ABSTRACT
RNA splicing refers to a process by which introns of a pre-mRNA are excised and the exons at both ends are joined together. Chloroplast introns are inherently self-splicing ribozymes, but over time, they have lost self-splicing ability due to the degeneration of intronic elements. Thus, the splicing of chloroplast introns relies heavily on nuclear-encoded splicing factors, which belong to diverse protein families. Different splicing factors and their shared intron targets are supposed to form ribonucleoprotein particles (RNPs) to facilitate intron splicing. As characterized in a previous review, around 14 chloroplast intron splicing factors were identified until 2010. However, only a few genetic and biochemical evidence has shown that these splicing factors are required for the splicing of one or several introns. The roles of splicing factors are generally believed to facilitate intron folding; however, the precise role of each protein in RNA splicing remains ambiguous. This may be because the precise binding site of most of these splicing factors remains unexplored. In the last decade, several new splicing factors have been identified. Also, several splicing factors were found to bind to specific sequences within introns, which enhanced the understanding of splicing factors. Here, we summarize recent progress on the splicing factors in land plant chloroplasts and discuss their possible roles in chloroplast RNA splicing based on previous studies.
KEYWORDS: Chloroplast, intron, RNA splicing, RNP, splicing factor
Introduction
Land plant chloroplasts evolved from endosymbiotic cyanobacteria, and chloroplast genome has retained some prokaryotic properties while also evolved some eukaryotic properties. The features of chloroplast genome have determined the complexity of the chloroplast gene expression regulation. The inconsistence of chloroplast gene transcription rates and steady-state levels of mature transcripts indicates that the post-transcriptional control is a critical step in regulating chloroplast gene expression [1–3]. The post-transcriptional processes include pre-mRNA cleavage, RNA splicing, RNA editing, and RNA stability [1] (see Fig. 1). Many nuclear-encoded proteins regulate these processes, which is essential to converting pre-mRNAs into mature mRNAs. In this review, we have focused on chloroplast intron splicing.
Figure 1.
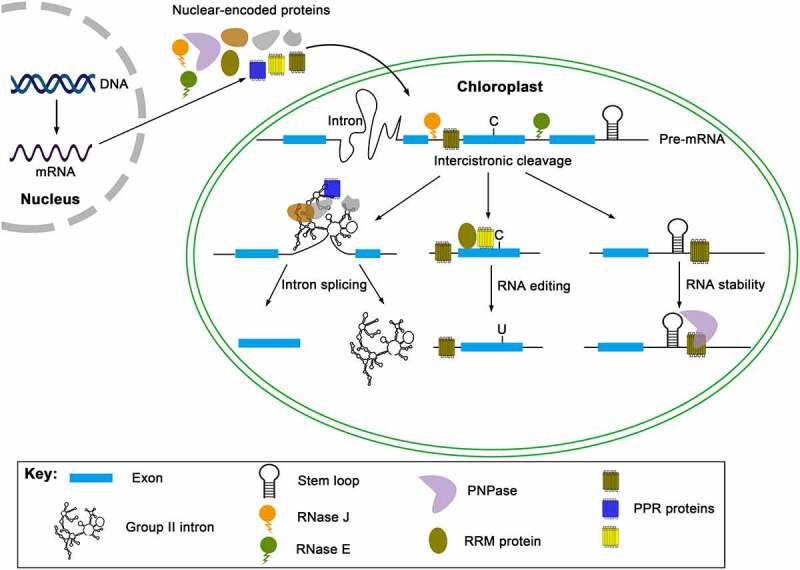
Post-transcriptional control of chloroplast gene expression in land plant chloroplasts. The post-transcriptional processing of chloroplast RNAs includes RNA cleavage of precursor mRNA (pre-mRNA), intron splicing, RNA editing, and RNA stability. Various nuclear-encoded proteins are implicated in these processes. RNase J and RNase E may act as endonucleases to cleave intercistronically. One or more sequence-specific RNA-binding proteins bind to intron RNA, and other splicing factors are recruited to intron RNAs, and the binding of these splicing factors assists the intron folding and splicing. The C-to-U editing is catalysed by PPR and RIP/MORF proteins, and a specific PPR protein targets an editing site by the cis sequence upstream of the editing site. The binding of RNA binding proteins can also protect RNA from exonucleolytic cleavage.
The chloroplast genome underwent intron gain during the evolution from cyanobacteria to land plants [4,5]. Based on the distinct structures and splicing mechanisms of chloroplast introns, these introns were categorized into group I and group II introns [6–8]. The presence of introns in some chloroplast genes that are essential for photosynthesis and chloroplast gene expression makes efficient RNA splicing necessary.
Group II introns, as mobile genetic elements, contain a catalytic RNA and an intron-encoded protein (IEP), the intron RNA catalyses RNA splicing reactions, which are similar to those of the spliceosomal introns, on the other hand, the role of IEP with reverse transcriptase activity is to assist RNA splicing by stabilizing the catalytically active RNA structure (known as maturase activity) and function in intron mobility [9–11]. However, group II introns in plant chloroplasts underwent degenerations in RNA structures and loss or degenerations of IEPs over evolution, thus they lost the ability to mobile and promote intron splicing [10]. So far, only one chloroplast group II intron within the trnK gene encodes a maturase, denoted MatK [12,13]. Certain group I introns are mobile genetic elements since they encode homing endonucleases (HEs). These HEs are site-specific DNA endonucleases that function in intron mobility and in some cases function in RNA splicing [14]. Only one group I intron in trnL gene is present in land plant chloroplasts; however, it fails to self-splice [15]. Therefore, the splicing of group I and group II introns in land plant chloroplasts relies on nuclear-encoded splicing factors. This review will discuss recent advances on the splicing of chloroplast introns in land plants, with emphasis on the splicing factors and their possible roles in RNA splicing.
Chloroplast introns
The presence of introns in chloroplast genes is a feature of the chloroplast genome. [6,8,12,16]. There are 17 to 20 group II introns and only one group I intron in trnL (UAA) in land plant chloroplast genomes. The basic set of these 20 introns (clpP-2 excluded) is also shared among bryophytes, indicating that these chloroplast introns were acquired before the emergence of land plants [17–20].
Group I introns
The structure of group I introns is relatively small and uniform. Each group I intron is folded into a secondary structure with 10 paired domains, P1 to P10, which then fold to form a tertiary structure with three domains [21]. Each domain functions specifically in RNA folding. Two helical domains constitute the central catalytic core of group I introns, which are stabilized via peripheral domains [8,22]. The splicing of group I introns is accomplished by a two-step trans-esterification reaction, first at 5’ and then at the 3’ splice sites. Firstly, exogenous guanosine attacks the 5’ splice site and attaches to the 5’ end of the intron, releasing the 5’ exon. Secondly, 3’-OH of the released exon attacks the 3’ splice site ligating the exons and releasing the intron. The prerequisite of successful splicing is the correct folding of the introns [1,8,16].
Group I introns are mobile genetic elements. This means that they can insert into other intronless genes, thus resulting in their spread [8]. The mobility of group I introns is accomplished by intron homing and reverse splicing [1]. Reverse splicing is the process by which an intron is integrated into other RNA molecules. Till now, no direct evidence is available on the spread of group I introns via reverse splicing. However, certain experimental and comparative data support this theory [8].
Intron homing refers to a process through which an intron spreads into the homologous position in an intronless allele. This process is catalysed by homing endonucleases (HEs), which are encoded by homing endonuclease genes (HEGs) within introns [8,23]. The HEG will lose its function of promoting intron mobility when the intron is immobilized in a population, and the HEG will be lost over time. Thus, HEG is absent in most of the group I introns [8,23]. Based on conserved amino acid motifs, HEs can be categorized into four families: LAGLIDADG, HNH, GIY-YIG, and His-Cys box [21,23,24]. Numerous HEs also possess maturase activity, which is postulated to be acquired to compensate for the reduced splicing efficiency caused by the presence of HEG [25]. As autocatalytic ribozymes, most group I introns have retained self-splicing ability. For instance, several group I introns in C. reinhardtii chloroplasts and other algae have been detected to have the ability of self-splicing in vitro [15,26]. Nevertheless, the only group I intron within the trnL gene in land plant chloroplasts has lost this ability, thus its splicing requires the assistance of additional proteins.
Group II introns
Group II introns are primarily present in eukaryotic organellar and bacterial genomes and rarely in eukaryotic nuclear genomes [9]. Group II introns in organelles may have degenerations in IEPs and RNA structures over time, such as mispairs in domains V and VI, or large insertions or deletions in regions that are vital for catalysis of splicing, which result in the introns’ inability to self-splice and loss of mobility [10,12,27–29]. Although group II introns do not contain a conserved primary nucleotide sequence, their secondary structure is conserved.
