, whereas combining the A107T and L192V mutations slightly altered the oxidation of 2-CP and variant A107T/L192V behaved more like the single variant L192V. For 3-CP oxidation, TouA variant A107G exhibited the largest shift in product distribution by producing the novel products 3-CC (33%) and CHQ (10%). Variants L192V and L192K also produced the novel product 3-CC (4% and 2%, respectively), whereas variant F176H produced the novel product CHQ (5%) from 3-CP. Combining the A107G and L192V mutations did not alter the oxidation of 3-CP very much; thus, double variant A107G/L192V behaved more like the single variant A107G. For 4-CP oxidation, all of the variants isolated exhibited the same regiospecificities as ToMO by only producing 4-CC. In terms of activity, variant L192V was a better catalyst than native ToMO for 2-CP, 3-CP, and 4-CP as well, exhibiting 1.4-, 2.4-, and 4.8-fold higher total product formation rates, respectively (Table 2). To investigate more fully the enhanced rate by L192V relative to the native ToMO, we also measured the apparent vmax values for 4-CP oxidation and found them to be 2.3 ± 0.25 and 0.58 ± 0.10 nmol/min/mg protein, whereas the apparent Km values were 0.17 ± 0.02 mM and 0.12 ± 0.02 mM, respectively. Hence, the apparent vmax/Km was enhanced 2.8-fold for variant L192V for 4-CP oxidation. Similar to chlorobenzene, TouA variants A107M, A107N, A107I, and A107V showed dramatically reduced rates toward 2-CP, 3-CP, and 4-CP as well (Table 2). No products were detected when using variant L192P. To confirm that the increase in the activity of the TouA variant L192V derived from the amino acid substitution rather than from increased enzyme expression levels, we used sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The expression levels of the TouA variant L192V were approximately the same as those of native ToMO; hence, the increase in activity appears to arise from the mutation and not from different expression levels.
DISCUSSION
In this paper, we show that native ToMO forms 2-CP, 3-CP, and 4-CP from chlorobenzene and 3CC, 4CC, 4CR, and CHQ from CPs (Tables 1 and 2). Further, the hydroxylation rate of substrates chlorobenzene, 2-CP, 3-CP, and 4-CP may be enhanced by introducing the TouA mutation L192V (Tables 1 and 2). Also, the regiospecific hydroxylation of substrate chlorobenzene may be changed by introducing the mutations A107G, A107T, A107M, A107N, A107I, A107V, L192V, L192A, L192C, and L192K (Table 1); the regiospecific hydroxylation of 2-CP may be changed by the mutations A107G, A107T, A107M, A107N, and F176H (Table 2); and the regiospecific hydroxylation of 3-CP may be changed by the mutations A107G, L192V, L192K, and F176H (Table 2). The activity of the enzyme is lost by introducing the TouA mutation L192P (no oxidation is observed, Tables 1 and 2). In this study, a total of eight novel substitutions of ToMO were isolated using saturation mutagenesis combined with a powerful agar plate screening method (9, 12, 15, 31, 33). To our knowledge, there are no previous data indicating the role of TouA position L192 in ToMO activity and regiospecificity. In addition, both TouA positions A107 and L192 have never been studied through the saturation mutagenesis approach of protein engineering.
