Abstract
The distribution of dye-linked l-amino acid dehydrogenases was investigated in several hyperthermophiles, and the activity of dye-linked l-proline dehydrogenase (dye-l-proDH, l-proline:acceptor oxidoreductase) was found in the crude extract of some Thermococcales strains. The enzyme was purified to homogeneity from a hyperthermophilic archaeon, Thermococcus profundus DSM 9503, which exhibited the highest specific activity in the crude extract. The molecular mass of the enzyme was about 160 kDa, and the enzyme consisted of heterotetrameric subunits (α2 β2) with two different molecular masses of about 50 and 40 kDa. The N-terminal amino acid sequences of the α-subunit (50-kDa subunit) and the β-subunit (40-kDa subunit) were MRLTEHPILDFSERRGRKVTIHF and XRSEAKTVIIGGGIIGLSIAYNLAK, respectively. Dye-l-proDH was extraordinarily stable among the dye-linked dehydrogenases under various conditions: the enzyme retained its full activity upon incubation at 70°C for 10 min, and ca. 40% of the activity still remained after heating at 80°C for 120 min. The enzyme did not lose the activity upon incubation over a wide range of pHs from 4.0 to 10.0 at 50°C for 10 min. The enzyme exclusively catalyzed l-proline dehydrogenation using 2,6-dichloroindophenol (Cl2Ind) as an electron acceptor. The Michaelis constants for l-proline and Cl2Ind were determined to be 2.05 and 0.073 mM, respectively. The reaction product was identified as Δ1-pyrroline-5-carboxylate by thin-layer chromatography. The prosthetic group of the enzyme was identified as flavin adenine dinucleotide by high-pressure liquid chromatography. In addition, the simple and specific determination of l-proline at concentrations from 0.10 to 2.5 mM using the stable dye-l-proDH was achieved.
A number of dye-linked dehydrogenases (dye-DHs) catalyze the oxidation of various kinds of amino acids, organic acids, amines, and alcohols in the presence of an artificial electron acceptor such as 2,6-dichloroindophenol (Cl2Ind) and ferricyanide. Dye-DHs have potential utilization as a specific element for biosensors (6). However, the practical application of dye-DHs is still limited because of their low stability.
On the other hand, thermophiles, especially hyperthermophiles, may produce much more stable enzymes than the counterparts of mesophiles (2, 5). We screened the stable dye-DHs which use various amino acids and alcohols as the substrate in hyperthermophiles. As the result, we have found a dye-linked l-proline dehydrogenase (dye-l-proDH), which catalyzes the reduction of Cl2Ind in the presence of l-proline, in several hyperthermophiles. The presence of dye-l-proDH catalyzing the oxidation of l-proline to Δ1-pyrroline-5-carboxylate has been reported in Escherichia coli and Salmonella enterica serovar Typhimurium cells (9, 18). However, information about the detailed structure and function of the enzyme is still lacking because of its low stability. In particular, there has been no report on the dye-l-proDH from hyperthermophilic archaea. We report here the purification and properties of dye-l-proDH from a hyperthermophilic archaeon, Thermococcus profundus DSM 9503, and its application to l-proline determination.
MATERIALS AND METHODS
Materials.
UnoQ was purchased from Bio-Rad, Superdex 200 was obtained from Pharmacia, and Butyl-Toyopearl 650M and TSKgel ODS-80Ts (4.6 by 150 mm) were purchased from Tosoh (Tokyo, Japan). Flavin adenine dinucleotide (FAD), flavin mononucleotide (FMN), and Cl2Ind were obtained from Sigma. The other chemicals were analytical-grade reagents from Nacalai Tesque (Kyoto, Japan).
Microorganisms and conditions of cell growth.
