Abstract
Fourier transform infrared and Raman microspectroscopy are currently being developed as new methods for the rapid identification of clinically relevant microorganisms. These methods involve measuring spectra from microcolonies which have been cultured for as little as 6 h, followed by the nonsubjective identification of microorganisms through the use of multivariate statistical analyses. To examine the biological heterogeneity of microorganism growth which is reflected in the spectra, measurements were acquired from various positions within (micro)colonies cultured for 6, 12, and 24 h. The studies reveal that there is little spectral variance in 6-h microcolonies. In contrast, the 12- and 24-h cultures exhibited a significant amount of heterogeneity. Hierarchical cluster analysis of the spectra from the various positions and depths reveals the presence of different layers in the colonies. Further analysis indicates that spectra acquired from the surface of the colonies exhibit higher levels of glycogen than do the deeper layers of the colony. Additionally, the spectra from the deeper layers present with higher RNA levels than the surface layers. Therefore, the 6-h colonies with their limited heterogeneity are more suitable for inclusion in a spectral database to be used for classification purposes. These results also demonstrate that vibrational spectroscopic techniques can be useful tools for studying the nature of colony development and biofilm formation.
In recent years, there has been much effort invested into the development of new techniques for the identification of microorganisms. Many of these methods are aimed at providing the clinician with more rapid identification of the microorganism responsible for infection in order to begin the appropriate course of antimicrobial treatment (1, 9, 15, 21, 27, 31, 44, 51). The emergence of these novel methods reflects the rise in drug-resistant microorganisms, which requires that antimicrobial treatment be more effectively managed (2, 12, 28, 52). Among the new methods are those based on vibrational spectroscopic techniques, namely Fourier transform infrared (FT-IR) and Raman spectroscopies. Vibrational spectroscopic methods are reagentless procedures in which there is no need to add dyes or labels for spectral measurement. These nondestructive techniques are based on the absorption (FT-IR) or scattering (Raman) of light directed onto a sample. The amount of light absorbed or scattered depends on the molecules found within the sample and the environment in which these molecules are found. With these highly sensitive techniques, the frequency of light in the resulting spectrum provides biochemical information regarding the molecular composition and molecular structure of and molecular interaction in cells and tissues (24, 55). Raman and infrared spectroscopies are complementary techniques which together can provide a more complete impression of the biochemical information within a sample. Furthermore, these two methods differ such that each is capable of providing information not easily obtainable by the other. For instance, with FT-IR spectroscopy, hydrated samples are difficult to measure since water absorbs so strongly that its signal masks other interesting peaks in the spectrum. On the other hand, water is less problematic in Raman spectroscopy, enabling measurement of hydrated samples. However, the signal-to-noise ratio of the resulting Raman spectrum is overall poorer than that obtained by FT-IR spectroscopy when spectra are measured for the same amount of time. When these sensitive techniques are coupled to a microscope, spectra can be acquired from microorganisms cultured for short periods of time (∼6 h) on or from solid culture media since large biomasses are not required for spectral measurement.
The application of various spectroscopic techniques to identify and characterize microorganisms has been explored previously (3, 9, 14–16, 20–23, 25–27, 29–35, 37, 42, 49, 54, 56). These studies have shown that it is possible to discriminate among various microorganisms at the genus, species, and strain level (14, 21–22, 25, 27, 29–32, 34, 49), and studies report the ability to differentiate microorganisms from various serogroups (21–23, 37). Furthermore, the use of FT-IR spectroscopy to identify drug resistance has also been investigated (3, 42). However, many of these previous studies are based on microorganisms cultured for 16 to 24 h or longer prior to spectral measurement. Our research is aimed at developing new rapid methods for the identification and characterization of clinically relevant microorganisms through the use of confocal Raman and FT-IR microspectroscopies. Current microbiological diagnostic methods require 2 to 3 days and involve culturing of microorganisms until a suitable biomass is obtained for subsequent tests. Such methods are inherently slow, especially in life-threatening situations such as cases of meningitis and sepsis and for critically ill patients in the intensive-care units of hospitals (52). With microspectroscopic methods, microorganisms can be cultured for as little as 6 h prior to spectral measurement. It is intended that these novel diagnostic methods quickly provide the clinician with results within the same day that patient samples are obtained.
A critical aspect of these rapid identification methods based on vibrational spectroscopy is the development of spectral reference databases against which clinical results can be compared in order to arrive at nonsubjective identification and classification schemes. Not only must the spectra contained within the database be derived from rigidly standardized protocols regarding the culturing and measurement conditions, the database needs to be comprehensive so that it reflects any kind of intrinsic biological diversity and heterogeneity found within microorganisms. Our interest in the development of microcolonies is in regard to the need to understand the heterogeneity of microorganism growth from the point of view of spectral variance. We have previously described a Raman method of characterizing cultures after 6 h of growth (27). In this paper, we present the investigation of spatial colony heterogeneity and its biochemical basis in 6-h and older cultures by Raman and FT-IR microspectroscopies. The findings based on five well-characterized microorganisms after growing on solid culture medium for various time periods are reported. For comparison, similar studies were performed using a conventional approach involving intrinsic fluorescence spectroscopy. From this investigation, the nature of the variance of spectra derived from these microorganisms provides insight on the development of microorganisms grown on solid culture medium.
MATERIALS AND METHODS
Strains and sample preparation.
A group of five well-characterized reference strains was obtained from the collection of the Pasteur Institute (Paris, France) (Staphyloccocus aureus CIP 4.83, S. aureus CIP 53.154, Escherichia coli CIP 53.126, E. coli CIP 54.8T) and the American Type Culture Collection (Candida albicans ATCC 90028). These strains were stored in brain-heart infusion broth (Becton Dickinson, Paramus, N.J.) containing 10% glycerol at −80°C until ready for use. Sample preparation involved one overnight passage on solid culture medium (to acclimatize the strain), followed by reculturing of the strain for various incubation times, typically for 6, 12, and 24 h to yield (micro)colonies ranging in size from less than 50 μm in the short incubation times to as large as 2,000 μm after the longer culture times. Typical culture conditions for the bacterial strains involved growing at 37°C on Mueller-Hinton medium (Merck, Darmstadt, Germany). The yeast strain was cultured at 37°C on Sabouraud-glucose 2% medium (Merck).
For the Raman studies, a biomass from the overnight passage was used to streak out the strains in four segments. Following incubation, spectra were typically acquired directly from microorganisms still growing on the culture plate of well-isolated (micro)colonies found in the third or fourth segment.