Each group II intron consists of 6 paired domains, domains I to VI, radiating from a central core [10,11,30,31]. Domain I is the largest domain of group II intron, it plays a role in recognizing and positioning the exon for catalysis due to the presence of two exon binding sites (EBS1 and EBS2). These EBS can form 5 ~ 6 base pairs with corresponding intron binding sites 1 and 2 (IBS1 and IBS2) located at the 3’ end of the upstream exon [9]. Domain II can recruit domain III into the core of intron RNA and contribute to RNA folding and enhance catalytic activity of intron RNA. The domain IV loop encodes IEP, which functions as a maturase that can bind to unspliced introns and change its conformation to facilitate splicing of the intron. Domain V is small and highly conserved in structure, and it contains a 34 nt long sequence that is part of the catalytic core, and thus the domain V also participates in ribozyme activity. Domain VI contains the branch point adenosine required for the first trans-esterification reaction [9–11,32]. Although group II introns possess conserved secondary structures, based on their structural features and mechanisms of exon recognition, they are subdivided into three subclasses: IIA, IIB, and IIC. IIC introns are found only in bacteria, and IIA and IIB only in plant organelles [11,31].
The splicing of group II introns is also accomplished by a two-step trans-esterification reaction, which is different from group I introns. Firstly, instead of an exogenous guanosine, the 2’-OH of a bulged adenosine within domain VI attacks the 5’ splice site and attaches to the 5’ end of the intron. This results in the release of the upstream exon and the formation of lariat intermediate. Secondly, the 3’-OH of the released upstream exon attacks the 3’ splice junction, yielding ligated exons and an excised intron in a lariat form [9,11,12,21].
Previous studies on the structure and splicing mechanism of each intron type have revealed that group II introns are closely related to spliceosomal introns [11,33–35]. The major similarities between the two intron types are their similar splicing mechanisms, both of which undergo a two-step transesterification reaction and form a lariat intermediate, and similar structures and functions of their active sites in the catalysis of splicing [31]. Besides, the similarities between group II intron reverse transcriptases (RTs) and the spliceosomal protein pre-mRNA processing protein 8 (Prp8) also support the notion that the spliceosome was derived from group II introns [36–38]. Prp8, located at the centre of the spliceosome, is an integral part of the U5 snRNP [38]. Due to the sequence and structure similarities between one of the Prp8 domains and a bacterial group II intron RT, Prp8 was considered to have evolved from the reverse transcriptase [37,38]. The crystal structure of Prp8 revealed that several domains, including the RT domain of Prp8, form a large cavity, which can accommodate the catalytic centre of group II intron RNA [37,38]. Like many group II introns, which do not have the endonuclease domain, Prp8 also lacks an active endonuclease domain [38]. The telomerase and group II introns perform telomere addition and intron mobility, respectively, via a target-primed reverse transcription (TPRT) mechanism. Thus, telomerase reverse transcriptase (TERT) also favours spliceosome origin from group II introns [39]. Thus, it is also believed that eukaryotic spliceosomes could have evolved from group II introns [11,31].
Splicing factors in land plant chloroplasts
As described above, the efficient splicing of degenerate group I and group II introns in land plants requires additional proteins. About twenty group II introns and one group I intron present in land plant chloroplasts have lost self-splicing ability and thus require additional splicing factors. These splicing factors can be categorized into two classes, maturase, and nuclear-encoded proteins. Their roles are summarized in Table S1. Maturases are encoded within the introns, and they have dual functions both in intron mobility and intron splicing. Nuclear-encoded splicing factors are recruited more recently from the nuclear genome.
Maturases
Maturases are intron-encoded proteins that are implicated in the splicing of their host intron [9,40,41]. Maturases aid intron splicing by helping folding the RNA into a catalytically active conformation or stabilizing the active structure [9,41]. Intron-encoded proteins (IEPs) are characterized into four domains: a) RT domain involved in intron mobility, b) maturase (also called X) domain involved in RNA binding and intron splicing, c) DNA binding domain (DBD), and d) endonuclease domain involved in retrotransposition [28,38]. IEPs are dual-function proteins that are involved in intron mobility and RNA splicing, and their role in splicing is known as maturase activity [11]. As per the previous hypothesis, IEPs were involved in intron mobility, and during evolution, they acquired maturase activity to reduce the adverse consequences of endonuclease gene invasion on intron self-splicing [25,42]. Based on previous studies on bifunctional proteins that act on both DNA and RNA, for instance, the study on E. coli maturase I-AniI, it seems likely that the maturase function was derived from its novel binding potential to RNA template [10,25].
Most group I intron-encoded maturases belong to the LAGLIDADG class of homing endonucleases, and some of these proteins lack DNA endonuclease activity and retain only the maturase activity [43]. trnL, the only group I intron in land plant chloroplasts, does not encode a maturase. There is only one group II intron-encoded maturase in land plant chloroplasts, denoted MatK, which is encoded by group II intron within trnK [12]. MatK contains a degenerate RT domain and lacks the endonuclease domain, but have a X domain, which shows that it has retained an essential splicing role [9]. An RNA immunoprecipitation assay in tobacco showed that MatK could capture seven chloroplast group II introns including the trnK intron [13]. In addition, the absence of MatK in barley mutant deficient in chloroplast ribosomes is related to the failure in splicing of trnK and other six introns [44]. These findings indicate that MatK is required for the splicing of the trnK intron, and the other six introns [13,44].
Four nuclear-encoded maturases (nMATs) have been found in Arabidopsis [45], three (nMAT1-3) of which are located in mitochondria and one (nMAT4) is dual localized in chloroplasts and mitochondria [46]. These nuclear-encoded mMATs probably derived from group II introns [9]. Similar to MatK, these nMATs all contain a conserved X domain. nMAT1 and nMAT2 have degenerate RT domains and lack the DBD and endonuclease domains. Interestingly, nMAT3 and nMAT4 contain all domains but have lost the endonuclease activity due to mutations [28]. In Arabidopsis, previous studies have shown that nMAT1, 2, 4 all participate in the splicing of several mitochondrion group II introns [46–49]. In maize, nMAT3 is required for the splicing of several mitochondrion group II introns [50]. Therefore, no nuclear-encoded maturases have been found to participate in intron splicing in land plant chloroplasts.
Nuclear-encoded splicing factors
Dozens of nuclear-encoded proteins from different protein families have been identified to participate in the splicing of chloroplast introns (see Fig. 2). Most of these splicing factors are RNA-binding proteins, some of which have long been reported to have RNA-binding domains, such as pentatricopeptide repeat (PPR) proteins, while others were first identified as RNA-binding proteins only by discovering their involvement in RNA splicing processes, such as proteins harbouring CRM, RNase III, PORR, Whirly, or DUF794 domains [51–55].
Figure 2.
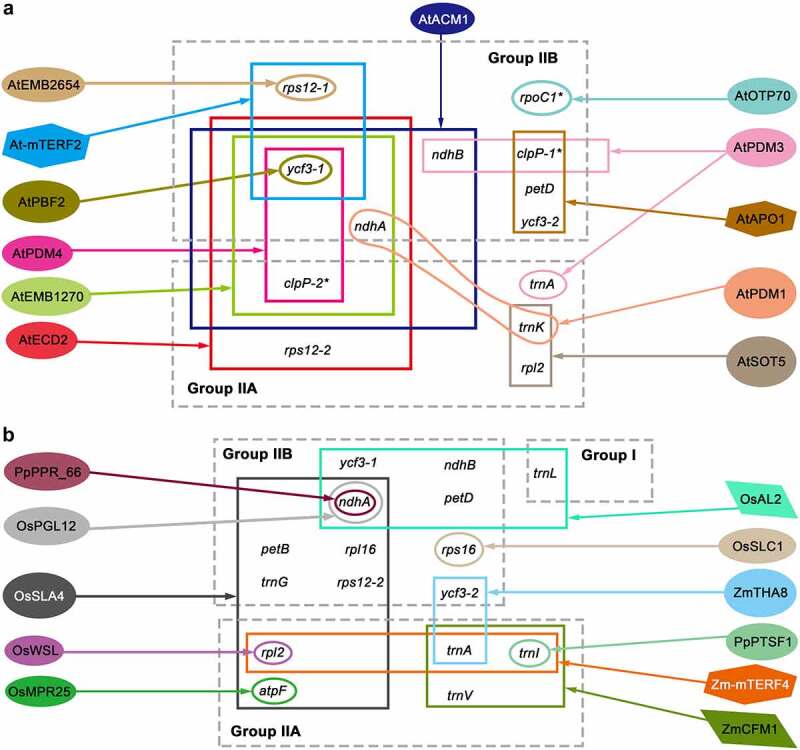
Nuclear-encoded proteins that promote chloroplast intron splicing in Arabidopsis (a) and other land plants (b). Most of the splicing factors that have been diagrammed in previous reviews (see de Longevialle et al., 2010; Stern et al., 2010; Khrouchtchova et al., 2012; Germain et al., 2013) are not shown in this figure. The chloroplast introns are classified into group I, IIA, and IIB. The oval represents PPR proteins, the quadrilateral represents CRM domain proteins, and the hexagon represents other domain proteins. The asterisk refers to introns present in Arabidopsis but absent in rice and maize.