Hyperthermophiles were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen. For the determination of enzyme distribution, hyperthermophiles were grown at temperatures between 80 and 90°C for about 24 h under anaerobic conditions (14). The medium (1 liter) consists of 5 g of tryptone, 1 g of yeast extract, 25 g of NaCl, 1 g of cysteine-HCl, 1.3 g of (NH4)2SO4, 0.28 g of KH2PO4, 0.25 g of MgSO4 · 7H2O, 0.07 g of CaCl2 · 2H2O, 0.02 g of FeCl3 · 6H2O, 1.8 mg of MnCl2 · 4H2O, 4.5 mg of Na2B4O7 · 10H2O, 0.22 mg of ZnSO4 · 7H2O, 0.05 mg of CuCl2 · 2H2O, 0.03 mg of Na2MoO4 · 2H2O, 0.03 mg of VOSO4 · 2H2O, 0.01 mg of CoSO4 · 7H2O, and 5 g of elemental sulfur. The pH of the medium was adjusted to 7.2 with 3 N NaOH. For enzyme purification, T. profundus DSM 9503 was anaerobically grown at 82°C for about 18 h using the same medium. The cells harvested by centrifugation (10,000 × g, 15 min) were washed with 3% NaCl and subsequently with 10 mM potassium phosphate buffer (pH 7.0) containing 1 mM EDTA and 10% glycerol. The washed cells were stored at −20°C until use.
Enzyme and protein assays.
Dye-l-proDH activity was assayed by measuring the reduction rate of Cl2Ind. The standard reaction mixture was composed of 100 mM l-proline, 0.1 mM Cl2Ind, 200 mM Tris-HCl buffer (pH 8.0), and enzyme in a total volume of 1.0 ml. The mixture without the substrate (l-proline) was previously incubated at 50°C for about 3 min in a cuvette with a 0.4-cm light path length, and then the reaction was started by the addition of l-proline. The initial decrease in absorbance at 600 nm was measured with a Shimadzu UV-160A recording spectrophotometer. One unit was defined as the amount of the enzyme catalyzing the reduction of 1 μmol of Cl2Ind/min at 50°C. The millimolar absorption coefficient (ɛ mM) of 21.5 mM−1 cm−1 at 600 nm was used for Cl2Ind (13). The reduction of ferricyanide, p-iodonitrotetrazolium violet (INT), nitroblue tetrazolium (NBT), and horse liver cytochrome c was monitored at 405 nm (ɛ mM = 1.04 mM−1 cm−1), 490 nm (ɛ mM = 15.0 mM−1 cm−1), 530 nm (ɛ mM = 36.0 mM−1 cm−1), and 553 nm (ɛ mM = 15.3 mM−1 cm−1), respectively. For the reduction of INT and NBT, phenazine methosulfate (PMS) was used as an electron-transfer intermediate. The protein concentration was determined by the method of Bradford with bovine serum albumin as a standard (3).
Purification of dye-l-proDH.
All steps in the purification were carried out at room temperature. Potassium phosphate buffer (10 mM, pH 7.0) containing 10% glycerol and 1 mM EDTA was basically used as the standard buffer system in the enzyme purification procedure unless otherwise stated.
(i) Preparation of crude extract.
The washed cells (ca. 20 g, wet weight) were suspended in 180 ml of the standard buffer system and were disrupted by sonication. The intact cells and cell debris were removed by centrifugation (10,000 × g, 10 min).
(ii) Red-Sepharose-CL4B affinity column chromatography.
The enzyme solution was applied on a Red-Sepharose-CL4B column (3.5 by 8 cm) previously equilibrated with the standard buffer system, and the enzyme was eluted with a linear gradient of 0 to 1.0 M NaCl in the same buffer. The active fractions were pooled, and then the enzyme solution was used for the next step.
(iii) Butyl-Toyopearl column chromatography.
Solid (NH4)2SO4 was added to the enzyme solution at up to a 10% saturation. The enzyme solution was applied on a Butyl-Toyopearl column (3.5 by 6 cm) which was previously equilibrated with the buffer supplemented with 10% (NH4)2SO4. After the column was washed with the same buffer (about 3 column-bed volumes), the enzyme was eluted with a linear gradient of 10 to 0% (NH4)2SO4 in the same buffer. The active fractions were pooled, and the solution was dialyzed for 18 h against 10 mM Tris-HCl buffer (pH 9.0) containing 10% glycerol and 1 mM EDTA. The enzyme solution was concentrated by ultrafiltration (UK-50 Ultrafilter; Advantec, Tokyo, Japan).
