Figure 5.
Translational silencing gates NRXN3 protein expression
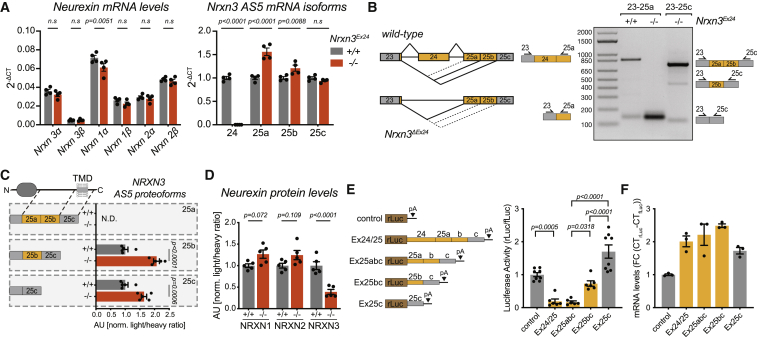
(A) Quantitative PCRs of major Nrxn transcript isoforms (left panel) and mRNAs containing exon 24, alternative accepters 25a, 25b, and the constitutive exon 25c (right panel, plotted relative to wild type) for wild type and Nrxn3ΔEx24 mice, normalized to Gapdh, hippocampus, P25-30, N = 4 mice per genotype.
(B) Schematic diagram illustrating alternative splicing events in wild-type and Nrxn3ΔEx24 hippocampus (left) and semiquantitative PCR visualizing Nrxn3 transcript variants arising from alternative splicing at AS5 in wild-type and Nrxn3ΔEx24 mice (right). Position of primer binding sites on alternative exon segments is illustrated. See STAR Methods for details.
(C and D) Detection of AS5 proteoforms by targeted proteomics with heavy peptides targeting alternative accepters 25a, 25b, and constitutive exon 25c (C) or targeting all NRXN1, NRXN2, and NRXN3 proteoforms (D). Ratios of light to heavy peptide detection are displayed in reference to wild-type samples of each peptide. One representative peptide shown, consistent results were obtained for multiple proteotypic peptides for the same proteoform (see Table S6), hippocampus, P25-20, N = 5 mice per genotype.
(E and F) Luciferase assay of dual-promoter plasmids expressing firefly (fLuc) and renilla (rLuc) luciferase, left panel: schematic representation of different Nrxn3 exonic sequences fused after the translational stop codon of rLuc, right panel: luciferase activity from renilla luciferase constructs normalized to firefly luciferase activity, N = 2–3 cell cultures, n = 2–3 replicates per culture (E) and mRNA levels determined by RT-qPCR of renilla luciferase constructs normalized to firefly luciferase, N = 3 cell cultures (F) (For sequences of luciferase constructs see Table S4).
Mean and SEM, with two-way or one-way ANOVA followed by Bonferroni’s test for both qPCR and proteomic analysis or luciferase assay, respectively.