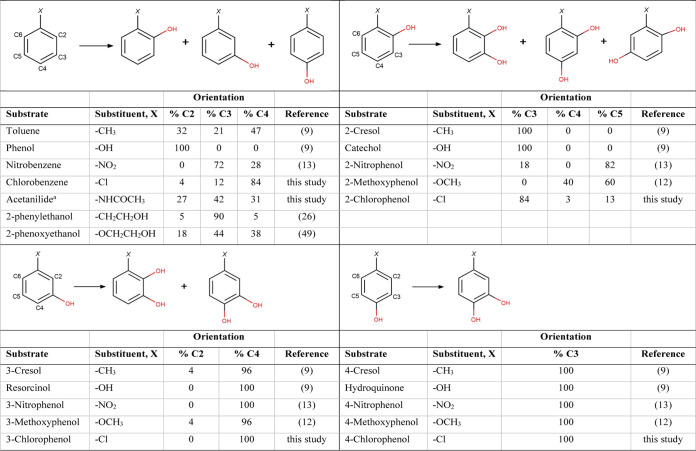
Using site-directed mutagenesis, Notomista et al. (24) previously studied two of the substitutions isolated here, A107V and A107I, for the oxidation of toluene and o-xylene, and these changes resulted in a preference for para-hydroxylation in ToMO. In this study, Notomista et al. also used computational modeling to predict that TouA mutations A107V and A107I would have significantly reduced kcat values for phenol, benzene, toluene, and o-xylene oxidation (24). Here, we report that TouA variants A107V and A107I also acted like para enzymes toward chlorobenzene at significantly reduced rates (Table 1), correlating well with the previous work. Using DNA shuffling, we had previously isolated double variant A107T/E214A with better para acting properties for toluene, nitrobenzene, 2-nitrophenol, o-xylene, and naphthalene oxidation, and concluded that A107T, rather than E214A, was most likely the amino acid responsible for making ToMO a para enzyme (13). Here, we report that single variant TouA A107T acted like a para enzyme toward chlorobenzene, correlating well with our previous prediction (11, 13). Furthermore, residue A107 in the related enzymes T4MO, TpMO, and TOM has also been studied and shown to control the regiospecificities of toluene, indole, o-cresol, o-methoxyphenol, naphthalene, fluorene, nitrobenzene, butadiene, and/or methoxybenzene oxidation (29–32). For example, it was found that changing alanine to glycine resulted in higher ortho hydroxylating capabilities and alanine to threonine resulted in higher para hydroxylating capabilities (30, 31); hence, our results with variants A107G and A107T (Table 1 and 2) are also in accord with those of previous studies of related enzymes. In addition, we also found that the regiospecificity of acetanilide (Table 3) was changed from 31% to 82% 4-acetamidophenol by variant A107T, and from 27% to 98% 2-acetamidophenol by variant A107G. TG1 expressing TouA variant A107G with enhanced ortho hydroxylation capability also changed the color of the cell suspension from native blue to brown, which is similar to the color of ortho-hydroxylating native TOM (29). The formation of blue- and brown-colored indigoid compounds, indigo and isoindigo, respectively, results from the oxidation of indole generated by tryptophanase activity in TG1 cells (29) (Fig. S1). Here, we also isolated novel substitutions A107M and A107N with altered but decreased activities (Tables 1 and 2). In terms of activity, alanine might be the preferred amino acid at TouA position 107 toward chloroaromatic compounds (Tables 1 and 2).
TABLE 3.
Oxidation of monosubstituted benzenes and phenols by native ToMO
To detect and separate acetanilide derivatives, an Ascentis C18 column (25 cm × 4.6 mm, 5 μm) was used with an isocratic method of 2-propanol: methanol: water, 8:18:74 (vol/vol), as described previously (50).
Using site-directed mutagenesis, L192M substitution has been previously isolated for the oxidation of toluene, trichloroethylene (TCE), and/or butadiene in the related enzyme T4MO (34). It was found that changing leucine to methionine resulted in a small change in the product distribution for the oxidation of butadiene (67% S- and 33% R-butadiene epoxide formation with native T4MO versus 73% S- and 27% R-butadiene epoxide with L192M variant). The same substitution in T4MO led to slightly increased activity for TCE (1.5-fold) and lowered activity for toluene (1.8-fold) oxidation (34). Here, we report that V192 of ToMO TouA had enhanced activity toward chlorobenzene, 2-CP, 3-CP, and 4-CP, exhibiting 1.8-, 1.4-, 2.4-, and 4.8-fold higher total product formation rates, respectively. Sazinsky et al. (18) previously suggested that having an isoleucine or valine at position 192 would create extra space in the active site cavity for toluene (Fig. 1). As toluene approaches the active site through the cavity, the bulk residue at 103 (E103) may steer the methyl group on the phenyl ring toward the FeB side of the active site pocket, where residues 180 and 192 reside. The result would be exposure of the toluene C-2 position to the diiron center with subsequent hydroxylation at the ortho position (18). Here, we report that the ortho hydroxylation capability of TouA variant L192V was enhanced by 3.3-fold toward chlorobenzene (Table 1), agreeing well with this previously reported prediction. Furthermore, we found that TouA variant L192V had the regiospecificity of acetanilide (Table 3) changed from 27% to 59% 2-acetamidophenol, also exhibiting 2.2-fold higher ortho hydroxylating capability.