(iv) Uno Q column chromatography on fast-performance liquid chromatography (FPLC) system.
The enzyme solution was applied on a Uno Q column (0.7 by 3.5 cm) equilibrated with 10 mM Tris-HCl buffer (pH 9.0) containing 10% glycerol and 1 mM EDTA. The column was washed with the same buffer (3 column-bed volumes), and the enzyme was eluted with a linear gradient of 0 to 0.25 M NaCl in the same buffer. The active fractions were pooled and the solution was concentrated by ultrafiltration (UK-50).
(v) Superdex 200 column chromatography on FPLC system.
The enzyme was applied on a Superdex 200 column (2.6 by 60 cm) equilibrated with 10 mM Tris-HCl buffer (pH 8.0) containing 10% glycerol, 1 mM EDTA, and 0.25 M NaCl and then eluted with the same buffer. The active fractions were pooled, and the solution was used for various experiments after dialysis against 10 mM potassium phosphate buffer (pH 7.0) containing 10% glycerol and 1 mM EDTA.
Molecular mass determination by native-gradient PAGE.
The apparent molecular mass of the native enzyme was determined on the native-gradient polyacrylamide gel electrophoresis (PAGE) using a premade gel system (Daiichi Chemical, Tokyo, Japan) by the method essentially as described by Slater (19) with some modifications. The protein samples were dissolved in 20 mM Tris-HCl sample buffer (pH 6.8) and subsequently applied onto a 2 to 15% slab polyacrylamide gradient gel. PAGE was carried out at a constant current of 20 mA per slab for 2 h. We used 50 mM Tris–380 mM glycine (pH 8.3) as a running buffer. Protein bands were visualized by staining with Coomassie brilliant blue R-250.
Electrophoresis of native enzyme and determination of subunit molecular weight by SDS-PAGE.
Disk PAGE with a 7.5% polyacrylamide gel was performed according to the method of Davis (4). Activity staining was performed at 37°C in a mixture containing 0.3 M Tris-HCl buffer (pH 8.0), 100 mM l-proline, 0.04 mM PMS, and 0.1 mM INT until a red band of sufficient intensity was visible. Protein was stained with 0.025% Coomassie brilliant blue G-250 in 50% methanol and 10% acetate. Sodium dodecyl sulfate (SDS)-PAGE was carried out with 12.5% polyacrylamide gel according to the method of Laemmli (8). Maltose-binding protein (MBP)–β-galactosidase (175 kDa), MBP-paramyosin (83 kDa), glutamate dehydrogenase (62 kDa), aldolase (47.5 kDa), triosephosphate isomerase (32.5 kDa), β-lactoglobulin A (25 kDa), lysozyme (16.5 kDa), and aprotinin (6.0 kDa) were used as the molecular mass standards (New England Biolabs). The protein sample was boiled for 5 min in 10 mM Tris-HCl buffer (pH 7.0) containing 1% SDS and 1% 2-mercaptoethanol. Protein bands were visualized by staining with Coomassie brilliant blue R-250.
Identification of product in the enzymatic oxidation of l-proline.
The reaction product in the oxidation of l-proline with dye-l-proDH was identified by thin-layer chromatography. As the product, Δ1-pyrroline-5-carboxylate or Δ1-pyrroline-2-carboxylate is postulated. The two products, Δ1-pyrroline-5-carboxylate and Δ1-pyrroline-2-carboxylate are converted into glutamate and 4-aminobutyrate, respectively, by H2O2 oxidation as described elsewhere (10). The reaction mixture (80 μl) containing 100 mM potassium phosphate buffer (pH 7.5), 30 mM l-proline, and 0.5 U of enzyme was prepared, and the reaction was started by the addition of 100 mM Cl2Ind (1 μl) to the mixture. The reaction mixture was incubated at 25°C until the blue color of Cl2Ind disappeared. This oxidization procedure of l-proline by the addition of Cl2Ind was periodically repeated 20 times. The product was further oxidized by the addition of 30% H2O2 (5 μl) to the mixture. After completion of the reaction, the enzyme was denatured by the addition of 30% (wt/vol) perchloric acid (5 μl); and the precipitate was removed by centrifugation (10,000 × g, 5 min). The supernatant was neutralized with 2 M K2CO3 (12 μl), and the aliquot was subjected to thin-layer chromatography (silica gel 60F254 plate, 20 by 20 cm; Merck) together with l-proline, 4-aminobutylate, and l-glutamate solutions using a developing solvent (phenol-H2O, 75:25). The spots of product and three other authentic amino acids were localized with ninhydrin (0.25% [wt/vol] in acetone).