For the infrared studies, following incubation, microcolonies were transferred from the agar plate to an infrared, transparent, zinc selenide optical plate using a specially designed stamping device (30–32). Imprints were allowed to air dry (approximately 15 min) prior to spectral measurement. Spectra were acquired from the imprinted microcolonies on this substrate.
For the fluorescence studies, from the incubated plates, colony imprints were done manually onto glass slides.
Spectral acquisition and data treatment. (i) Raman spectroscopy.
Raman spectra were acquired using a Renishaw System 1000 Raman microspectrometer (Renishaw PLC, Gloucestershire, United Kingdom) equipped with a 300-line/mm grating as described previously (27). The accompanying Leica microscope was fitted with an 80× near-infrared objective (MIR Plan 80×/0.75; Olympus). Raman signal was collected in the spectral interval from 250 to 2,150 cm−1, with a spectral resolution of 8 cm−1. Measurements were performed using 830-nm excitation from a titanium-sapphire laser (model 3900; Spectra Physics, Mountain View, Calif.) pumped by an argon ion laser (series 2000; Spectra Physics) delivering 100 mW of laser power on the sample.
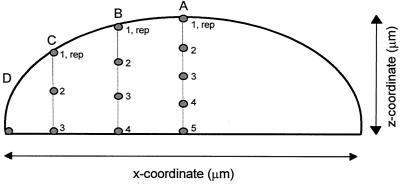
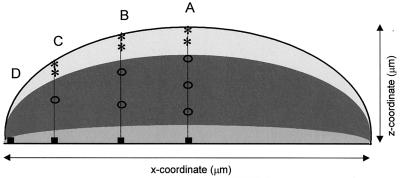
The plates with cultures were taken from the incubator and were placed directly under the microscope objective for measurements. Firstly, for each (micro)colony, the diameter and depth were estimated. Thereafter, various measurement positions were determined to give four equally spaced lateral positions from the center to the edge of the colony. At the thicker lateral positions, various depths within the colony were also selected (Fig. 1). At each measurement position, five spectra at 30 s were acquired and averaged. At the very edge and at the bottom of the colony closest to the culture medium, 10 spectra each at 30 s were acquired and averaged in order to improve the spectral signal-to-noise ratio. The depth spectra were acquired beginning from the surface and working towards the bottom of the colony. Following the deepest measurement, a repeat measurement of the surface was taken as a duplicate check (labeled “rep” in Fig. 1). With the use of a computer-controlled xyz stage, it was possible to determine the lateral and focusing positions reproducibly.
FIG. 1.
Schematic representation of the various measuring positions within a colony for spectra acquired by Raman microspectroscopy. Letters refer to the lateral measuring positions, while numbers refer to the depth measuring positions; “rep” refers to a measurement of the surface position as described in Materials and Methods.
Following spectral acquisition, the constant background signal contribution originating from optical elements in the laser light delivery pathway was subtracted from all spectra. The reference spectrum of a tungsten-band lamp of known temperature was used to correct for the wavelength-dependent signal detection efficiency of the Raman setup (36, 55). Spectral treatment also involved taking the first derivative of the Raman spectra, scaling to standard normal variance (i.e., zero mean and unit variance) and using the spectral region between 400 and 1,800 cm−1 for further analysis. Despite the use of a confocal arrangement in which typical measuring volumes were approximately 1.5 μm in the lateral direction and 7 to 8 μm along the optical axis, there exists the possibility of sampling the underlying culture medium especially for measurements toward the bottom of the colony. Therefore, it was necessary to correct the acquired spectra for the variable underlying signal of the culture medium using a vector correction approach. (Details are provided in reference 27. As stated therein, it should be stressed once again that care must be taken in making biochemical interpretations of spectra that were treated with the vector correction method.) Reduction of data was then performed through principal-component analysis using the Matlab PLS Toolbox (Eigenvector Research Inc., Manson, Wash.). The principal-component analysis scores accounting for 99.9% of the total variance captured were used in a hierarchical cluster analysis (SPSS, Chicago, Ill.) using Ward's clustering method and squared Euclidean distance measure.
(ii) Infrared spectroscopy.
FT-IR spectra were recorded on an FT-IR microscope, model IR Scope II, which was interfaced to an IFS 28/B spectrometer (Bruker Optics, Karlsruhe, Germany) and was equipped with a motor-driven xy stage. For each spectrum, 256 interferrograms were coadded and averaged. Fourier transformation was done using a Blackmann-Harris three-term apodization function and a zero-filling factor of 4, resulting in a nominal resolution of 6 cm−1. Spatial heterogeneity of the microcolonies was examined by linearly mapping the microcolony in 10-μm steps in the x and y directions and using an aperture size of 30 μm. The first derivative spectra within the range of 820 to 1,780 cm−1 were vector normalized as described previously (4). Hierarchical cluster analysis was performed using Pearson's product moment correlation coefficient and Ward's algorithm.
The infrared measurements were done independently at two different laboratories. Alternatively, FT-IR absorption spectra were collected using a UMA 500 infrared microscope coupled to an FTS-40A spectrometer (Bio-Rad Laboratories, Spectroscopy Division, Hemel-Hampstead, United Kingdom) and equipped with a mercury-cadmium telluride narrow-band detector and a microscope diaphragm varying from 10 × 10 to 500 × 500 μm2. Measurements on single microcolonies were performed by setting the microscope aperture from 60 × 60 up to 100 × 100 μm2. Absorption spectra were obtained in the microscope transmission mode using the following parameters: 4-cm−1 resolution, 5-kHz scan speed, 32- to 64-scan coaddition, triangular apodization, and spectral range of 800 to 4,000 cm−1. No baseline correction or smoothing was applied to the data. The data was first normalized using the z-score function (i.e., zero mean and unit variance) in Matlab (The Math Works, Inc., Natick, Mass.) and then subjected to cluster analysis using Matlab's Statistics Toolbox employing Ward's algorithm and Euclidean distance measure.
(iii) Intrinsic fluorescence.
Fluorescence emission signals from (micro)colony imprints on glass slides were measured by excitation in the UV wavelength (360 nm) with laser power of 1 μW, using an argon ion laser (model 2065A; Spectra Physics). Spectra were recorded using a UV confocal laser microspectrofluorometer (Dilor, Lille, France). For the UV measurements, an Olympus BH2 microscope containing a 100× objective was employed. Point-by-point analysis was done using the spectral imaging acquisition mode, which consisted of scanning the laser over the (micro)colony by moving the computer-controlled xy stage. For each (micro)colony, 100 points were collected. The data gave an emission spectral profile, which in turn produced a spectral image that can be compared to the conventional optical image. All spectral manipulations were done with the LabSpec software (Dilor). The spectral profiles obtained were used in a hierarchical cluster analysis constructed with Ward's method and Euclidean distance measure (Statistica; Statsoft, Tulsa, Okla.).