Position A107 is located in the TouA helix B, whereas position L192 is located in the TouA helix E. Both of the positions are closer to the FeB site of the diiron center than to the FeA site, and the closest distances between the FeB site and residues A107 and L192 are around 5.6 and 6.0 Å, respectively. Modeling of the side chains of variants TouA A107 and L192 (Fig. 1) suggests that substitutions at these positions may result in changes in the accessibility, character, and/or size of the active site pocket and may also change the distances of the side chains with respect to iron atoms. Previous modeling-docking calculations, as well as structure-mechanism studies, have provided valuable insights for substrate/product docking at the substrate binding cavity (19, 20, 24, 27, 31, 35, 36). Among all of the A107 variants, only alanine to glycine substitution causes this residue to move further away from both FeA and FeB sites by ~1.4 Å. All of the other substitutions at A107 cause this residue to move closer to the FeB site by ~1.0 Å. The changes in the decreased size of the side chain of residue 107 (A107G), which increases the size of the active site cavity, might have caused the benzene ring of chlorobenzene to shift such that the C-2 is directed more toward the diiron center. On the other hand, the increased size of the side chains of residue 107 (A107T, M, N, I and V) might have changed the location of the chloride group of chlorobenzene to promote more 4-CP formation by oxidation of the C-4. Here, we also observed that the residue in position L192 may need to be hydrophobic in order for ToMO to be efficient catalysts, whereas position A107 can accommodate either hydrophobic or hydrophilic substitutions. Among all mutations, only TouA mutation A107T may introduce an additional hydrogen bond with the main chain of residue E103, whereas other variants do not cause any H-bond formation with its neighbors (Fig. 1).
Unlike the other family enzymes, native ToMO has a relaxed regiospecificity toward the natural substrate toluene (9, 16, 28, 33, 37) and the type of substitutions on the aromatic ring affects its regiospecificity. Table 3 summarizes the influence of ring substituents on regioselective hydroxylation by native ToMO. For example, native ToMO hydroxylates nitrobenzene primarily at the meta position and produces 72% of 3-nitrophenol (3-NP), whereas it hydroxylates chlorobenzene primarily at the para position and produces 84% of 4-CP. In this study, we have shown that native ToMO hydroxylates 2-CP faster than 3- and 4-CP, and hydroxylates 3-CP faster than 4-CP (Table 2), which follows a similar trend for the hydroxylation of NPs (13). The total product formation rates from 500 μM 2-CP (Table 2) and 2-NP (13) were also comparable (1.1 and 0.91 nmol/min/mg protein, respectively).
In this study, we also report that ToMO can successfully oxidize acetanilide, which was once a commonly used antipyretic and analgesic drug (38) (Table 3). Our initial results with acetanilide and other typical substrates used in drug-metabolizing human P450 monooxygenase studies (10, 11, 13, 39–41) show the potential of nonhuman ToMO and its variants in drug metabolism applications; hence, their applicability to selectively hydroxylate other drugs (42, 43), including those which are structurally related to chlorobenzene, is currently being investigated. Given its engineering-friendly properties and the strong expression of its multiple components, whole-cell transformations with ToMO are easy and more reflective of how the biocatalysts may be used (14, 44, 45). Whole cells expressing ToMO perform aromatic hydroxylations at room temperature in water (Tris·HNO3 [pH 7.0]) using molecular oxygen and the NADH cofactor which is provided by the cells (Fig. S1). Our journey began about 18 years ago (9), probing the plasticity of ToMO though protein engineering for the synthesis of fine chemicals and bioremediation. We are excited to continue exploring and expanding the substrate repertoire of this wonderful enzyme for a variety of whole-cell oxidative transformations.
MATERIALS AND METHODS
E. coli strain TG1 was utilized as the host for gene cloning and expression. The relative expressions of the TouA, TouE, and TouF from TG1 expressing native ToMO and its variants were evaluated using SDS-PAGE, as described previously (9). Chemicals were from Sigma-Aldrich (St. Louis, MO), Thermo Fisher Scientific (Waltham, MA), and TCI America (Portland, OR).
