Figure 2.
Selective expression of NRXN3 AS5+ proteoforms
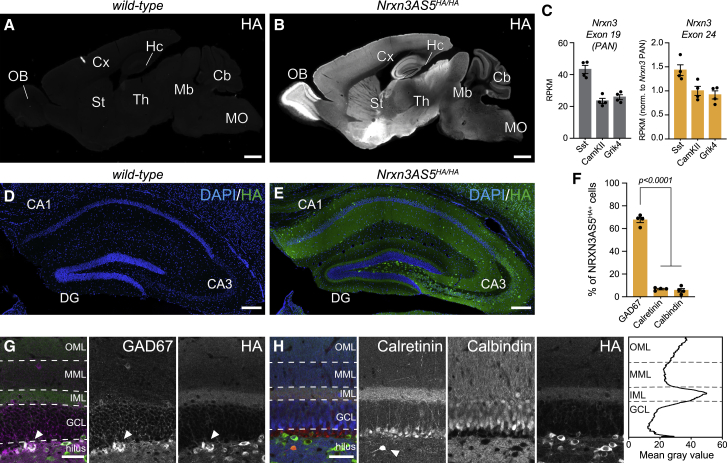
(A and B) Detection of NRXN3-AS5HA proteins in 28 day old wild-type (A) and homozygous Nrxn3-AS5HA mice (B); OB, olfactory bulb, Cx, cortex, St, striatum, Hc, hippocampus, Th, thalamus, Mb, midbrain, Cb, cerebellum, and MO, medulla oblongata.
(C) RNA sequencing reads for constitutive (exon 19) and alternative exon 24 of Nrxn3, from ribosome-associated mRNAs isolated from SSTcre, CamKIIcre (CA1), and Grik4cre (CA3)—defined hippocampal cell populations. Read counts extracted from published data (Furlanis et al., 2019).
(D and E) Detection of NRXN3-AS5HA proteins in hippocampus of wild-type (D) and Nrxn3-AS5HA/HA mice (E).
(F) Fraction of NRXN3-AS5HA protein expressing cells in the hilus of dentate gyrus coexpressing GAD67, calbindin (CB), or calretinin (CR), N = 4 mice, n = 3–4 brain slices per mouse, P25-30.
(G and H) Colocalization analysis of NRXN3-AS5HA protein (HA, green) with (G) GAD67 (magenta), arrowheads indicate colocalized example cell and (H) calretinin (CR, red) and calbindin (CB, blue) in dentate gyrus in Nrxn3-AS5HA/HA knockin mice. Note that calretinin is expressed in glutamatergic mossy cells as well as (at higher level) in Cajal-Retzius cells (indicated with arrow head in [H]). Right panel in (H) shows a line scan of HA staining intensity across layers of dentate gyrus (Average of N = 3 animals, n = 2 ROIs per animal). OML, outer molecular layer, MML, middle molecular layer, IML, inner molecular layer, and GCL, granule cell layer.
Mean and SEM, two-way ANOVA followed by Bonferroni’s test. Scale bars, 1 mm in (A and B); 200 μm in (D and E); and 50 μm in (G and H).