RESULTS AND DISCUSSION
In the development of new routine methods based on vibrational spectroscopic techniques for the rapid identification of microorganisms, spectra derived from rigidly standardized protocol are used to establish a spectral database for the nonsubjective classification and identification of clinically relevant microorganisms. Thus, it is imperative that the database be comprehensive so that the natural variance of the microorganism is captured within the spectral database. A potential problem is that in recent years, it is becoming more widely accepted that microorganisms are not necessarily unicellular organisms but rather multicellular organisms able to form complex communities with specific division of tasks and population differentiation (39–41). Furthermore, it is known that biofilms are elaborate structures composed of microcolonies attached to a surface and that within these microcolonies, the bacteria are organized into communities with functional heterogeneity (6). Given that colonies of microorganisms are complex multicellular communities, it is necessary to establish at what growth stage infrared and Raman spectra should be acquired from such (micro)colonies. In so doing, any heterogeneity which can interfere with the discrimination of microorganisms can be minimized. As the aim of these new methods is to provide the clinician with laboratory results on the same day that patient material is acquired, the culture time should be kept short: for example, approximately 6 h of growth time. To gain an understanding of the spectral heterogeneity, the development of (micro)colonies was monitored over several culture times and at various positions within the (micro)colonies.
In the discussion to follow, a large part of the analysis is based on the results of subjecting the data to hierarchical clustering analysis. This is a procedure that nonsubjectively groups the input cases (i.e., the spectra) based on similarities of their properties (the spectral characteristics). When graphically displayed, the result of the analysis forms a dendrogram; the relationship between the input cases is represented by the distance at which they connect on a dissimilarity scale (e.g., Fig. 2A). The more similar the cases are, the smaller their connecting distance on the dissimilarity scale. Dendrograms resemble the phylogenetic trees that arise from taxonomic classification. Groups of similar members can be readily visualized. Since the spectral information reflects the biochemistry of the sample measured, the distance in the dendrograms can be interpreted as a measure of how biochemically different the various spectra are and, hence, the measurement positions within a colony.
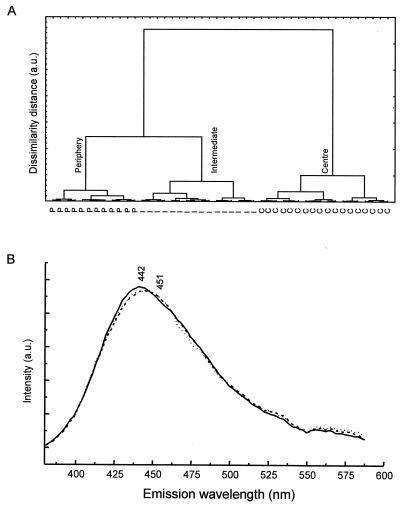
FIG. 2.
(A) Dendrogram of hierarchical cluster analysis of UV (360 nm)-excited intrinsic fluorescence spectra (derived from the spectral image after filtering) of an E. coli CIP 54.8T microcolony imprint. Spectra were acquired from various positions within the peripheral, intermediate, and central regions of the microcolony imprint. (B) Averaged intrinsic fluorescence spectra corresponding to spectra acquired from the central (——), intermediate (- - - -), and peripheral (. . . . . .) regions of the microcolony imprint. a.u., arbitrary units.
Fluorescence of colony imprints.
As a starting point in the investigation of (micro)colony heterogeneity, an approach involving fluorescence microspectrometry was first employed. A series of UV-excited intrinsic fluorescence spectra were acquired from a microcolony imprint of E. coli CIP 54.8T cultured for 6 h. The spectra were normalized to account for possible differences in intensity due to variation in the thickness of the microcolony imprint. Subjecting the data to hierarchical cluster analysis revealed that the intrinsic fluorescence was not homogeneously distributed over the microcolony. As shown in the dendrogram in Fig. 2A, the spectral profiles from the various positions within the imprint form their own subclusters (central, intermediate, and peripheral). When the averaged fluorescence spectrum from each subcluster is examined (Fig. 2B), the various spectra are quite similar. The average spectrum from the center of the microcolony has an emission wavelength maximum of 442 nm. In contrast, the spectra corresponding from the intermediate and peripheral regions have a broader spectral profile with a flatter maximum, possibly corresponding to the superposition of two maxima at 442 and 451 nm. Despite these differences, further information regarding the compositional heterogeneity of the various regions could not be directly obtained from the intrinsic fluorescence data. Hence, other methods were required.
Infrared spectra of (micro)colony imprints.