Saturation mutagenesis of TouA positions A107 and L192 was performed using three-step overlap extension PCR, as described previously (9, 16, 46). To introduce all amino acids at TouA position A107, a 491-bp DNA fragment which included the MluI restriction site upstream of codon A107 was amplified using primers ToMO-KpnI-front and A107-rear, and a 1,161-bp DNA fragment which included the SalI restriction site downstream of codon A107 was amplified using primers A107-front and ToMO- SalI-rear (Table 4). Similarly, to introduce all amino acids at TouA position L192, a 750-bp DNA fragment which included the MluI restriction site upstream of codon L192 was amplified using primers ToMO-KpnI-front and L192-rear, and a 909-bp DNA fragment which included the SalI restriction site downstream of codon L192 was amplified using primers L192-front and ToMO-SalI-rear. The two sets of DNA fragments were combined with the ToMO-KpnI-front and ToMO-SalI-rear primers to obtain the full-length products (1,616 bp), and cloning was done using restriction enzymes MluI and SalI. A total of 500 colonies from both A107 and L192 libraries were screened on chlorobenzene and 2-CP using a nylon membrane assay (9) to ensure that all of the 64 possible codons sampled, as described previously (9, 28). Briefly, the variant libraries were first plated on LB agar plates containing 100 μg/mL kanamycin and 1% glucose. After overnight growth at 37°C, the colonies were transferred to LB plates containing 100 μg/mL kanamycin and 1 mM substrate (chlorobenzene or 2-CP) using the nylon membrane. Colonies exhibiting a different color around their cell mass or a more intense color than that of native ToMO were further evaluated. To create the TouA A107G/L192V and A107T/L192V double variants, we performed site-directed mutagenesis using the TouA L192V variant as a PCR template to add A107G or A107T to TouA. We used a similar approach as described above using the primers A107G-front and A107G-rear or A107T-front and A107T-rear (Table 4). Oxidation activity levels and regiospecificities were determined as described previously (9, 16, 17) using reverse-phase high-performance liquid chromatography (HPLC). One milliliter of a concentrated exponential-phase cell suspension in Tris-HNO3 buffer (50 mM [pH 7.0]) was contacted with substrates for 5 min to 24 h, and then the supernatant was analyzed by HPLC. An Ascentis C18 column (25 cm × 4.6 mm, 5 μm) (Supelco, Bellefonte, PA) was used with a Shimadzu Prominence solvent delivery system (LC-20AT) coupled to a UV detector (SPD20A; Shimadzu, Kyoto, Japan) and injected by an autosampler (SIL-20A, Shimadzu). A gradient elution was performed with H2O and acetonitrile (70:30 for 0 to 8 min, 40:60 for 15 min, 70:30 for 25 min) as the mobile phase at a flow rate of 1 mL/min. Under these conditions, the retention times for chlorobenzene, 2-CP, 3-CP, 4-CP, CHQ, 4-CR, 3-CC, and 4-CC standards were 19.6, 15.3, 16.4, 16.0, 5.9, 8.1, 10.2, and 11.8 min, respectively. Product identifications were analyzed at 275 nm and 285 nm, confirmed by comparison of retention times and by co-elution with standards. Experiments were performed with at least three replicates of native enzyme and the selected variants for each substrate, and at least four injections were made for each substrate. To determine the kinetic parameters, 4-CC formation was measured at six different time points (2.5, 5, 10, 15, 30, and 45 min) from five different 4-CP concentrations (0.05, 0.125, 0.25, 0.5, and 1.0 mM). At least three time points from the linear parts of the curves were used to calculate specific initial product formation rates. Kinetic constants (apparent vmax, Km) were determined from the specific initial product formation rates at each concentration using a double reciprocal Lineweaver-Burk plot. DNA sequencing was performed using the dideoxy chain termination method (ElimsBio, Hayward, CA) with the primers listed in Table 4. Mutations at TouA A107 and L192 were modeled and visualized from the X-ray crystallography data of ToMO hydroxylase (PDB: 1T0Q) using PyMOL (47) and Swiss-Pdb Viewer (48), as described previously (15). The surface of cavities (culled) was detected using PyMOL. The selected cavity detection radius and cutoff values were 3 and 5 solvent radii, respectively. All distance measurements (expressed in angstroms) and H-bond analyses were done using PyMOL.
TABLE 4.