PPR domain family
PPR protein family is a major group of proteins involved in organellar RNA splicing. PPR proteins are typically defined as tandem arrays of the approximately 35-amino-acid long repeat (PPR motif), and each member of the PPR protein family consists of 2 to 30 PPR repeats [56]. The PPR protein specifically binds to its RNA target based on the one-repeat:one-nucleotide recognition mechanism [57–60]. PPR proteins are widely distributed in chloroplasts, mitochondria or both of eukaryotes, especially in land plants and moss, but less in other eukaryotes, such as algae and animals [61].
PPR proteins are divided into P and PLS subfamilies based on the characteristics of their PPR repeats. The P-class PPR proteins consist of tandem arrays of only P repeats, while PLS-class proteins contain triplets of P, L (Long, generally 36 amino acids), and S (Short, generally 31 amino acids) repeats [56,62,63]. Some P-class proteins also have extra domains other than P repeats at their C-terminal, such as the small MutS-related (SMR) domain [64]. PLS-class PPR proteins are further divided into four subgroups (denoted PLS, E, E+, and DYW) based on their C-terminal domains [62,65]. These domains are closely related to RNA editing in organelles [66]. On the other hand, P-class proteins are required for diverse RNA maturation processes, including RNA stability, intergenic RNA cleavage, and RNA splicing [62].
Till now, around twenty PPR proteins have been reported to participate in RNA splicing in land plant chloroplasts, and most of these PPR proteins belong to the P subfamily. Moreover, most of these PPR proteins splice one or two specific intron targets. For instance, Arabidopsis Organelle Transcript Processing 51 (OTP51), containing seven PPR motifs and two LAGLIDADG motifs, is primarily required for the splicing of ycf3 intron 2 [67]. Zm-OTP51 was shown to bind the first 197 nt of this intron with high affinity [68]. Maize THYLAKOID ASSEMBLY 8 (THA8) is a PPR protein with only four PPR motifs and no other domains [68]. RIP-chip assay and slot-blot hybridizations demonstrated that THA8 is responsible for the splicing of maize ycf3 intron 2 and trnA intron, a conserved function in Arabidopsis. Weak binding of recombinant THA8 to the first 197 nt of ycf3 intron 2 was detected via electrophoretic mobility shift assays (EMSAs). The results of this assay were in line with the short PPR repeats of THA8 [68]. Bioinformatics and structural analyses identified that Brachypodium distachyon THA8 dimer binds to two RNA fragments in ycf3 intron 2, moreover, it is the binding of RNA that induces the dimerization of THA8 [69]. PpPPR_66, a Physcomitrella patens P-class PPR protein, is implicated in the ndhA intron splicing, it has been demonstrated to bind to the 115-nt region extending from part of exon 1 to the ndhA intron. In addition, the splicing of ndhA pre-mRNA was also defected in Arabidopsis [70]. Notably, the recognition sequence of PpPPR_66 predicted by the PPR code is not within ndhA intron, suggesting that PpPPR_66 might recognize the RNA structure [70]. In the in vitro analysis, OTP51, THA8, and PpPPR_66 were found to bind to RNA; however, their precise binding sites should be determined via further analyses, such as RNA footprint analysis and EMSAs. Fortunately, four P-class PPR proteins from different species have defined their precise binding site in introns. For instance, in Arabidopsis, the qRT-PCR analysis revealed that EMB2654 is involved in rps12 intron splicing. Furthermore, RNA footprint analysis and EMSAs revealed that EMB2654 binds directly to the 3’ end of rps12 intron 1a [71,72]. Maize PPR4, which contains 16 PPR motifs and an RNA recognition motif (RRM), has been demonstrated to be involved in the trans-splicing of rps12 intron 1 via a series of biochemical assays [73]. Further studies have shown that PPR4 plays the same role in Arabidopsis and rice [72]. ppr4 mutants in Arabidopsis and rice are embryo-lethal and seedling-lethal, respectively, suggesting that PPR4 is indispensable for the development and growth of both dicot and monocot plants. RNA coimmunoprecipitation (RIP) assay combined with RNA footprint analysis and EMSAs revealed that PPR4 could bind directly to the 5’ end of rps12 intron 1b [72]. The binding of PPR4 and EMB2654 to rps12 introns 1b and 1a, respectively, may have altered the structure of domain III within the intron, causing it to fold into a structure that facilitates effective trans-splicing of rps12 intron 1 [72]. In moss Physcomitrella patens, a P-class PPR protein PpPPR_4, also named plastid tRNA splicing factor 1 (PpPTSF1), was shown to be responsible for the splicing of pre-tRNAIle. EMSA revealed that recombinant PpPPR_4 binds to a 25-nt region in domain III of the tRNAIle intron [74]. qRT-PCR and RT-PCR analyses revealed that Arabidopsis PPR protein PHOTOSYSTEM I BIOGENESIS FACTOR2 (PBF2) is responsible for the splicing of ycf3 intron 1. Prediction of recognition sequence, bioinformatics analysis as well as EMSA showed that PBF2 bound to a specific sequence within intron 1 of ycf3 [75]. Unlike those PPR proteins characterized above, which have been shown to bind intron RNA in vitro, some P-class proteins reported to participate in RNA splicing have not. For example, Arabidopsis SUPPRESSOR OF THF1 5 (SOT5) is a PPR protein with 11 PPR repeats, prediction of binding target combined with qRT-PCR and RT-PCR assays have identified that SOT5 is implicated in the splicing of chloroplast rpl2 and trnK introns [76]. SOT5 is predicted to bind to a specific sequence in rpl2 and trnK introns, but it has not been demonstrated to bind intron RNA in vitro as the expression of recombinant SOT5 is not available. In rice, OsPPR6 is involved in splicing ycf3 introns, but which one is not determined. Besides, it also participates in editing of the ndhB transcript [77]. The Oryza sativa SEEDLING-LETHAL CHLOROSIS 1 (OsSLC1) contains 12 PPR motifs, RT-PCR and qRT-PCR data demonstrated that it mainly participate in the intron splicing of rps16 in rice [78]. However, more biochemical evidence is required to support this hypothesis; however, it is challenging to isolate functional PPR protein in vitro.
Certain P-class PPR proteins affect multiple intron targets, unlike the above-mentioned P-class PPR proteins that affect one or two introns. Among them, a recently reported Arabidopsis P-class PPR protein, EMB1270, cooperate with CFM2 to promote the splicing of ycf3 intron 1, clpP intron 2, and ndhA intron. Moreover, EMSA analysis demonstrated the direct binding of truncated EMB1270 with these three introns containing predicted consensus sequences [79]. Pigment-Defective Mutant3 (PDM3) is a P-class PPR protein harbouring 12 PPR motifs, it might be involved in the intron splicing of ndhB, trnA, and clpP intron 1 in Arabidopsis [80]. Arabidopsis PDM4 is implicated in the splicing of ycf3 intron 1 and clpP intron 2 [81,82], Albino Cotyledon Mutant1 (ACM1) in ndhA intron, ndhB intron, clpP intron 2, and ycf3 intron 1 [83], Early Chloroplast Development 2 (ECD2) in ndhA intron, rps12 intron 2, ycf3 intron 1, and clpP intron 2 [84], rice WHITE STRIPE LEAF 4 (WSL4) in rps12 intron 2, ndhA, atpF, and rpl2 introns [85]. Another rice P-class PPR protein WHITE STRIPE LEAF 5 (WSL5) harbours 15 PPR motifs and an RRM motif at its N-terminal. According to a previous report, it affects the splicing of rpl2 and rps12 intron 2, and also the editing of two editing sites [86]. In maize, EMB-7 L can splice rps12 intron 1, rpl2, atpF, and ndhA [87]. EMB-7 L is predicted to target rps12 intron 1 based on PPR code, that is, the 5th and 35th amino acids of each PPR motif determine its recognition specificity of nucleotides [57–60]. But biochemical data in vivo and in vitro are needed to identify the target intron of EMB-7 L. The splicing function of most of these proteins was validated via RT-PCR and qRT-PCR evidence, more biochemical data are required to identify their specific intron targets. These PPR proteins affecting multiple introns might recognize the RNA structure specific to these introns, or bind to the common sequence of their target introns, or alternatively, one PPR protein just directly targets one specific intron, and other introns it affects are direct targets of other proteins associated with it. It remains to be tested which hypothesis is reliable.