Extraction and determination of flavin.
The flavin compound from the enzyme was extracted with 1% perchloric acid as described elsewhere (13, 21). After the removal of the precipitate formed by centrifugation, the supernatant was used to identify the flavin compound by high-pressure liquid chromatography (HPLC) with a TSKgel ODS-80Ts column (4.6 by 150 mm; Tosoh). A linear gradient between 10 mM potassium phosphate buffer (pH 6.0) containing methanol (20% [vol/vol]) and methanol was used for the elution. The flow rate was 1.0 ml/min, and the total elution time was 15 min. FAD and FMN were monitored by determining the absorbance at 260 nm.
N-terminal amino acid analysis.
The N-terminal amino acid sequence of the enzyme was analyzed with an automated Edman degradation protein sequencer. The phenylthiohydantoin derivatives (Pth-Xaa) were separated and identified using the Protein Sequencer PPSQ-10 (Shimadzu).
l-Proline determination with dye-l-proDH.
l-Proline was determined by the rate assay method of dye-l-proDH using PMS and INT as electron carrier and electron acceptor, respectively. The reaction mixture for the calibration curve for the enzymatic determination of l-proline consisted of 200 mM Tris-HCl (pH 7.5), 0.10 mM PMS, 0.10 mM INT, dye-l-proDH (about 0.1 U in the assay with Cl2Ind), and various concentrations of l-proline in a total volume of 1.00 ml. The reaction mixture was incubated for 5 min at 50°C, and then the reaction was stopped by cooling it in ice water. The increase in the absorbance at 490 nm was measured.
RESULTS
Distribution of dye-l-proDH in hyperthermophilic archaea.
To find organisms that produce thermostable dye-dependent amino acid dehydrogenases, we screened enzymes which catalyze the reduction of C12Ind in the presence of amino acids such as l-proline, l-glutamate, l-aspartate, l-leucine, l-isoleucine, l-valine, l-lysine, l-phenylalanine, l-methionine, l-arginine, l-histidine, l-serine, l-threonine, and l-tyrosine among several strains of hyperthermophilic archaea from culture collections. As the result, we found a dye-l-proDH in four strains of the Thermococcales order: T. profundus DSM9503 (specific activity, 25 mU/mg), T. peptonophilus DSM 10343 (specific activity, 8.0 mU/mg), Pyrococcus furiosus DSM 3638 (specific activity, 2.0 mU/mg), and P. horikoshii OT-3 (specific activity, 5.0 mU/mg). Since T. profundus DSM 9503 exhibited the highest activity, this strain was chosen for the enzyme purification.
Purification of dye-l-proDH.
Many dye-DHs are known to be membrane-bound enzymes. The cells of T. profundus were disrupted by sonication and fractionated into particulate and supernatant fractions by ultracentrifugation with a Beckman Ultracentrifuge at 140,000 × g for 90 min. The activity of dye-l-proDH in the two fractions was measured. More than 90% of the activity was found in the supernatant fraction. This shows that the enzyme may be loosely bound to the cytoplasmic membrane and easily solubilized. Thus, we omitted the step for solubilization and fractionation of the enzyme from its purification procedures. The purification of dye-l-proDH from T. profundus is summarized in Table 1. In the purification procedure, dye-l-proDH bound weakly to the resin of Red-Sepharose-CL4B, and the chromatography was effective in removing contaminants such as NAD-dependent dehydrogenases and ATP-dependent kinases which may exhibit high affinity for the resin. Finally, the enzyme was purified about 110-fold, with an overall yield of about 10%. The purified enzyme was found to be homogeneous: the native PAGE of the enzyme gave a single protein band, which corresponded to that stained with the proline dehydrogenase activity.