Primers used for saturation mutagenesis of TouA A107 and L192, site-directed mutagenesis of TouA A107G/L192V and A107T/L192V, and sequencing of the A107 and L192 regions of the touA gene in TG1/pBS(Kan)ToMO (9)
| Primer | Nucleotide sequence |
|---|---|
| Mutagenesis | |
| ToMO-KpnI-front | 5′-CCGGCTCGTATGTTGTGTGGAATTGTGAGCGG-3′ |
| ToMO-SalI-rear | 5′-CCCACTCATAATCATGAGCGTCG-3′ |
| L192-front | 5′-CCGTGGCCGTCTCTATCATGNNNACCTTCGCATTCGAAACAGG-3′ |
| L192-rear | 5′-CCTGTTTCGAATGCGAAGGTNNNCATGATAGAGACGGCCACGG-3′ |
| A107-front | 5′-CACTTGAAGAATACGCCNNNAGCACTGCTGAAGCCCG-3′ |
| A107-rear | 5′-CGGGCTTCAGCAGTGCTNNNGGCGTATTCTTCAAGTG-3′ |
| A107G-front | 5′-CTTGAAGAATACGCCGGAAGCACTGCTGAAGCC-3′ |
| A107G-rear | 5′-GGCTTCAGCAGTGCTTCCGGCGTATTCTTCAAG-3′ |
| A107T-front | 5′-CTTGAAGAATACGCCACAAGCACTGCTGAAGCC-3′ |
| A107T-rear | 5′-GGCTTCAGCAGTGCTTGTGGCGTATTCTTCAAG-3′ |
| Sequencing | |
| L192_check_1 | 5′-GCTGGGTTAGCACTATGCAANNNCACTTCGGAGCGATCGCA-3′ |
| L192_check_2 | 5′-CCGGTGAGTACCGANNNAAGCTTCCAAGATCGCCAGATTGCG-3′ |
| A107_check_1 | 5′-CAATCGCTGCACGGTCTTTCNNNGACGACATGATGATGACC-3′ |
| A107_check_2 | 5′-GCGGCCAAACCGAGAAANNNCATATTGGTGAAGCCTGTTTCG-3′ |
ACKNOWLEDGMENTS
This study was supported by the National Institutes of Health Support of Competitive Research Program (project number: 1SC3GM136524-01), a California State University Program for Education and Research in Biotechnology New Investigator Grant, and a U.S. Department of Education STEM Success Program Title III grant.
Footnotes
Supplemental material is available online only.
Contributor Information
Gönül Vardar-Schara, Email: gschara@csustan.edu.
Pablo Ivan Nikel, Novo Nordisk Foundation Center for Biosustainability.
REFERENCES
- 1.Agency for Toxic Substances and Disease Registry (ATSDR). 2017. Agency for Toxic Substances and Disease Registry’s Substance Priority List. ATSDR, US Department of Health and Human Services, Washington, DC. Available from https://www.atsdr.cdc.gov/spl/index.html. [Google Scholar]
- 2.Larrañaga MD, Lewis RJ, Lewis RA. 2016. Hawley’s condensed chemical dictionary, 16th edition. John Wiley & Sons, Inc., Hoboken, NJ. [Google Scholar]
- 3.U.S. Environmental Protection Agency. 2009. National Primary Drinking Water Regulations, p 7. US EPA, Washington, DC. Available from https://www.epa.gov/sites/production/files/2016-06/documents/npwdr_complete_table.pdf. [Google Scholar]
- 4.Beck U, Löser E. 2011. Chlorinated benzenes and other nucleus-chlorinated aromatic hydrocarbons. Ullmann’s encyclopedia of industrial chemistry, Wiley-VCH Verlag GmbH & Co, Weinheim, Germany. [Google Scholar]
- 5.Olaniran AO, Igbinosa EO. 2011. Chlorophenols and other related derivatives of environmental concern: properties, distribution, and microbial degradation processes. Chemosphere 83:1297–1306. 10.1016/j.chemosphere.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 6.França SAd, Dario MF, Esteves VB, Baby AR, Velasco MVR. 2015. Types of hair dye and their mechanisms of action. Cosmetics 2:110–126. 10.3390/cosmetics2020110. [DOI] [Google Scholar]
- 7.Carraher CE, Jr, Roner MR, Shahi K, Barot G. 2011. Structural consideration in designing organotin polyethers to arrest the growth of breast cancer cells in vitro. Materials (Basel) 4:801–815. 10.3390/ma4040801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tao JA, Kazlauskas RJ. 2011. Biocatalysis for green chemistry and chemical process development. John Wiley & Sons, Inc, Hoboken, NJ. [Google Scholar]