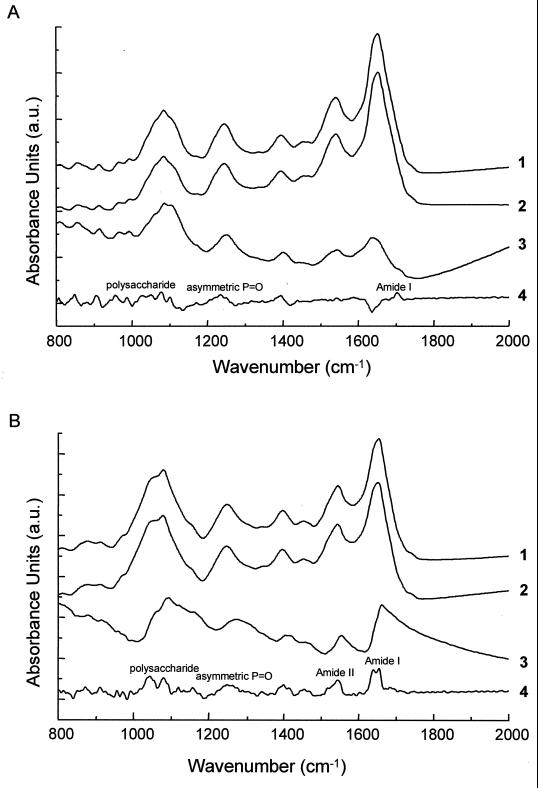
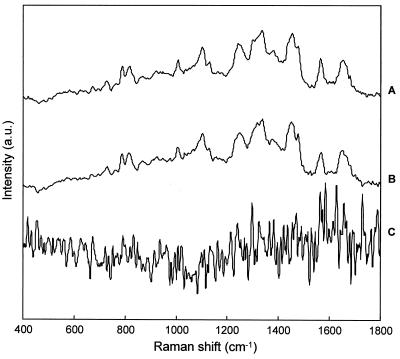
In order to gain a further understanding of the biochemical heterogeneity of (micro)colonies, vibrational spectroscopic techniques were employed. Hierarchical cluster analysis of FT-IR spectra acquired from colonies of E. coli CIP 54.8T which were 100 μm or larger in diameter produced a dendrogram similar to that shown in Fig. 2A, with the spectra tending to cluster into different groups depending upon the measurement position within the colony (not shown for brevity). By examining the individual infrared spectra and calculating difference spectra, it is possible to gain a better understanding of the source of the clustering scheme observed. In Fig. 3A, the FT-IR spectra acquired from the center and edge of an E. coli CIP 54.8T colony are shown. Although to the untrained eye these two spectra look remarkably similar, any differences that exist can be highlighted by taking difference spectra. The difference spectrum which results from subtracting the spectrum of the edge from that of the center is shown as well as the difference obtained from subtracting the first-derivative spectra from the two measuring positions. Since infrared bands tend to be quite broad, thereby potentially masking peak differences, the differences are more apparent in the derivative spectra. Comparison of the peak positions with those from empirical studies in the literature (24) reveals differences in the spectral region around 1,230 cm−1 which can be assigned to the phosphate double-bond asymmetric stretching vibration of phosphodiester, free phosphate, and monoester phosphate functional groups. Smaller alterations are also observed in the protein amide I regions (approximately 1,620 to 1,670 cm−1). This band arises predominantly from the C⩵O stretching vibration of the amide C⩵O groups of proteins. Furthermore, changes visible around 1,400 cm−1 may be attributed to the symmetric stretching vibrations of COO− functional groups, and very weak changes were observed in the carbohydrate region around 900 to 1,200 cm−1. Similar changes were observed for the other bacterial and yeast strains (35), although the changes were more pronounced in the yeast strains (Fig. 3B).
FIG. 3.
Original FT-IR spectra from a 100-μm-diameter E. coli CIP 54.8T colony (A) and a 100-μm-diameter C. albicans ATCC 90028 colony (B) measured at the center (1) and edge (2) positions of the colony. The corresponding difference spectrum (magnification, ×5) (3) between center and edge is shown, as well as the difference spectrum (magnification, ×2) (4) obtained from the first-derivative spectra. The difference spectra were obtained by 1-to-1 subtraction of vector-normalized spectra (normalization between 820 and 1,780 cm−1). a.u., arbitrary units.
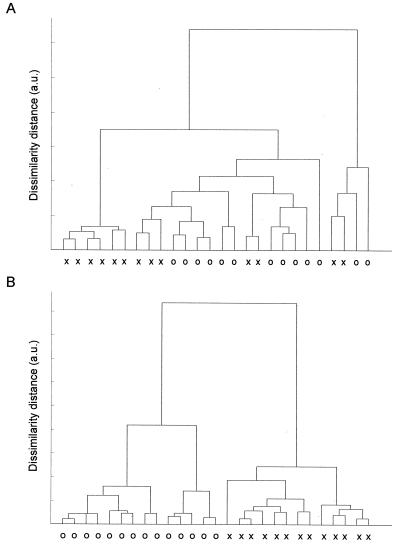
Interestingly, in a separate study measuring FT-IR spectra from 12-h colonies of the two E. coli strains (CIP 54.8T and 53.126), heterogeneity between spectra acquired from the center and edge of a colony was large enough to influence the discrimination between the different strains. As shown in Fig. 4A, the spectra arising from the two strains formed mixed clusters. However, when spectra acquired from younger colonies (approximately 7 h of culture time) were subjected to cluster analysis, two major clusters were formed corresponding to the different strains (Fig. 4B). These results suggest that with the older colonies, there is significant heterogeneity in the spectra from various positions within the colony. Similar studies performed with 50-μm-diameter colonies or growth time of about 6 to 7 h revealed that there was very little variance in the infrared spectra sampled from the center or periphery of the colony. Therefore, it appears that until 6 to 7 h of growth, there is very little heterogeneity observed in the composition of microcolonies. However, beyond this time frame, marked biochemical differences which vary from the center to the periphery of the colony, such as changes in the protein amide I bands, phosphate moieties likely arising from nucleic acids, protein constitution, and carbohydrate moieties, are noted. These differences likely influence the classification results observed (Fig. 4A and B).
FIG. 4.
Dendrogram from hierarchical clustering analysis of FT-IR spectra of 12-h cultures (A) and 7-h cultures (B) of E. coli CIP 54.8T (denoted by x) and E. coli CIP 53.126 (denoted by o). a.u., arbitrary units.
Raman spectra directly from (micro)colonies.
With infrared microspectroscopy, spectra were acquired through the entire depth of the colony at the central, intermediate, and peripheral regions. However, with this approach, any heterogeneity arising from various depths within the colony would not be readily revealed. Insight into the heterogeneity of microcolonies can also be obtained from confocal Raman microspectroscopy, in which spectra can be acquired from the various lateral positions throughout the colony as well as at various depths within the colony. The Raman spectra acquired in this manner were subjected to hierarchical cluster analysis. In Fig. 5A, the results are shown for spectra acquired from 6-h cultures. Visual inspection of the dendrogram revealed no obvious groupings or clusters. This observation is in accordance with the infrared findings for 6-h colonies, thereby suggesting that at this growth stage, the cultures are quite homogeneous overall. Such observations were apparent irrespective of the strain studied.
FIG. 5.
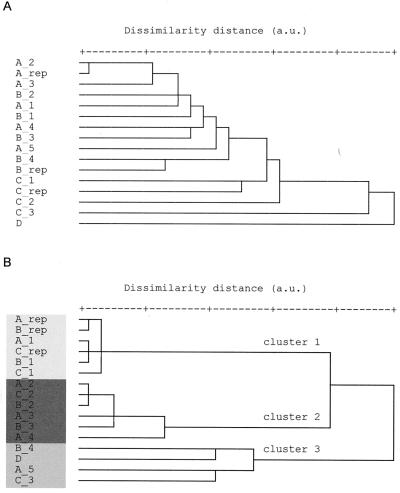
Dendrograms from hierarchical cluster analysis of Raman spectra from various measurement positions within a 6-h E. coli CIP 53.126 microcolony (A) and a 24-h E. coli CIP 53.126 colony (B). Shading highlights the various clusters and corresponds to the shading in Fig. 6. The labels correspond to the measuring positions in the schematic diagram of Fig. 1 and 6. a.u., arbitrary units.