TABLE 1.
Purification of dye-l-proDH from T. profundus
| Step | Total activity (U) | Total protein (mg) | Sp act (U/mg) | Yield (%) |
|---|---|---|---|---|
| Crude extract | 23.0 | 816 | 0.028 | 100 |
| Red-Sepharose-CL4B | 21.4 | 185 | 0.116 | 93 |
| Butyl-Toyopearl | 12.2 | 9.18 | 1.34 | 53 |
| Uno Q | 5.93 | 0.945 | 6.28 | 26 |
| Superdex 200 | 2.34 | 0.760 | 3.08 | 10 |
Molecular mass and subunit structure.
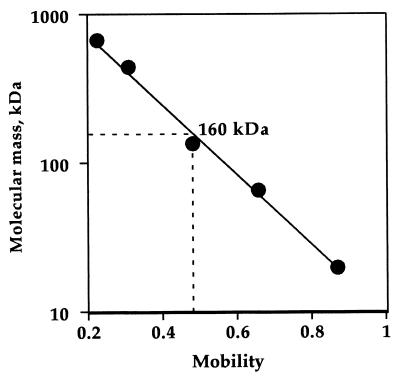
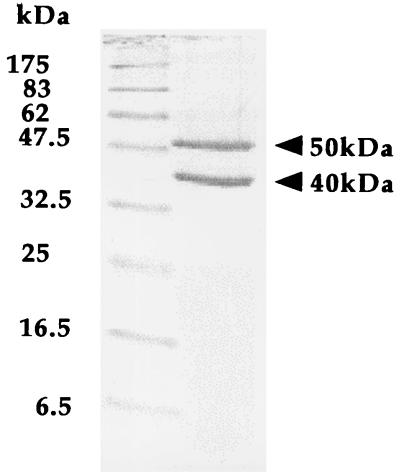
The molecular mass of the enzyme was determined to be about 160 kDa by native-gradient PAGE (Fig. 1). The subunit structure was examined by SDS-PAGE. The electrophoresis showed two different kinds of bands indicating that the enzyme consists of heterogeneous subunits (Fig. 2). The masses of the two subunits were estimated to be about 50 and 40 kDa from the standard curve obtained from SDS-PAGE. This shows that the enzyme molecule may be composed of a heterotetrameric structure of subunits, i.e., α2β2.
FIG. 1.
Molecular mass determination of the native dye-l-proDH. The molecular mass was determined on the native PAGE using a 2 to 15% gradient gel as described in the text. Cycloglobulin (669 kDa), ferritin (443 kDa), lactate dehydrogenase (140 kDa), bovine serum albumin (66 kDa), and trypsin inhibitor (20 kDa) were used as the molecular mass standards (Daiichi Chemical, Tokyo, Japan).
FIG. 2.
SDS-PAGE of the purified enzyme. Left column, molecular marker proteins; right column, the enzyme.
pH and temperature optima and thermostability.