The above-mentioned PPR proteins involved in RNA splicing belong to the P subfamily. In addition, certain PLS-class PPR proteins are also reported to participate in RNA splicing, and most of them target a single intron. Among them, some have lost the ability of RNA editing but gained the ability to splice. For example, the Arabidopsis ORGANELLE TRANSCRIPT PROCESSING 70 (OTP70) with an E domain at its C-terminal is a member of the E-subgroup PPR proteins. E domain has been proved to be indispensable for RNA editing by deletion assays [88,89]. However, the E domain of OTP70 is truncated and has been validated to lack the ability of RNA editing. Nonetheless, OTP70 is identified to be responsible for the splicing of rpoC1 intron, and this function is independent of the E domain [90]. WHITE STRIPE LEAF (WSL) is a rice PPR protein belonging to the PLS subfamily, and notably, it consists of 14 PPR motifs and no other functional domains, such as E, E+ or DYW at its C-terminal. Studies showed that WSL is responsible for the splicing of chloroplast rpl2, and has no effect on RNA editing [91]. Rice PLS-class PPR protein Seedling-Lethal Albino 4 (SLA4) has 15 PPR motifs and an atypical DYW-like motif, it was reported to affect the intron splicing of petB, ndhA, atpF, rpl2, rpl16, trnG, and rps12 intron 2, but have no effect on RNA editing [92]. PALE-GREEN LEAF12 (PGL12) is also a PLS-class PPR protein with no functional domain at its C-terminal, findings showed that it was not implicated in RNA editing, but in the splicing of ndhA transcript in rice [93]. There are also PLS-class PPR proteins play roles in both RNA editing and intron splicing. Arabidopsis Pigment-Deficient Mutant 1/ Seedling Lethal1 (PDM1/SEL1), as a PLS-class PPR protein, lacks the E, E+ or DYW domain at its C-terminus that is essential for RNA editing, but it is still demonstrated to affect RNA editing of accD [94]. In addition, PDM1 was reported to participate in splicing ndhA and trnK introns [95]. Arabidopsis ATPF EDITING FACTOR 1 (AEF1) and its rice orthologue MITOCHONDRIAL PPR25 (MPR25) are PLS-class PPR proteins with an E domain at their C-terminal. They are reported to be implicated in RNA editing, as well as atpF splicing in Arabidopsis and rice [96].
Two other PPR proteins in chloroplasts, HCF152 and PPR5, might be responsible for RNA splicing. Arabidopsis HCF152 is a PPR protein composed of 12 PPR repeats. It participates in the processing of chloroplast psbB-psbT-psbH-petB-petD polycistronic transcripts [97]. Previous studies have shown reduced accumulation of spliced petB intron in hcf152 mutant. However, HCF152 is identified to bind specifically to the untranslated region between psbH and petB. Therefore, HCF152 may participate directly in splicing petB intron, or alternatively, it may participate in stabilizing the splicing products [98]. In maize, PPR5 is responsible for stabilizing the unspliced precursor of trnG-UCC, as it binds to this region in vivo [99]. PPR5 may also participate in splicing trnG-UCC directly in vitro, as its binding site in trnG-UCC intron contains two crucial group II functional elements [100].
CRM domain family
Another protein family involved in intron splicing is the protein family containing the chloroplast RNA splicing and ribosome maturation (CRM) domains. The CRM domain originated from an ancient ribosome-associated protein, it exists as a stand-alone protein in Archaea and Bacteria, and the CRM proteins have developed multiple CRM domains in plants [51]. The previous study in maize has shown that the CRM domain can bind RNA in vitro, and a conserved ‘GxxG’ motif contributes to its RNA binding activity [51]. So far, all the characterized CRM proteins are required for RNA splicing in land plant chloroplasts or mitochondria.
There have been 10 CRM domain proteins identified to participate in RNA splicing in maize, Arabidopsis, and rice: Chloroplast RNA Splicing 1 (CRS1), CRS2-associated factors 1 (CAF1), CAF2, CRM Family Member 2 (CFM2), CFM3, CFM1, and their orthologs. Most CRM proteins are initially studied in maize and Arabidopsis, and then characterized in rice. Their functions in maize and Arabidopsis are almost conserved, but distinct in rice. Maize CRS1 was the first characterized CRM protein, it contains three CRM domains [101,102]. CRS1 is involved in the splicing of atpF intron (group IIA) [102]. Futher studies revealed that CRS1 binds to two non-conserved intron segments in domains I and IV of atpF intron in vitro, this further demonstrated that CRS1 is involved solely in the splicing of atpF intron [103]. Unlike the splicing function of orthologous CRS1 in maize, rice protein OsAL2 is not only involved in the splicing of four group II introns, including ndhA, petD, ndhB, and ycf3 intron 1, but also the group I intron, trnL [104]. CRS2 is closely related to peptidyl-tRNA hydrolases (PTHs), but has lost the PTH activity due to the substitution of several amino acids [105]. CRS2 is responsible for the splicing of several group II introns, and generally, introns requiring CRS2 to assist splicing also require the involvement of CAF1 and CAF2 [106]. CAF1 and CAF2 are identified through yeast two-hybrid screen using CRS2 as bait. The two proteins can interact with the CRS2 protein respectively, forming two complexes CRS2-CAF1 and CRS2-CAF2. In maize, the CRS2-CAF1 complex is involved in the intron splicing of petD, trnG, rps16, rpl16, ndhA, and ycf3 intron 1. Besides these introns, Arabidopsis CAF1 is also involved in the intron splicing of rpoC1 and clpP intron 1, two introns absent in maize [106,107]. The CRS2-CAF2 complex is involved in the splicing of a subset of group IIB introns, including petB, ndhB, ndhA, rps12 intron 1 and ycf3 intron 1 [106]. CRM Family Member 2 (CFM2) is another CRM protein with four CRM domains. RIP-chip assay combined with coimmunoprecipitation and cosedimentation assays in maize together showed that CFM2 and CAF1 and/or CAF2 are present in large ribonucleoprotein particle (RNP) complexes that contain trnL intron, ndhA intron and ycf3 intron 1. In addition, the Arabidopsis AtCFM2 also influences the splicing of clpP intron 2, which is absent in maize [108]. Unlike its orthologs in Arabidopsis and maize, rice proteins OsCAF1 and OsCAF2 are implicated in the splicing of group IIA as well as IIB introns. Similarly, both OsCAF1 and OsCAF2 interact with OsCRS2 to form a complex, respectively [109,110]. OsCFM2 in rice participates in the splicing of one group I intron, trnL, and five group II introns, including rps12, rpl2, ndhA, atpF, and ycf3 intron 1 [111]. CFM3, harbouring three CRM domains, is implicated in splicing introns of rps16, rpl16, ndhB, petB, petD, and trnG in Arabidopsis, and rps16, rpl16, petD, and ndhB in rice [112]. Thus, the introns that require CRS2/CAF complexes for their splicing also require CFM2 or CFM3, but not both. CFM1 is a recently characterized CRM domain protein in Setaria viridis and maize [113]. RIP-chip data demonstrated that Zm-CFM1 promotes the intron splicing of trnA, trnI and trnV, and these introns have not been shown to require any previous reported CRM domain proteins. The function of CFM1 is conserved in Setaria viridis and maize, as the splicing defects of these introns were also identified in Sv-cfm1 [113]. In conclusion, the CRM proteins function in splicing of almost all the chloroplast introns, and each subset of introns spliced by each CRM protein is overlapping but distinct.