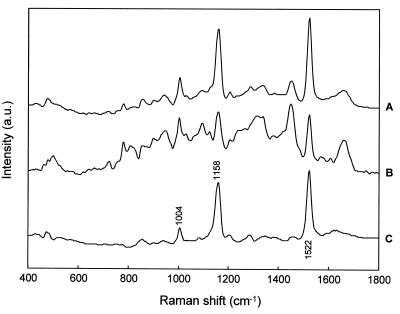
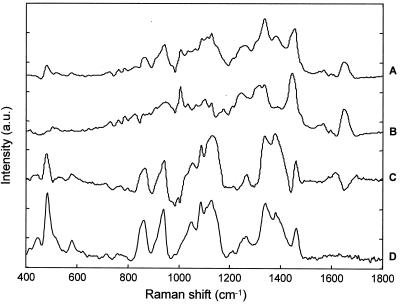
Unlike the dendrograms obtained for 6-h cultures, the dendrograms of spectra acquired from microorganisms grown for 12 and 24 h showed distinct subclusters (Fig. 5B). When the members of the same cluster were assigned to a group and the various groups were projected onto a schematic diagram depicting the measurement location of each spectrum, it appears that there are different layers in the 12- and 24-h colonies (Fig. 6). Similar findings were observed for the various strains studied for 12- and 24-h cultures. Further examination of the spectra indicates that for S. aureus CIP 4.83, the clustering differences arise from distinct spectral peaks at 1,004, 1,158, and 1,522 cm−1, which can be assigned to the various C—C vibrations found in carotenoids (Fig. 7) (29, 30, 33, 54). The clustering reveals that the carotenoid concentration is higher within the upper layers of the colony and less prominent in the deeper layers. Carotenoids are responsible for the yellow-orange pigmentation observed in the 12- and 24-h colonies and are one of the classical characteristics of this species. Studies have shown that S. aureus is very sensitive to the bactericidal effects of fatty acids, such as oleic acid. The incorporation of such lipophilic agents into the membranes results in increased membrane fluidity and thus in a decrease in membrane-associated functions (5). It is believed that the production of carotenoids might help S. aureus stabilize its cell membrane, thereby preventing potentially lethal fatty acid-induced changes in the fluidity of its membrane (5). Other studies have also shown that pigmented S. aureus strains are far more resistant to singlet oxygen lethality than are carotenoidless S. aureus mutants (8). Hence, the bacterium might use the carotenoid pigmentation as a mechanism to resist killing by fatty acids and to quench singlet oxygen, thus protecting against lethal effects of photosensitization. Previous studies (18) have shown that the carotenoid production is mainly correlated with the time of growth, and this finding has also been observed by FT-IR spectroscopy (29). Therefore, it might signify that older cells which produce significant pigmentation are found towards the surface layers of the colony. Alternatively, our finding of higher carotenoid concentration in the upper layers of older colonies might suggest a means by which the colony protects itself from its environment.
FIG. 6.
Diagrammatic projection of the various clusters (∗, cluster 1; ○, cluster 2; ▪, cluster 3; as determined from Fig. 5) from the hierarchical cluster analysis of Raman spectra from various measurement positions within a 24-h E. coli CIP 53.126 colony. Shading is used to highlight the various layers within the colony.
FIG. 7.
Averaged Raman spectra of the members of cluster 1 (surface layer) (A) and cluster 2 (layer beneath surface) (B) and the corresponding difference spectrum (A and B) from a 24-h colony of S. aureus CIP 4.83 (C) are shown. a.u., arbitrary units.
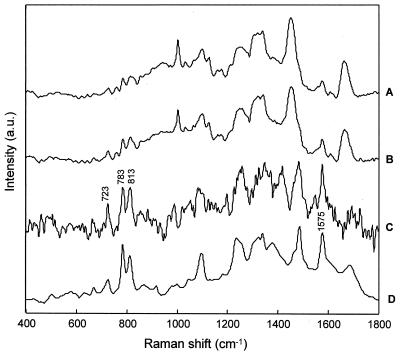
Interestingly, the Raman spectra of the other S. aureus strain, CIP 53.154, showed that this strain does not produce the characteristic pigmentation. Nonpigmented derivatives of S. aureus are known to exist and are often found in subcultures of stored organisms (53). The cluster analysis shows a similar sort of distinction, with spectra acquired from the surface layers clustering together and those within deeper layers clustering as a group. However, the lack of pigmentation suggests that another spectral feature is responsible for the formation of distinct clusters. This clustering trend was found for S. aureus CIP 53.154, E. coli CIP 53.126, E. coli CIP 54.8T, and C. albicans ATCC 90028. Closer examination of the Raman difference spectra showed that in the deeper layers of 12- and 24-h colonies, there are characteristic spectral peaks at 723, 783, 813, and 1,575 cm−1 (Fig. 8). These features all arise from the nucleotide and phosphate backbone vibration found in RNA (19). It appears that the RNA concentration is higher in the deeper layers of the colony. Observations of the decrease in RNA content in older cells which have transitioned to the stationary phase from the logarithmic phase have been reported with FT-IR spectra of Bacillus subtilis (29). Hence, this finding again suggests that the colony is composed of older cells in the surface layers and younger cells in the deeper layers which are more actively dividing, thus reflecting a higher RNA content.
FIG. 8.
Averaged Raman spectra of the members of cluster 2 (layer beneath surface) (A) and cluster 3 (deeper layer) (B) and the corresponding difference spectrum (A and B) from a 24-h colony of E. coli CIP 54.8T (C) are shown. The Raman spectrum of RNA (D) is also shown. a.u., arbitrary units.