- 9.Vardar G, Wood TK. 2004. Protein engineering of toluene-o-xylene monooxyganase from Pseudomonas stutzeri OX1 for synthesizing 4-methylresorcinol, methylhydroquinone, and pyrogallol. Appl Environ Microbiol 70:3253–3262. 10.1128/AEM.70.6.3253-3262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vardar G, Wood TK. 2005. Protein engineering of toluene-o-xylene monooxygenase from Pseudomonas stutzeri OX1 for enhanced chlorinated ethene degradation and o-xylene oxidation. Appl Microbiol Biotechnol 68:510–517. 10.1007/s00253-005-1923-4. [DOI] [PubMed] [Google Scholar]
- 11.Vardar G, Wood TK. 2005. Alpha-subunit positions methionine 180 and glutamate 214 of Pseudomonas stutzeri OX1 toluene-o-xylene monooxygenase influence catalysis. J Bacteriol 187:1511–1514. 10.1128/JB.187.4.1511-1514.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vardar G, Tao Y, Lee J, Wood TK. 2005. Alanine 101 and alanine 110 of the alpha subunit of Pseudomonas stutzeri OX1 toluene-o-xylene monooxygenase influence the regiospecific oxidation of aromatics. Biotechnol Bioeng 92:652–658. 10.1002/bit.20637. [DOI] [PubMed] [Google Scholar]
- 13.Vardar G, Ryu K, Wood TK. 2005. Protein engineering of toluene-o-xylene monooxygenase from Pseudomonas stutzeri OX1 for oxidizing nitrobenzene to 3-nitrocatechol, 4-nitrocatechol, and nitrohydroquinone. J Biotechnol 115:145–156. 10.1016/j.jbiotec.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 14.Fishman A, Tao Y, Vardar G, Rui L, Wood TK. 2006. Controlling regiospecific oxidation of aromatics and the degradation of chlorinated aliphatics via active site engineering of toluene monooxygenases. In Ramos J-L, Levesque RC (ed), Pseudomonas: molecular biology of emerging issues, vol 4, p 237–286. Springer US, Netherlands. [Google Scholar]
- 15.Kurt C, Sönmez B, Vardar N, Yanık-Yıldırım KC, Vardar-Schara G. 2016. Cavity residue leucine 95 and channel residues glutamine 204, aspartic acid 211, and phenylalanine 269 of toluene-o-xylene monooxygenase influence catalysis. Appl Microbiol Biotechnol 100:7599–7609. 10.1007/s00253-016-7658-6. [DOI] [PubMed] [Google Scholar]
- 16.Yanık-Yıldırım KC, Vardar-Schara G. 2014. Saturation mutagenesis of Bradyrhizobium sp. BTAi1 toluene 4-monooxygenase at alpha-subunit residues proline 101, proline 103, and histidine 214 for regiospecific oxidation of aromatics. Appl Microbiol Biotechnol 98:8975–8986. 10.1007/s00253-014-5913-2. [DOI] [PubMed] [Google Scholar]
- 17.Sönmez B, Yanık-Yıldırım KC, Wood TK, Vardar-Schara G. 2014. The role of substrate binding pocket residues phenylalanine 176 and phenylalanine 196 on Pseudomonas sp. OX1 toluene o-xylene monooxygenase activity and regiospecificity. Biotechnol Bioeng 111:1506–1512. 10.1002/bit.25212. [DOI] [PubMed] [Google Scholar]
- 18.Sazinsky MH, Bard J, Donato AD, Lippard SJ. 2004. Crystal structure of the toluene/o-xylene monooxygenase hydroxylase from Pseudomonas stutzeri OX1. Insight into the substrate specificity, substrate channeling, and active site tuning of multicomponent monooxygenases. J Biol Chem 279:30600–30610. 10.1074/jbc.M400710200. [DOI] [PubMed] [Google Scholar]
- 19.McCormick MS, Lippard SJ. 2011. Analysis of substrate access to active sites in bacterial multicomponent monooxygenase hydroxylases: X-ray crystal structure of xenon-pressurized phenol hydroxylase from Pseudomonas sp. OX1. Biochemistry 50:11058–11069. 10.1021/bi201248b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song WJ, Gucinski G, Sazinsky MH, Lippard SJ. 2011. Tracking a defined route for O2 migration in a dioxygen-activating diiron enzyme. Proc Natl Acad Sci USA 108:14795–14800. 10.1073/pnas.1106514108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sazinsky MH, Dunten PW, McCormick MS, DiDonato A, Lippard SJ. 2006. X-ray structure of a hydroxylase-regulatory protein complex from a hydrocarbon-oxidizing multicomponent monooxygenase, Pseudomonas sp. OX1 phenol hydroxylase. Biochemistry 45:15392–15404. 