Other domain proteins
In addition to the PPR and CRM protein families, some proteins containing other domains also are reported to participate in chloroplast intron splicing. RNC1 was identified in CAF1 and CAF2 coimmunoprecipitates through mass spectrometry analysis [54]. RNC1 has two ribonuclease III (RNase III) domains, but lacks endonuclease activity due to the loss of several essential amino acids. It has been identified that RNC1 promotes the splicing of a subset of group IIA introns, including rps12 intron 2, atpF, trnK, trnV, trnI, trnA introns, and group IIB introns, including petB, petD, ndhB, and trnG introns [54]. Moreover, recombinant RNC1 can bind RNA with high affinity in vitro, indicating that RNC1 is an RNA binding protein, and it may promote intron splicing via binding directly to RNA [54]. What’s This Factor? (WTF1) is another splicing factor identified from CAF1 and CAF2 coimmunoprecipitates through mass spectrometry analysis [52]. It has been shown that WTF1 and RNC1 form a heterodimer to facilitate splicing most of chloroplast group II introns. WTF1 has a plant-specific domain of unknown function 860 (DUF860), also known as Plant Organelle RNA Recognition (PORR). Both WTF1 and DUF860 can bind RNA in vitro, suggesting that DUF860 is a newly identified RNA-binding domain [52]. Thus, DUF860 expands the repertoire of RNA-binding domains specific to plants. Maize ZmWHY1 is a member of the Whirly protein family, which is plant-specific. ZmWHY1 associates with CRS1 to facilitate the splicing of atpF intron. EMSAs demonstrated that ZmWHY1 can bind single-stranded DNA and RNA [53]. ACCUMULATION OF PHOTOSYSTEM ONE1 (APO1) was initially thought to promote the assembly of [4Fe-4S] cluster-containing chloroplast complexes, such as the photosystem I (PSI) and NADH dehydrogenase complexes [114]. In maize, APO1 coimmunoprecipitated with CAF1, which led to the speculation of the involvement of APO1 in intron splicing. Subsequent studies have shown that APO1 promotes the splicing of ycf3 intron 2, petD intron, and clpP intron 1 [55]. APO1 contains an unknown functional domain DUF794, which is a plant-specific domain containing two motifs that resemble zinc fingers. EMSAs showed that maize and Arabidopsis recombinant APO1 binds RNA in ycf3 intron 2 with high affinity [55]. Thus, DUF794 was termed as an RNA-binding domain. RH3 in maize and Arabidopsis is a member of the DEAD box RNA helicase family. It is responsible for the intron splicing of trnA, trnI, rpl2, and rps12, and may also contribute to 50S ribosome biogenesis [115]. In addition, one later study demonstrated that AtRH3 also participates in splicing trnL, trnK, and atpF introns [116]. The mitochondrial transcription termination factor (mTERF) protein family in metazoan mainly affects mitochondrial transcription, ribosome biogenesis and DNA replication [117]. Maize Zm-mTERF4 is a member of this family, and it was identified to participate in the intron splicing of trnI-GAU, trnA-UGC and rpl2 [118]. Another mTERF protein in Arabidopsis, mTERF2, was shown to participate in the splicing of ycf3 intron 1 and rps12 intron 1 through RNA-seq and RNA gel-blot analyses [119]. These findings extend the functional repertoire of mTERF family in plants. Taken together, characterizations of these domains derived from diverse protein families extend the repertoire of RNA-binding domains. Some of these protein factors have lost their ancestral activities, and they are recruited to participate in RNA splicing, such as RNC1. While some splicing factors might also have DNA binding activity, such as WHY1. These proteins were demonstrated to be associated with RNAs, but not specifically with a certain intron. Thus, maybe these proteins are recruited to participate in splicing via protein–protein interactions and they function in intron splicing together with other splicing factors.
The roles of splicing factors in RNA splicing
Although lots of RNA-binding proteins have been identified to participate in chloroplast intron splicing in land plants through reverse genetic screen or coimmunoprecipitation assay combined with mass spectrometry analysis, the specific roles of these splicing factors during RNA splicing remain ambiguous. The possible roles of splicing factors are speculated based on existing evidence. The common feature of these splicing factors including matuarases is that most of them are bifunctional moonlighting proteins, since they participate in other cellular functions besides serving as splicing factors. For instance, bacterial peptidyl-tRNA hydrolase-related protein CRS2 moonlights as a splicing factor. WHY1, which belongs to a family that has been described as DNA-binding proteins also moonlights as a splicing factor. Similarly, RNC1 with two RNase III domains also was recruited to function in splicing. Maturases have dual functions both on DNA and RNA. In addition, many factors have been identified based on genetic screens or limited biochemical assays, and they affect multiple introns, but their exact role is not clear, these proteins may have other important functions, and maybe they just moonlight as a splicing factor, or maybe they are recruited via protein–protein interactions and function in splicing together with other splicing factors [9].
Given previous findings, each organellar intron requires more than one splicing factor, and these splicing factors are found in one or more intron-containing RNP complexes. Cosedimentation via a sucrose gradient assay showed that THA8 is present in large RNP complexes containing trnA intron and ycf3 intron 2. In addition, the communioprecipitation assay showed that THA8 associates with WTF1 and RNC1 [68]. Therefore, it is likely that THA8 associates with WTF1/RNC1 heterodimer to advance the splicing of their shared intron target trnA. Meanwhile, THA8 may also cooperate with OTP51 and APO1 to facilitate the splicing of their shared intron target, ycf3 intron 2. Sucrose gradients and coimmunoprecipitation assays showed that CFM2 is associated with CAF1, CAF2, ndhA intron, ycf3 intron 1, and trnL intron in large chloroplast stromal RNP complexes. The intron splicing of ndhA requires CFM2 and CRS2/CAF2 complex, and the splicing of ycf3 intron 1 requires CFM2, CRS2/CAF1, and CRS2/CAF2 complexes [108]. Cosedimentation and coimmunoprecipitation data validated that CFM3 coimmunoprecipitated with CAF1, CAF2, and RNC1, and these splicing factors associated simultaneously with their shared intron targets in large RNPs [112]. The splicing of atpF intron requires CRS1, ZmWHY1, WTF1 and RNC1, and CRS1 interacts with ZmWHY1 in an RNA-dependent manner [52–54,102]. WTF1 coimmunoprecipitated with CRS1, CAF1, CAF2 and RNC1, and they are found in stromal RNP complexes of ~ 600 to 700 kD including a set of group II introns [52]. The trans-splicing of rps12 intron 1 requires PPR4, EMB2654, and CRS2/CAF2 complex [71,72]. Thus, there are diverse chloroplast group II intron RNP complexes containing several splicing factors and a set of introns to advance the splicing of distinct group II introns, and the roles of these splicing factors are considered to assist the intron folding into its catalytically active structure favourable for splicing by stabilizing its inherently native structure, preventing the formation of nonnative structures, or destabilizing nonnative structures [54].
Nonetheless, the precise function of these splicing factors in intron folding or sequence recognition in the process of splicing remains unclear. This is due to the lack of identification of precise and direct binding sites of most splicing factors. Identifying specific binding sites of some splicing factors could improve our current understanding of their roles in RNA splicing. CRS1 was the first protein explored to reveal its direct binding site. In vitro biochemical analyses have shown that CRS1 facilitates the correct folding of atpF intron through binding with its two regions in domains I and IV [103]. Later, PPR proteins OTP51, THA8, PTSF1, PpPPR_66, EMB2654, PPR4, and PBF2 were also shown to bind RNA directly in vitro. For instance, OTP51 and THA8 were shown to bind the first 197 nt of ycf3 intron 2 with high and weak affinity, respectively [68]. PTSF could bind to a 25-nt region within the domain III of tRNAIle intron, forming a nanoloop [74]. Domain III functions as a catalytic effector, thus, the function of PTSF might promote the interaction of domain III with other domains PpPPR_66 binds preferentially to a 115-nt region at the 5’ half of the domain I of the ndhA intron, and this region encompasses from part of exon 1 to the intron, which can form a long stem-loop structure [70]. EMB2654 and PPR4 were demonstrated to bind to rps12 intron 1a and 1b, respectively, which is speculated to help the folding of domain III of rps12 intron 1 [72]. PBF2 was shown to bind a specific sequence within domain II of ycf3 intron 1 [75]. Although almost all the splicing factors are considered RNA-binding proteins, only PPR proteins were validated to bind RNA directly in vitro using RNA footprinting analysis and EMSAs. Moreover, most PPR proteins are specific to only one target intron, which was determined using the structure and PPR codes of PPR motifs, and the CRM domain does not possess this specificity for RNA. Since each intron generally requires several splicing factors, we speculate that splicing factors in intron splicing are involved in the binding of PPR protein to its target intron with sequence specificity. Thus, they assist the intron in folding into a structure necessary for recruiting relevant general splicing factors, such as CAF1 and CAF2. Also, some general splicing factors are recruited by the PPR protein via direct protein–protein interactions. These splicing factors as well as their shared introns form RNP complexes to promote the intron splicing. Nevertheless, further studies are needed to test this hypothesis in the future.