Aside from RNA differences, it was noted that 12- and 24-h colonies from the bacterial strains contain a relatively higher glycogen concentration in the surface layers (Fig. 9). This glycogen difference is not observed with younger 6-h bacterial microcolonies (Fig. 10) or with the yeast strain. At present it is unknown whether the glycogen is contained within the cells of the surface layers or found extracellularly in the form of a film. Previous FT-IR studies have also found increases in the carbohydrate C—O stretching mode of 24-h cultures of Bradyrhizobium japonicum strains which have been transferred from liquid to solid culture medium. From transmission electron micrographs, the authors ascribed such changes to alteration of the bacterial wall component, possibly the formation of glycocalyx (56). The organization of colonies into distinct layers has also been observed with E. coli strains (cultured for 2 weeks) in which vertical sections through colonies revealed a stratification of different cell types, as could be seen with standard microscopic reagents, such as staining with toluidine blue (41). Previous reports in the literature using scanning electron microscopy to study the surface structure of E. coli colonies growing for over 24 h on agar medium in normal petri dishes have revealed that each colony secretes extracellular materials, some of which form a skin or framework over its surface (38–39). Other studies have shown that at later stages of colony development (20 to 24 h), the surface film of E. coli colonies became thicker. On the other hand, the film was not observed for colonies cultured for 6 to 16 h of growth (45–48). Therefore, it is possible that the glycogen-rich surface layer observed with Raman microspectroscopy is the polysaccharide-rich extracellular coat, commonly known as the glycocalyx, of bacterial cells. These exopolysaccharides are mainly composed of homopolysaccharides (cellulose, levans, dextrans, and glucans) and heteropolysaccharides (monosaccharides including a uronic acid) (43). It is thought that the formation of the glycocalyx serves as an integral matrix for a biofilm and that following the adhesion of bacteria to a substrate, the glycocalyx forms a protective milieu for cell division and microcolony formation and growth (7, 11). Some studies propose that the glycocalyx either acts as a diffusion barrier or, by complexing antibacterial agents, excludes and/or influences the penetration of antimicrobial agents to the underlying cells (10, 13). Modern medicine increasingly relies on the use of indwelling medical devices such as catheters and prosthetic joints for multiple purposes. These so-called foreign bodies are implanted for a short period, intermittently or permanently. One of the most frequently encountered complications of these devices is the development of infections. The ability of bacteria to adhere to the surface of these indwelling devices by binding to biofilm layers is still not completely understood (H. P. Endtz, personal communication). Hence, there is much interest in the development of biofilms, associated with disease in humans due to the increasing use of medical devices and the difficulty, resulting from resistance to antimicrobial agents, of effectively controlling infection (6, 13, 17).
FIG. 9.
Averaged Raman spectra of the members of cluster 1 (surface layer) (A) and cluster 2 (layer beneath surface) (B) and the corresponding difference spectrum (A and B) from a 24-h colony of E. coli CIP 53.126 (C) are shown. The Raman spectrum of glycogen (D) is also shown. a.u., arbitrary units.
FIG. 10.
Raman spectra are shown from different depths corresponding to the measuring positions in the schematic diagram of Fig. 1, with A1 from the surface (A) and A3 from deeper within the colony (B) and the corresponding difference spectrum (A and B) from a 6-h microcolony of E. coli CIP 53.126 (C). a.u., arbitrary units.
Overall, these infrared and Raman studies of the development of microorganisms cultured for various growth times reveal that there is significant colony heterogeneity in the strains cultured for 12 and 24 h. These differences can be attributed to higher glycogen content in the surface layers and to increased levels of carotenoid pigmentation in certain S. aureus strains. Furthermore, a relatively higher RNA content was observed in the deeper layers of the colony. Therefore, spectra derived from these older colonies are quite variable, indicating the need to sample spectra from a multitude of positions within these colonies in order to capture the biological variance of the various cell types. The lack of group clusters and absence of obvious spectral differences in the various spectra obtained from 6-h cultures suggest that the microcolonies at this growth stage are very homogenous in terms of molecular composition. Thus, these spectra are suitable for inclusion in and building of spectral libraries of microorganisms. With the development of a comprehensive spectral database, it should be possible to use Raman and FT-IR microspectroscopies to provide rapid identification and classification of clinically relevant microorganisms. Moreover, the present study demonstrates that vibrational microspectroscopy can be applied to further understand the heterogeneity of microorganism growth. For example, the attachment and microcolony formation of biofilms, as well as the actual mechanisms of biofilm resistance to antimicrobial agents, still remain unclear (13). FT-IR spectroscopy, including attenuated total reflectance spectroscopy, has been used previously to study bacterial growth and biofilm formation (56). The use of Raman microspectroscopy to probe various layers within a colony can be extended to study the formation of sessile communities found at the base of the biofilm. These sessile cells are believed to be the root of many persistent and chronic bacterial infections since they can withstand host immune responses, unlike their nonattached planktonic counterparts which are killed by antibiotic therapy (6). The knowledge gained from such studies can be used to develop new strategies for the treatment of infection, especially those associated with indwelling medical devices.
ACKNOWLEDGMENT
We gratefully acknowledge financial support from the European Union Biomed II program, Project No. BMH4–97-2054.
REFERENCES