10.1021/bi0618969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Canada KA, Iwashita S, Shim H, Wood TK. 2002. Directed evolution of toluene ortho-monooxygenase for enhanced 1-naphthol synthesis and chlorinated ethene degradation. J Bacteriol 184:344–349. 10.1128/JB.184.2.344-349.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tao Y, Fishman A, Bentley WE, Wood TK. 2004. Oxidation of benzene to phenol, catechol, and 1,2,3-trihydroxybenzene by toluene 4-monooxygenase of Pseudomonas mendocina KR1 and toluene 3-monooxygenase of Ralstonia pickettii PKO1. Appl Environ Microbiol 70:3814–3820. 10.1128/AEM.70.7.3814-3820.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Notomista E, Cafaro V, Bozza G, Donato AD. 2009. Molecular determinants of the regioselectivity of toluene/o-xylene monooxygenase from Pseudomonas sp. strain OX1. Appl Environ Microbiol 75:823–836. 10.1128/AEM.01951-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cafaro V, Notomista E, Capasso P, Donato AD. 2005. Mutation of glutamic acid 103 of toluene o-xylene monooxygenase as a means to control the catabolic efficiency of a recombinant upper pathway for degradation of methylated aromatic compounds. Appl Environ Microbiol 71:4744–4750. 10.1128/AEM.71.8.4744-4750.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Notomista E, Scognamiglio R, Troncone L, Donadio G, Pezzella A, Donato AD, Izzo V. 2011. Tuning the specificity of the recombinant multicomponent toluene o-xylene monooxygenase from Pseudomonas sp. strain OX1 for the biosynthesis of tyrosol from 2-phenylethanol. Appl Environ Microbiol 77:5428–5437. 10.1128/AEM.00461-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell KH, Studts JM, Fox BG. 2002. Combined participation of hydroxylase active site residues and effector protein binding in a para to ortho modulation of toluene 4-monooxygenase regiospecificity. Biochemistry 41:3176–3188. 10.1021/bi012036p. [DOI] [PubMed] [Google Scholar]
- 28.Rui L, Kwon YM, Fishman A, Reardon KF, Wood TK. 2004. Saturation mutagenesis of toluene ortho-monooxygenase of Burkholderia cepacia G4 for enhanced 1-naphthol synthesis and chloroform degradation. Appl Environ Microbiol 70:3246–3252. 10.1128/AEM.70.6.3246-3252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rui L, Reardon KF, Wood TK. 2005. Protein engineering of toluene ortho-monooxygenase of Burkholderia cepacia G4 for regiospecific hydroxylation of indole to form various indigoid compounds. Appl Microbiol Biotechnol 66:422–429. 10.1007/s00253-004-1698-z. [DOI] [PubMed] [Google Scholar]
- 30.Tao Y, Fishman A, Bentley WE, Wood TK. 2004. Altering toluene 4-monooxygenase by active-site engineering for the synthesis of 3-methoxycatechol, methoxyhydroquinone, and methylhydroquinone. J Bacteriol 186:4705–4713. 10.1128/JB.186.14.4705-4713.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fishman A, Tao Y, Rui L, Wood TK. 2005. Controlling the regiospecific oxidation of aromatics via active site engineering of toluene para-monooxygenase of Ralstonia pickettii PKO1. J Biol Chem 280:506–514. 10.1074/jbc.M410320200. [DOI] [PubMed] [Google Scholar]
- 32.McClay K, Boss C, Keresztes I, Steffan RJ. 2005. Mutations of toluene-4-monooxygenase that alter regiospecificity of indole oxidation and lead to production of novel indigoid pigments. Appl Environ Microbiol 71:5476–5483. 10.1128/AEM.71.9.5476-5483.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fishman A, Tao Y, Bentley WE, Wood TK. 2004. Protein engineering of toluene 4-monooxygenase of Pseudomonas mendocina KR1 for synthesizing 4-nitrocatechol from nitrobenzene. Biotechnol Bioeng 87:779–790. 10.1002/bit.20185. [DOI] [PubMed] [Google Scholar]
- 34.Steffan RJ, McClay KR. 2000. Preparation of enantio-specific epoxides. US patent WO/2000/073425.