Conclusions and prospects
RNA splicing is a complex process involving multiple splicing factors from diverse protein families. Some of these splicing factors are inherently considered to be RNA-binding proteins, such as PPR proteins, and others (CRM, PORR, RNase III, APO protein families, and so on) are first identified as RNA-binding proteins by identifying their functions in chloroplast RNA splicing. Diverse splicing factors are involved in the splicing of distinct but overlapping intron subsets. However, most PPR proteins are implicated in splicing single introns as they bind RNA in a sequence-specific manner. Several splicing factors, which share at least one intron target, cooperate to form splicing complexes, the RNP complexes, which assist intron folding and promote intron splicing. However, only a few PPR proteins have identified the exact binding sequence within intron domains. Thus, the precise role of each splicing factor within RNP complexes in facilitating splicing is still unclear. Crystal structure analysis and prediction of recognition sequences by PPR code and bioinformatics analysis will be needed to obtain precise binding sequences of more splicing factors. Furthermore, in vivo and in vitro biochemical data are also required to determine whether splicing factors with only RT-PCR and qRT-PCR evidence are directly involved in intron splicing. Thus, it seems likely that PPR proteins specifically binding to intron target are more likely involved in splicing directly, and proteins affecting multiple introns are more likely working with other proteins and having an indirect involvement in splicing. In the future, more potential splicing factors can be screened by reverse genetic screening of mutants or mass spectrometry analysis after coimmunoprecipitation with known splicing factors. All of these investigations can contribute to the clarification of the splicing mechanism of splicing factors.
Supplementary Material
Acknowledgments
This research was supported by Shandong Province Key Research and Development Program (2019GSF107079), the Development Plan for Youth Innovation Team of Shandong Provincial (2019KJE012), the Science and Technology Demonstration Project of “Bohai Granary” of Shandong Province (2019BHLC002).
Funding Statement
This work was supported by the Shandong Province Key Research and Development Program [2019GSF107079]; the Science and Technology Demonstration Project of “Bohai Granary” of Shandong Province [2019BHLC002]; the Development Plan for Youth Innovation Team of Shandong Provincial [2019KJE012; the Science and Technology Demonstration Project of “Bohai Granary” of Shandong Provincethe Science and Technology Demonstration Project of ”Bohai Granary” of Shandong Province [2019BHLC002];
Author contributions
Xuemei Wang, Jingyi Wang and Simin Li wrote the paper. Na Sui and Congming Lu revised the paper. All authors read and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15476286.2022.2096801
References
- [1].Del Campo EM. Post-transcriptional control of chloroplast gene expression. Gene Regul Syst Bio. 2009;3:31–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sugita M, Sugiura M. Regulation of gene expression in chloroplasts of higher plants. Plant Mol Biol. 1996;32(1–2):315–326. [DOI] [PubMed] [Google Scholar]
- [3].Zoschke R, Bock R. Chloroplast translation: structural and functional organization, operational control, and regulation. Plant Cell. 2018;30(4):745–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rogozin IB, Carmel L, Csuros M, et al. Origin and evolution of spliceosomal introns. Biol Direct. 2012;7:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Tillich M, Beick S, Schmitz-Linneweber C. Chloroplast RNA-binding proteins: repair and regulation of chloroplast transcripts. RNA Biol. 2010;7(2):172–178. [DOI] [PubMed] [Google Scholar]
- [6].Fedorova O, Zingler N. Group II introns: structure, folding and splicing mechanism. Biol Chem. 2007;388(7):665–678. [DOI] [PubMed] [Google Scholar]
- [7].Germain A, Hotto AM, Barkan A, et al. RNA processing and decay in plastids. Wiley Interdiscip Rev RNA. 2013;4(3):295–316. [DOI] [PubMed] [Google Scholar]
- [8].Haugen P, Simon DM, Bhattacharya D. The natural history of group I introns. Trends Genet. 2005;21(2):111–119. [DOI] [PubMed] [Google Scholar]
- [9].Lambowitz AM, Zimmerly S. Mobile group II introns. Annu Rev Genet. 2004;38:1–35. [DOI] [PubMed] [Google Scholar]
- [10].Lambowitz AM, Zimmerly S. Group II introns: mobile ribozymes that invade DNA. Cold Spring Harb Perspect Biol. 2011;3(8):a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zimmerly S, Semper C. Evolution of group II introns. Mob DNA. 2015;6(1). DOI: 10.1186/s13100-015-0037-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Stern DB, Goldschmidt-Clermont M, Hanson MR. Chloroplast RNA metabolism. Annu Rev Plant Biol. 2010;61(1):125–155. [DOI] [PubMed] [Google Scholar]
- [13].Zoschke R, Nakamura M, Liere K, et al. An organellar maturase associates with multiple group II introns. Proc Natl Acad Sci U S A. 2010;107(7):3245–3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hausner G, Hafez M, Edgell DR. Bacterial group I introns: mobile RNA catalysts. Mob DNA. 2014;5(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Simon D, Fewer D, Friedl T, et al. Phylogeny and self-splicing ability of the plastid tRNA-Leu group I Intron. J Mol Evol. 2003;57(6):710–720. [DOI] [PubMed] [Google Scholar]
- [16].de Longevialle AF, Small ID, Lurin C. Nuclearly encoded splicing factors implicated in RNA splicing in higher plant organelles. Mol Plant. 2010;3(4):691–705. [DOI] [PubMed] [Google Scholar]
- [17].Kugita M, Kaneko A, Yamamoto Y, et al. The complete nucleotide sequence of the hornwort (Anthoceros formosae) chloroplast genome: insight into the earliest land plants. Nucleic Acids Res. 2003;31(2):716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sugiura C, Kobayashi Y, Aoki S, et al. Complete chloroplast DNA sequence of the moss Physcomitrella patens: evidence for the loss and relocation of rpoA from the chloroplast to the nucleus. Nucleic Acids Res. 2003;31(18):5324–5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Turmel M, Otis C, Lemieux C. The chloroplast and mitochondrial genome sequences of the charophyte Chaetosphaeridium globosum: insights into the timing of the events that restructured organelle DNAs within the green algal lineage that led to land plants. Proc Natl Acad Sci U S A. 2002;99(17):11275–11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wakasugi T, Tsudzuki T, Sugiura M. The genomics of land plant chloroplasts: gene content and alteration of genomic information by RNA editing. Photosynth Res. 2001;70(1):107–118. [DOI] [PubMed] [Google Scholar]
- [21].Jacobs J, Kück U. Function of chloroplast RNA-binding proteins. Cell Mol Life Sci. 2011;68(5):735–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Woodson SA. Structure and assembly of group I introns. Curr Opin Struct Biol. 2005;15(3):324–330. [DOI] [PubMed] [Google Scholar]
- [23].Chevalier BS, Stoddard BL. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 2001;29(18):3757–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Stoddard BL. Homing endonuclease structure and function. Q Rev Biophys. 2005;38(1):49–95. [DOI] [PubMed] [Google Scholar]
- [25].Belfort M. Two for the price of one: a bifunctional intron-encoded DNA endonuclease-RNA maturase. Genes Dev. 2003;17(23):2860–2863. [DOI] [PubMed] [Google Scholar]
- [26].Herrin DL, Nickelsen J. Chloroplast RNA processing and stability. Photosynth Res. 2004;82(3):301–314. [DOI] [PubMed] [Google Scholar]
- [27].Bonen L. Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion. 2008;8(1):26–34. [DOI] [PubMed] [Google Scholar]
- [28].Brown GG, Colas Des Francs-Small C, Ostersetzer-Biran O. Group II intron splicing factors in plant mitochondria. Front Plant Sci. 2014;5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Robart AR, Zimmerly S. Group II intron retroelements: function and diversity. Cytogenet Genome Res. 2005;110(1–4):589–597. [DOI] [PubMed] [Google Scholar]
- [30].Pyle AM, Fedorova O, Waldsich C. Folding of group II introns: a model system for large, multidomain RNAs? Trends Biochem Sci. 2007;32(3):138–145. [DOI] [PubMed] [Google Scholar]
- [31].Zhao C, Pyle AM. Structural insights into the mechanism of group II intron splicing. Trends Biochem Sci. 2017;42(6):470–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Fedorova O, Mitros T, Pyle AM. Domains 2 and 3 interact to form critical elements of the group II intron active site. J Mol Biol. 2003;330(2):197–209. [DOI] [PubMed] [Google Scholar]
- [33].Agrawal RK, Wang HW, Belfort M. Forks in the tracks: group II introns, spliceosomes, telomeres and beyond. RNA Biol. 2016;13(12):1218–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Cech TR. The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell. 1986;44(2):207–210. [DOI] [PubMed] [Google Scholar]