- 1.Belgrader P, Benett W, Hadley D, Richards J, Stratton P, Mariella R, Jr, Milanovich F. PCR detection of bacteria in seven minutes. Science. 1999;284:449–450. doi: 10.1126/science.284.5413.449. [DOI] [PubMed] [Google Scholar]
- 2.Binder S, Levitt A M, Sacks J J, Huges J M. Emerging infectious diseases: public health issues for the 21st century. Science. 1999;284:1311–1313. doi: 10.1126/science.284.5418.1311. [DOI] [PubMed] [Google Scholar]
- 3.Bouhedja W, Sockalingum G D, Pina P, Allouch P, Bloy C, Labia R, Millot J M, Manfait M. ATR-FTIR spectroscopic investigation of E. coli transconjugants β-lactams-resistance phenotype. FEBS Lett. 1997;412:39–42. doi: 10.1016/s0014-5793(97)00725-4. [DOI] [PubMed] [Google Scholar]
- 4.Bruker Optics. Opus I. R. handbook. Karlsruhe, Germany: Bruker Optics; 1991. [Google Scholar]
- 5.Chamberlain N R, Mehrtens B G, Xiong Z, Kapral F A, Boardman J L, Rearick J I. Correlation of carotenoid production, decreased membrane fluidity, and resistance to oleic acid killing in Staphylococcus aureus 18Z. Infect Immun. 1991;59:4332–4337. doi: 10.1128/iai.59.12.4332-4337.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costerton J W, Stewart P S, Greenberg E P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 7.Costerton J W, Irvin R T, Cheng K-J. The bacterial glycocalyx in nature and disease. Annu Rev Microbiol. 1981;35:299–324. doi: 10.1146/annurev.mi.35.100181.001503. [DOI] [PubMed] [Google Scholar]
- 8.Dahl T A, Midden R, Hartman P E. Comparison of killing of gram-negative and gram-positive bacteria by pure singlet oxygen. J Bacteriol. 1989;171:2188–2194. doi: 10.1128/jb.171.4.2188-2194.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards-Jones V, Claydon M A, Evason D J, Walker J, Fox A J, Gordon D B. Rapid discrimination between methicillin-sensitive and methicillin-resistant Staphylococcus aureus by intact cell mass spectrometry. J Med Microbiol. 2000;49:295–300. doi: 10.1099/0022-1317-49-3-295. [DOI] [PubMed] [Google Scholar]
- 10.Farber B F, Kaplan M H, Clogston A G. Staphylococcus epidermidis extracted slime inhibits the antimicrobial action of glycopeptide antibiotics. J Infect Dis. 1990;161:37–40. doi: 10.1093/infdis/161.1.37. [DOI] [PubMed] [Google Scholar]
- 11.Fassel T A, Edmiston C E. Ruthenium red and the bacterial glycocalyx. Biotech Histochem. 1999;74:194–212. doi: 10.3109/10520299909047974. [DOI] [PubMed] [Google Scholar]
- 12.File T M., Jr Overview of resistance in the 1990s. Chest. 1999;115(Suppl. 3):3S–8S. doi: 10.1378/chest.115.suppl_1.3s. [DOI] [PubMed] [Google Scholar]
- 13.Gander S. Bacterial biofilms: resistance to antimicrobial agents. J Antimicrob Chemother. 1996;37:1047–1050. doi: 10.1093/jac/37.6.1047. [DOI] [PubMed] [Google Scholar]
- 14.Goodacre R, Timmins E M, Rooney P J, Rowland J J, Kell D B. Rapid identification of Streptococcus and Enterococcus species using diffuse reflectance-absorbance Fourier transform infrared spectroscopy and artificial neural networks. FEMS Microbiol Lett. 1996;140:233–239. doi: 10.1016/0378-1097(96)00186-3. [DOI] [PubMed] [Google Scholar]
- 15.Goodacre R, Rooney P J, Kell D B. Discrimination between methicillin-resistant and methicillin-susceptible Staphylococcus aureus using pyrolysis mass spectrometry and artificial neural networks. J Antimicrob Chemother. 1998;41:27–34. doi: 10.1093/jac/41.1.27. [DOI] [PubMed] [Google Scholar]
- 16.Goodacre R, Shann B, Gilbert R J, Timmins E M, McGovern A C, Alsberg B K, Kell D B, Logan N A. Detection of the dipicolinic acid biomarker in Bacillus spores using Curie-point pyrolysis mass spectrometry and Fourier transform infrared spectroscopy. Anal Chem. 2000;72:119–127. doi: 10.1021/ac990661i. [DOI] [PubMed] [Google Scholar]
- 17.Habash M, Reid G. Microbial biofilms: their development and significance for medical device-related infections. J Clin Pharmacol. 1999;39:887–898. doi: 10.1177/00912709922008506. [DOI] [PubMed] [Google Scholar]
- 18.Hammond R K, White D C. Carotenoid formation by Staphyloccus aureus. J Bacteriol. 1970;103:191–198. doi: 10.1128/jb.103.1.191-198.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartman K A, Thomas G J. The identification, interactions and structure of viruses by Raman spectroscopy. In: Nelson W H, editor. Instrumental methods for rapid microbiological analysis. Deerfield Beach, Fla: VCH Publishers; 1985. pp. 91–134. [Google Scholar]
- 20.Helm D, Naumann D. Identification of some bacterial cell components by FT-IR spectroscopy. FEMS Microbiol Lett. 1995;126:75–80. [Google Scholar]
- 21.Helm D, Labischinski H, Naumann D. Elaboration of a procedure for identification of bacteria using Fourier-Transform IR spectral libraries: a stepwise correlation approach. J Microbiol Methods. 1991;14:127–142. [Google Scholar]
- 22.Helm D, Labischinski H, Schallehn G, Naumann D. Classification and identification of bacteria by Fourier-transform infrared spectroscopy. J Gen Microbiol. 1991;137:69–79. doi: 10.1099/00221287-137-1-69. [DOI] [PubMed] [Google Scholar]
- 23.Horbach I, Naumann D, Fehrenbach F J. Simultaneous infections with different serogroups of Legionella pneumophila investigated by routine methods and Fourier transform infrared spectroscopy. J Clin Microbiol. 1988;26:1106–1110. doi: 10.1128/jcm.26.6.1106-1110.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson M, Mantsch H H. Biomedical infrared spectroscopy. In: Mantsch H H, Chapman D, editors. Infrared spectroscopy of biomolecules. Toronto, Canada: Wiley-Liss, Inc.; 1996. pp. 311–340. [Google Scholar]
- 25.Kümmerle M, Scherer S, Seiler H. Rapid and reliable identification of food-borne yeasts by Fourier-transform infrared spectroscopy. Appl Environ Microbiol. 1998;64:2207–2214. doi: 10.1128/aem.64.6.2207-2214.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manoharan R, Ghiamanti E, Dalterio R A, Britton K A, Nelson W H, Sperry J F. UV resonance Raman spectra of bacteria, bacterial spores, protoplasts and calcium dipicolinate. J Microbiol Methods. 1990;11:1–15. [Google Scholar]
- 27.Maquelin K, Choo-Smith L-P, van Vreeswijk T, Endtz H P, Smith B, Bennett R, Bruining H A, Puppels G J. Raman spectroscopic method for identification of clinically relevant microorganisms growing on solid culture medium. Anal Chem. 2000;72:12–19. doi: 10.1021/ac991011h. [DOI] [PubMed] [Google Scholar]
- 28.Moellering R C., Jr Antibiotic resistance: lessons for the future. Clin Infect Dis. 1998;27(Suppl. 1):S135–S140. doi: 10.1086/514902. [DOI] [PubMed] [Google Scholar]