- 35.Bailey LJ, Acheson JF, McCoy JG, Elsen NL, George N, Phillips J, Fox BG. 2012. Crystallographic analysis of active site contributions to regiospecificity in the diiron enzyme toluene 4-monooxygenase. Biochemistry 51:1101–1113. 10.1021/bi2018333. [DOI] [PubMed] [Google Scholar]
- 36.Acheson JF, Bailey LJ, Brunold TC, Fox BG. 2020. Toluene 4-monooxygenase reaction intermediates. In Scott RA (ed), Encyclopedia of Inorganic and Bioinorganic Chemistry. John Wiley & Sons, Inc., Hoboken, NJ. 10.1002/9781119951438. [DOI] [Google Scholar]
- 37.Fishman A, Tao Y, Wood TK. 2004. Physiological relevance of successive hydroxylations of toluene by toluene para-monooxygenase of Ralstonia pickettii PKO1. Biocatal Biotransformation 22:283–289. 10.1080/10242420400012008. [DOI] [Google Scholar]
- 38.Mahmud S, Rosen N. 2019. History of NSAID use in the treatment of headaches pre and post-industrial revolution in the United States: the rise and fall of antipyrine, salicylic acid, and acetanilide. Curr Pain Headache Rep 23:6. 10.1007/s11916-019-0744-6. [DOI] [PubMed] [Google Scholar]
- 39.Sun-Ha P, Dong-Hyun K, Dooil K, Dae-Hwan K, Heung-Chae J, Jae-Gu P, Taeho A, Donghak K, Chul-Ho Y. 2010. Engineering bacterial cytochrome P450 (P450) BM3 into a prototype with human P450 enzyme activity using indigo formation. Drug Metab Dispos 38:732–739. 10.1124/dmd.109.030759. [DOI] [PubMed] [Google Scholar]
- 40.Nardo GD, Fantuzzi A, Sideri A, Panicco P, Sassone C, Giunta C, Gilardi G. 2007. Wild-type CYP102A1 as a biocatalyst: turnover of drugs usually metabolised by human liver enzymes. J Biol Inorg Chem 12:313–323. 10.1007/s00775-006-0188-4. [DOI] [PubMed] [Google Scholar]
- 41.Sakurai H, Hatayama E, Nishida M. 1983. Aromatic hydroxylation of acetanilide and aniline by hemin-thiolester complex as a cytochrome P450-model. Inorganica Chim Acta 80:7–12. 10.1016/S0020-1693(00)91254-4. [DOI] [Google Scholar]
- 42.Rentmeister A, Brown TR, Snow CD, Carbone MN, Arnold FH. 2011. Engineered bacterial mimics of human drug metabolizing enzyme CYP2C9. ChemCatChem 3:1065–1071. 10.1002/cctc.201000452. [DOI] [Google Scholar]
- 43.Chul-Ho Y, Keon-Hee K, Dong-Hyun K, Heung-Chae J, Jae-Gu P. 2007. The bacterial P450 BM3: a prototype for a biocatalyst with human P450 activities. Trends Biotechnol 25:289–298. 10.1016/j.tibtech.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 44.Zöllner A, Buchheit D, Meyer MR, Maurer HH, Peters FT, Bureik M. 2010. Production of human phase I and 2 metabolites by whole-cell biotransformation with recombinant microbes. Bioanalysis 2:1277–1290. 10.4155/bio.10.80. [DOI] [PubMed] [Google Scholar]
- 45.Wachtmeister J, Rother D. 2016. Recent advances in whole cell biocatalysis techniques bridging from investigative to industrial scale. Curr Opin Biotechnol 42:169–177. 10.1016/j.copbio.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 46.Özgen FF, Vardar-Yel N, Roth OS, Shahbaz LS, Vardar-Schara G. 2020. Surface residues serine 69 and arginine 194 of metagenome-derived lipase influence catalytic activity. Biochem Eng J 154:107442. 10.1016/j.bej.2019.107442. [DOI] [Google Scholar]
- 47.DeLano WL. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA. [Google Scholar]
- 48.Guex N, Peitsch MC, Schwede T. 2009. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30:S162–S173. 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- 49.Donadio G, Sarcinelli C, Pizzo E, Notomista E, Pezzella A, Cristo CD, Lise FD, Donato AD, Izzo V. 2015. The toluene o-xylene monooxygenase enzymatic activity for the biosynthesis of aromatic antioxidants. PLoS One 10:e0124427. 10.1371/journal.pone.0124427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mancilla J, Valdes E, Gil L. 1989. A novel isocratic HPLC method to separate and quantify acetanilide and its hydroxy aromatic derivatives: 2-, 3-, and 4-hydroxyacetanilide (paracetamol or acetaminophenol). Eur J Drug Metab Pharmacokinet 14:241–244. 10.1007/BF03190105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Download aem.00358-22-s0001.pdf, PDF file, 0.2 MB (251.6KB, pdf)