- [35].Zhao C, Pyle AM. Crystal structures of a group II intron maturase reveal a missing link in spliceosome evolution. Nat Struct Mol Biol. 2016;23(6):558–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Dlakic M, Mushegian A. Prp8, the pivotal protein of the spliceosomal catalytic center, evolved from a retroelement-encoded reverse transcriptase. RNA. 2011;17(5):799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Galej WP, Oubridge C, Newman AJ, et al. Crystal structure of Prp8 reveals active site cavity of the spliceosome. Nature. 2013;493(7434):638–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Smathers CM, Robart AR. The mechanism of splicing as told by group II introns: ancestors of the spliceosome. BBA Gene Regul Mech. 2019;1862(11–12):194390. [DOI] [PubMed] [Google Scholar]
- [39].Qu G, Kaushal PS, Wang J, et al. Structure of a group II intron in complex with its reverse transcriptase. Nat Struct Mol Biol. 2016;23(6):549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Moran JV, Mecklenburg KL, Sass P, et al. Splicing defective mutants of the COXI gene of yeast mitochondrial DNA: initial definition of the maturase domain of the group II intron AI2. Nucleic Acids Res. 1994;22(11):2057–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lambowitz AM, and Caprara MG. Group I and group II ribozymes as RNPs: clues to the past and guides to the future. NY: Cold Spring Harbor Laboratory Press.The RNA World. 451–485. 1999. [Google Scholar]
- [42].Kennell JC, Moran JV, Periman PS, et al. Reverse transcriptase activity associated with maturase-encoding group II lntrons in yeast mitochondria. Cell. 1993;73(1):133–146. [DOI] [PubMed] [Google Scholar]
- [43].Schmitz-Linneweber C, and Barkan A. RNA splicing and RNA editing in chloroplasts . In Cell and Molecular Biology of Plastids. Heidelberg, Germany: Springer. 2007. pp. 213–248. [Google Scholar]
- [44].Vogel J, Borner T, Hess WR. Comparative analysis of splicing of the complete set of chloroplast group II introns in three higher plant mutants. Nucleic Acids Res. 1999;27(19):3866–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Mohr G, Lambowitz AM. Putative proteins related to group II intron reverse transcriptase/maturases are encoded by nuclear genes in higher plants. Nucleic Acids Res. 2003;31(2):647–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Keren I, Bezawork-Geleta A, Kolton M, et al. AtnMat2, a nuclear-encoded maturase required for splicing of group-II introns in Arabidopsis mitochondria. RNA. 2009;15(12):2299–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Cohen S, Zmudjak M, Colas Des Francs-Small C, et al. nMAT4, a maturase factor required for nad1 pre-mRNA processing and maturation, is essential for holocomplex I biogenesis in Arabidopsis mitochondria. Plant J. 2014;78(2):253–268. [DOI] [PubMed] [Google Scholar]
- [48].Keren I, Tal L, Des Francs-Small CC, et al. nMAT1, a nuclear-encoded maturase involved in the trans-splicing of nad1 intron 1, is essential for mitochondrial complex I assembly and function. Plant J. 2012;71(3):413–426. [DOI] [PubMed] [Google Scholar]
- [49].Nakagawa N, Sakurai N. A mutation in At-nMat1a, which encodes a nuclear gene having high similarity to group II intron maturase, causes impaired splicing of mitochondrial NAD4 transcript and altered carbon metabolism in Arabidopsis thaliana. Plant Cell Physiol. 2006;47(6):772–783. [DOI] [PubMed] [Google Scholar]
- [50].Chen W, Cui Y, Wang Z, et al. Nuclear-encoded maturase protein 3 is required for the splicing of various group II introns in mitochondria during maize (Zea mays L.) seed development. Plant Cell Physiol. 2021;62(2):293–305. [DOI] [PubMed] [Google Scholar]
- [51].Barkan A, Klipcan L, Ostersetzer O, et al. The CRM domain: an RNA binding module derived from an ancient ribosome-associated protein. RNA. 2007;13(1):55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kroeger TS, Watkins KP, Friso G, et al. A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing. Proc Natl Acad Sci U S A. 2009;106(11):4537–4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Prikryl J, Watkins KP, Friso G, et al. A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res. 2008;36(16):5152–5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Watkins KP, Kroeger TS, Cooke AM, et al. A ribonuclease III domain protein functions in group II intron splicing in maize chloroplasts. Plant Cell. 2007;19(8):2606–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Watkins KP, Rojas M, Friso G, et al. APO1 promotes the splicing of chloroplast group II introns and harbors a plant-specific zinc-dependent RNA binding domain. Plant Cell. 2011;23(3):1082–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Barkan A, Small I. Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol. 2014;65:415–442. [DOI] [PubMed] [Google Scholar]
- [57].Barkan A, Rojas M, Fujii S, et al. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012;8(8):e1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Shen C, Zhang D, Guan Z, et al. Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins. Nat Commun. 2016;7(1):11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Yin P, Li Q, Yan C, et al. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature. 2013;504(7478):168–171. [DOI] [PubMed] [Google Scholar]
- [60].Zhou W, Lu Q, Li Q, et al. PPR-SMR protein SOT1 has RNA endonuclease activity. Proc Natl Acad Sci U S A. 2017;114(8):E1554–E1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 2008;13(12):663–670. [DOI] [PubMed] [Google Scholar]
- [62].Shikanai T, Fujii S. Function of PPR proteins in plastid gene expression. RNA Biol. 2013;10(9):1446–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wang X, An Y, Xu P, et al. Functioning of PPR proteins in organelle RNA metabolism and chloroplast biogenesis. Front Plant Sci. 2021;12:627501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Liu S, Melonek J, Boykin LM, et al. PPR-SMRs: ancient proteins with enigmatic functions. RNA Biol. 2013;10(9):1501–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Lurin C, Andres C, Aubourg S, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16(8):2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Small ID, Schallenberg-Rudinger M, Takenaka M, et al. Plant organellar RNA editing: what 30 years of research has revealed. Plant J. 2020;101(5):1040–1056. [DOI] [PubMed] [Google Scholar]
- [67].de Longevialle AF, Hendrickson L, Taylor NL, et al. The pentatricopeptide repeat gene OTP51 with two LAGLIDADG motifs is required for the cis-splicing of plastid ycf3 intron 2 in Arabidopsis thaliana. Plant J. 2008;56(1):157–168. [DOI] [PubMed] [Google Scholar]
- [68].Khrouchtchova A, Monde RA, Barkan A. A short PPR protein required for the splicing of specific group II introns in angiosperm chloroplasts. RNA. 2012;18(6):1197–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Ke J, Chen RZ, Ban T, et al. Structural basis for RNA recognition by a dimeric PPR-protein complex. Nat Struct Mol Biol. 2013;20(12):1377–1382. [DOI] [PubMed] [Google Scholar]
- [70].Ito A, Sugita C, Ichinose M, et al. An evolutionarily conserved P-subfamily pentatricopeptide repeat protein is required to splice the plastid ndhA transcript in the moss Physcomitrella patens and Arabidopsis thaliana. Plant J. 2018;94(4):638–648. [DOI] [PubMed] [Google Scholar]
- [71].Aryamanesh N, Ruwe H, Sanglard LV, et al. The pentatricopeptide repeat protein EMB2654 is essential for trans-splicing of a chloroplast small ribosomal subunit transcript. Plant Physiol. 2017;173(2):1164–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Lee K, Park SJ, Colas Des Francs-Small C, et al. The coordinated action of PPR4 and EMB2654 on each intron half mediates trans-splicing of rps12 transcripts in plant chloroplasts. Plant J. 2019;100(6):1193–1207. [DOI] [PubMed] [Google Scholar]
- [73].Schmitz-Linneweber C, Williams-Carrier RE, Williams-Voelker PM, et al. A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell. 2006;18(10):2650–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Goto S, Kawaguchi Y, Sugita C, et al. P-class pentatricopeptide repeat protein PTSF1 is required for splicing of the plastid pre-tRNA(I) (le) in Physcomitrella patens. Plant J. 2016;86(6):493–503. [DOI] [PubMed] [Google Scholar]
- [75].Wang X, Yang Z, Zhang Y, et al. Pentatricopeptide repeat protein PHOTOSYSTEM I BIOGENESIS FACTOR2 is required for splicing of ycf3. J Integr Plant Biol. 2020;62(11):1741–1761. [DOI] [PubMed] [Google Scholar]
- [76].Huang W, Zhu Y, Wu W, et al. The pentatricopeptide repeat protein SOT5/EMB2279 is required for plastid rpl2 and trnK intron splicing. Plant Physiol. 2018;177(2):684–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Tang J, Zhang W, Wen K, et al. OsPPR6, a pentatricopeptide repeat protein involved in editing and splicing chloroplast RNA, is required for chloroplast biogenesis in rice. Plant Mol Biol. 2017;95(4–5):345–357. [DOI] [PubMed] [Google Scholar]