- 29.Naumann D. FT-IR and FT-NIR Raman spectroscopy in biomedical research. In: De Haseth J A, editor. Fourier transform spectroscopy: Eleventh International Conference. Woodbury, N.Y: American Institute of Physics; 1998. pp. 96–109. [Google Scholar]
- 30.Naumann D. Infrared and NIR Raman spectroscopy in medical microbiology. In: Mantsch H H, Jackson M, editors. Infrared spectroscopy. 3257. New tool in medicine. Bellingham, Wash: SPIE—International Society for Optical Engineering; 1998. pp. 245–257. [Google Scholar]
- 31.Naumann D, Helm D, Labischinski H. Microbiological characterizations by FT-IR spectroscopy. Nature. 1991;35:81–82. doi: 10.1038/351081a0. [DOI] [PubMed] [Google Scholar]
- 32.Naumann D, Labischinski H, Giesbrecht P. The characterization of microorganisms by Fourier-Transform Infrared Spectroscopy (FT-IR) In: Nelson W H, editor. Modern techniques for rapid microbial analysis. New York, N.Y: Wiley-VCH; 1991. pp. 43–96. [Google Scholar]
- 33.Naumann D, Keller S, Helm D, Schultz C, Schrader B. FT-IR spectroscopy and FT-Raman spectroscopy are powerful analytical tools for the non-invasive characterization of intact microbial cells. J Mol Struct. 1995;347:399–406. [Google Scholar]
- 34.Nelson W H, Sperry J F. UV resonance Raman spectroscopic detection and identification of bacteria and other microorganisms. In: Nelson W, editor. Modern techniques for rapid microbiological analysis. New York, N.Y: VCH Publishers; 1991. pp. 97–143. [Google Scholar]
- 35.Orsini F, Ami D, Villa A M, Sala G, Bellotti M G, Doglia S M. FT-IR microspectroscopy for microbiological studies. J Microbiol Methods. 2000;42:17–27. doi: 10.1016/s0167-7012(00)00168-8. [DOI] [PubMed] [Google Scholar]
- 36.Puppels G J, Colier W, Olminkof J H F, Otto C, de Mul F F M, Greve J. Description and performance of a highly sensitive confocal Raman spectrometer. J Raman Spectrosc. 1991;22:217–225. [Google Scholar]
- 37.Seltmann G, Voigt W, Beer W. Application of physico-chemical typing methods for the epidemiological analysis of Salmonella enteritidis strains of phage type 25/17. Epidemiol Infect. 1994;113:411–424. doi: 10.1017/s0950268800068424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shapiro J A. Organization of developing Escherichia coli colonies viewed by scanning electron microscopy. J Bacteriol. 1987;169:142–156. doi: 10.1128/jb.169.1.142-156.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shapiro J A. Bacteria as multicellular organisms. Sci Am. 1988;256:82–89. [Google Scholar]
- 40.Shapiro J A. Thinking about bacterial populations as multicellular organisms. Annu Rev Microbiol. 1988;52:81–104. doi: 10.1146/annurev.micro.52.1.81. [DOI] [PubMed] [Google Scholar]
- 41.Shapiro J A. Pattern and control in bacterial colony development. Sci Prog. 1992;76:399–424. [PubMed] [Google Scholar]
- 42.Sockalingum G D, Bouhedja W, Pina P, Allouch P, Mandray C, Labia R, Millot J M, Manfait M. ATR-FTIR spectroscopic investigation of imipenem-susceptible and -resistant Pseudomonas aeruginosa isogenic strains. Biochem Biophys Res Commun. 1997;232:240–246. doi: 10.1006/bbrc.1997.6263. [DOI] [PubMed] [Google Scholar]
- 43.Sutherland I W. Bacterial exopolysaccharides—their nature and production. In: Sutherland I W, editor. Surface carbohydrates of the prokaryotic cell. London, United Kingdom: Academic Press; 1977. pp. 27–96. [Google Scholar]
- 44.Tang Y-W, Procop G W, Persing D H. Molecular diagnostics of infectious diseases. Clin Chem. 1997;43:2021–2038. [PubMed] [Google Scholar]
- 45.Tetz V V, Rybalchenko O V. Ultrastructure of colony-like communities of bacteria. APMIS. 1997;105:99–107. doi: 10.1111/j.1699-0463.1997.tb00547.x. [DOI] [PubMed] [Google Scholar]
- 46.Tetz V V, Rybalchenko O V, Savkova G A. Ultrastructural features of microbial colony organization. J Basic Microbiol. 1990;30:597–607. doi: 10.1002/jobm.3620300819. [DOI] [PubMed] [Google Scholar]
- 47.Tetz V V, Rybalchenko O V, Savkova G A. Surface films of Escherichia coli colonies. FEMS Microbiol Lett. 1993;107:231–239. doi: 10.1016/0378-1097(93)90315-s. [DOI] [PubMed] [Google Scholar]
- 48.Tetz V V, Rybalchenko O V, Savkova G A. Ultrastructure of the surface film of bacterial colonies. J Gen Microbiol. 1993;139:855–858. doi: 10.1099/00221287-139-4-855. [DOI] [PubMed] [Google Scholar]
- 49.Timmins E A, Howell S A, Alsberg B K, Noble W C, Goodacre R. Rapid differentiation of closely related Candida species and strains by pyrolysis-mass spectrometry and Fourier transform-infrared spectroscopy. J Clin Microbiol. 1998;36:367–374. doi: 10.1128/jcm.36.2.367-374.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tintelnot K, Haase G, Seibold M, Bergmann F, Staemmler M, Franz T, Naumann D. Evaluation of phenotypic markers for selection and identification of Candida dubliniensis. J Clin Microbiol. 2000;38:1599–1608. doi: 10.1128/jcm.38.4.1599-1608.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vaneechoutte M, Van Eldere J. The possibilities and limitations of nucleic acid amplification technology in diagnostic microbiology. J Med Microbiol. 1997;46:188–194. doi: 10.1099/00222615-46-3-188. [DOI] [PubMed] [Google Scholar]
- 52.Weber D J, Raasch R, Rutala W A. Nosocomial infections in the ICU: the growing importance of antibiotic-resistant pathogens. Chest. 1999;115(Suppl. 3):34S–41S. doi: 10.1378/chest.115.suppl_1.34s. [DOI] [PubMed] [Google Scholar]
- 53.Wieland B, Feil C, Gloria-Maercker E, Thumm G, Lechner M, Bravo J-M, Poralla K, Götz F. Genetic and biochemical analyses of the biosynthesis of the yellow carotenoid 4,4′-diaponeurosporene of Staphylococcus aureus. J Bacteriol. 1994;176:7719–7726. doi: 10.1128/jb.176.24.7719-7726.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams A C, Edwards H G M. Fourier transform Raman spectroscopy of bacterial cell walls. J Raman Spectrosc. 1994;25:673–678. [Google Scholar]
- 55.Wolthuis R, Bakker Schut T C, Caspers P J, Buschman H P J, Romer T J, Bruining H A, Puppels G J. Raman spectroscopic methods for in vitro and in vivo tissue characterization. In: Mason W T, editor. Fluorescent and luminescent probes for biological activity. 2nd ed. London, United Kingdom: Academic Press; 1999. pp. 433–455. [Google Scholar]
- 56.Zeroual W, Choisy C, Doglia S M, Bobichon H, Angiboust J-F, Manfait M. Monitoring of bacterial growth and structural analysis as probed by FT-IR spectroscopy. Biochim Biophys Acta. 1994;1222:171–178. doi: 10.1016/0167-4889(94)90166-x. [DOI] [PubMed] [Google Scholar]