- [78].Lv J, Shang L, Chen Y, et al. OsSLC1 encodes a pentatricopeptide repeat protein essential for early chloroplast development and seedling survival. Rice (N Y). 2020;13(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Zhang L, Chen J, Zhang L, et al. The pentatricopeptide repeat protein EMB1270 interacts with CFM2 to splice specific group II introns in Arabidopsis chloroplasts. J Integr Plant Biol. 2021;63(11):1952–1966. [DOI] [PubMed] [Google Scholar]
- [80].Zhang J, Xiao J, Li Y, et al. PDM3, a pentatricopeptide repeat-containing protein, affects chloroplast development. J Exp Bot. 2017;68(20):5615–5627. [DOI] [PubMed] [Google Scholar]
- [81].Chen J, Zhu H, Huang J, et al. A new method for functional analysis of plastid EMBRYO-DEFECTIVE PPR genes by efficiently constructing cosuppression lines in Arabidopsis. Plant Methods. 2020;16(1):154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Wang X, Zhao L, Man Y, et al. PDM4, a pentatricopeptide repeat protein, affects chloroplast gene expression and chloroplast development in Arabidopsis thaliana. Front Plant Sci. 2020;11:1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Wang X, An Y, and Li Y, et al. A PPR protein ACM1 is involved in chloroplast gene expression and early plastid development in arabidopsis. Int J Mol Sci. 2021;22(5): 2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Wang X, An Y, Qi Z, et al. PPR protein early chloroplast development 2 is essential for chloroplast development at the early stage of Arabidopsis development. Plant Sci. 2021;308:110908. [DOI] [PubMed] [Google Scholar]
- [85].Wang Y, Ren Y, Zhou K, et al. WHITE STRIPE LEAF4 encodes a novel P-type PPR protein required for chloroplast biogenesis during early leaf development. Front Plant Sci. 2017;8:1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Liu X, Lan J, Huang Y, et al. WSL5, a pentatripeptide repeat protein, is essential for chloroplast biogenesis in rice under cold stress. J Exp Bot. 2018;69(16):3949–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Yuan N, Wang J, Zhou Y, et al. EMB-7L is required for embryogenesis and plant development in maize involved in RNA splicing of multiple chloroplast genes. Plant Sci. 2019;287:110203. [DOI] [PubMed] [Google Scholar]
- [88].Okuda K, Chateigner-Boutin AL, Nakamura T, et al. Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. Plant Cell. 2009;21(1):146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Okuda K, Myouga F, Motohashi R, et al. Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proc Natl Acad Sci U S A. 2007;104(19):8178–8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Chateigner-Boutin AL, Des Francs-Small CC, Delannoy E, et al. OTP70 is a pentatricopeptide repeat protein of the E subgroup involved in splicing of the plastid transcript rpoC1. Plant J. 2011;65(4):532–542. [DOI] [PubMed] [Google Scholar]
- [91].Tan J, Tan Z, Wu F, et al. A novel chloroplast-localized pentatricopeptide repeat protein involved in splicing affects chloroplast development and abiotic stress response in rice. Mol Plant. 2014;7(8):1329–1349. [DOI] [PubMed] [Google Scholar]
- [92].Z-w W, Lv J, S-z X, et al. OsSLA4 encodes a pentatricopeptide repeat protein essential for early chloroplast development and seedling growth in rice. Plant Growth Regul. 2018;84(2):249–260. [Google Scholar]
- [93].Chen L, Huang L, Dai L, et al. PALE-GREEN LEAF12 encodes a novel pentatricopeptide repeat protein required for chloroplast development and 16S rRNA processing in rice. Plant Cell Physiol. 2019;60(3):587–598. [DOI] [PubMed] [Google Scholar]
- [94].Pyo YJ, Kwon KC, Kim A, et al. Seedling Lethal1, a pentatricopeptide repeat protein lacking an E/E+ or DYW domain in Arabidopsis, is involved in plastid gene expression and early chloroplast development. Plant Physiol. 2013;163(4):1844–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Zhang HD, Cui YL, Huang C, et al. PPR protein PDM1/SEL1 is involved in RNA editing and splicing of plastid genes in Arabidopsis thaliana. Photosynth Res. 2015;126(2–3):311–321. [DOI] [PubMed] [Google Scholar]
- [96].Yap A, Kindgren P, Colas Des Francs-Small C, et al. AEF1/MPR25 is implicated in RNA editing of plastid atpF and mitochondrial nad5, and also promotes atpF splicing in Arabidopsis and rice. Plant J. 2015;81(5):661–669. [DOI] [PubMed] [Google Scholar]
- [97].Meierhoff K, Felder S, Nakamura T, et al. HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell. 2003;15(6):1480–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Nakamura T, Meierhoff K, Westhoff P, et al. RNA-binding properties of HCF152, an Arabidopsis PPR protein involved in the processing of chloroplast RNA. Eur J Biochem. 2003;270(20):4070–4081. [DOI] [PubMed] [Google Scholar]
- [99].Beick S, Schmitz-Linneweber C, Williams-Carrier R, et al. The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol Cell Biol. 2008;28(17):5337–5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Williams-Carrier R, Kroeger T, Barkan A. Sequence-specific binding of a chloroplast pentatricopeptide repeat protein to its native group II intron ligand. RNA. 2008;14(9):1930–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Jenkins BD, Kulhanek DJ, Barkan A. Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell. 1997;9:283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Till B, Schmitz-Linneweber C, Williams-Carrier R, et al. CRS1 is a novel group II intron splicing factor that was derived from a domain of ancient origin. RNA. 2001;7(9):1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Ostersetzer O, Cooke AM, Watkins KP, et al. CRS1, a chloroplast group II intron splicing factor, promotes intron folding through specific interactions with two intron domains. Plant Cell. 2005;17(1):241–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Liu C, Zhu H, Xing Y, et al. Albino Leaf 2 is involved in the splicing of chloroplast group I and II introns in rice. J Exp Bot. 2016;67(18):5339–5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Jenkins BD, Barkan A. Recruitment of a peptidyl-tRNA hydrolase as a facilitator of group II intron splicing in chloroplasts. EMBO J. 2001;20(4):872–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Ostheimer GJ, Williams-Carrier R, Belcher S, et al. Group II intron splicing factors derived by diversification of an ancient RNA-binding domain. EMBO J. 2003;22(15):3919–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Asakura Y, Barkan A. Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns. Plant Physiol. 2006;142(4):1656–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Asakura Y, Barkan A. A CRM domain protein functions dually in group I and group II intron splicing in land plant chloroplasts. Plant Cell. 2007;19(12):3864–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Shen L, Zhang Q, Wang Z, et al. OsCAF2 contains two CRM domains and is necessary for chloroplast development in rice. BMC Plant Biol. 2020;20(1):381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Zhang Q, Shen L, and Wang Z, et al. OsCAF1, a CRM domain containing protein, influences chloroplast development. Int J Mol Sci. 2019;20(18):4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Zhang Q, Shen L, Ren D, et al. Characterization of the CRM gene family and elucidating the function of OsCFM2 in rice. Biomolecules. 2020;10(2):327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Asakura Y, Bayraktar OA, Barkan A. Two CRM protein subfamilies cooperate in the splicing of group IIB introns in chloroplasts. RNA. 2008;14(11):2319–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Feiz L, Asakura Y, Mao L, et al. CFM1, a member of the CRM-domain protein family, functions in chloroplast group II intron splicing in Setaria viridis. Plant J. 2021;105(3):639–648. [DOI] [PubMed] [Google Scholar]
- [114].Amann K, Lezhneva L, Wanner G, et al. ACCUMULATION OF PHOTOSYSTEM ONE1, a member of a novel gene family, is required for accumulation of [4Fe-4S] cluster-containing chloroplast complexes and antenna proteins. Plant Cell. 2004;16(11):3084–3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Asakura Y, Galarneau E, Watkins KP, et al. Chloroplast RH3 DEAD box RNA helicases in maize and Arabidopsis function in splicing of specific group II introns and affect chloroplast ribosome biogenesis. Plant Physiol. 2012;159(3):961–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Gu L, Xu T, Lee K, et al. A chloroplast-localized DEAD-box RNA helicaseAtRH3 is essential for intron splicing and plays an important role in the growth and stress response in Arabidopsis thaliana. Plant Physiol Biochem. 2014;82:309–318. [DOI] [PubMed] [Google Scholar]
- [117].Roberti M, Polosa PL, Bruni F, et al. The MTERF family proteins: mitochondrial transcription regulators and beyond. Biochim Biophys Acta. 2009;1787(5):303–311. [DOI] [PubMed] [Google Scholar]
- [118].Hammani K, Barkan A. An mTERF domain protein functions in group II intron splicing in maize chloroplasts. Nucleic Acids Res. 2014;42(8):5033–5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Lee K, Leister D, and Kleine T. Arabidopsis mitochondrial transcription termination factor mTERF2 promotes splicing of group IIB introns. Cells. 2021;10(2):315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