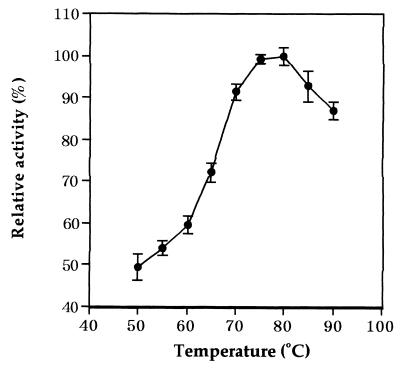
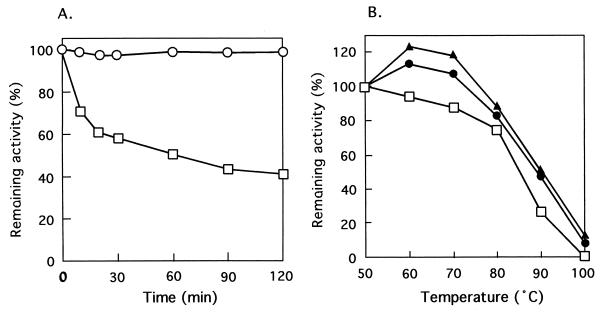
The enzyme activity was measured at various pHs at 50°C. Acetate, potassium phosphate, Tris-HCl, and glycine-KOH buffers were used in the assays for pH 5.0 to 6.0, pH 6.0 to 8.5, pH 8.0 to 9.0, and pH 9.0 to 10.0, respectively. After the assay, the pH of the reaction mixture was measured. The optimum pH was ca. 7.5 for the l-proline dehydrogenation. We were able to detect the enzyme activity at temperatures from 50 to 90°C using a ferricyanide as an electron acceptor (see below) in 200 mM Tris-HCl buffer (pH 7.5). The assay was started by the addition of enzyme after preincubation for 3 min at various temperatures. The reaction mixture minus l-proline was used as the control at each temperature. As shown in Fig. 3, the optimum temperature was ca. 80°C. The thermostability of dye-l-proDH in 10 mM potassium phosphate (pH 7.0) containing 1 mM EDTA and 10% glycerol was examined. The enzyme retained its full activity upon heating at 50°C for at least 120 min (Fig. 4A), but the activity gradually decreased with increasing temperature (Fig. 4B), The half-life at 80°C was about 60 min, and the enzyme was stabilized by the addition of 1 M NaCl and KCl. In the presence of 1 M NaCl and KCl, the enzyme did not lose its activity even with incubation at 70°C for 10 min as shown in Fig. 4B. In addition, when the enzyme was incubated between pH 4.0 and 10.0 at 50°C for 10 min, the activity totally remained. These results indicate that the hyperthermophile dye-l-proDH is very stable under various conditions.
FIG. 3.
Effect of temperature on the enzyme activity. The enzyme activity was measured at various temperatures in 200 mM Tris-HCl buffer (pH 7.5) by using a ferricyanide as an electron acceptor. Vertical bars represent the means and standard deviations from three independent experiments.
FIG. 4.
Thermostability of dye-l-proDH. (A) Effect of incubation time on thermostability of dye-l-proDH. The enzyme in potassium phosphate buffer (pH 7.0) containing 1 mM EDTA and 10% glycerol was incubated at 50°C (○) and 80°C (□), and the remaining activity of the aliquot was assayed at 50°C. (B) Effects of incubation temperature and the addition of two salts on the thermostability of dye-l-proDH. The enzyme in the buffer solution described in panel A was incubated at various temperatures for 10 min, and the aliquot was assayed at 50°C under the standard assay conditions (□) or with 1 M NaCl (▴) or 1 M KCl (●) added to the enzyme solution.
Substrate and electron acceptor specificity.
The ability of dye-l-proDH to catalyze the dehydrogenation of various amino acids was examined. The enzyme acted exclusively on l-proline. The following substrates were inert: d-proline, l-hydroxyproline, l-ornithine, l-glutamate, l-aspartate, l-arginine, l-lysine, l-serine, glycine, l-leucine, l-valine, and l-alanine. The electron acceptor specificity of the enzyme was examined. Ferricyanide, PMS-INT, and PMS-NTB, as well as Cl2Ind, exhibited electron acceptor activity (Table 2). Ferricyanide was the most preferred electron acceptor of the enzyme. NAD, NADP, and bovine heart cytochrome c were inert as the electron acceptor.
TABLE 2.
Electron acceptor specificity of dye-l-proDH
| Acceptor | Concn (mM) | Relative activity (%) |
|---|---|---|
| DCIP | 0.1 | 100 |
| PMS-INTa | 0.1 | 51 |
| PMS-NBTa | 0.1 | 24 |
| Ferricyanide | 1.5 | 207 |
| NAD | 1.25 | 0 |
| NADP | 1.25 | 0 |
| Cytochrome c | 0.1 | 0 |
A PMS concentration of 0.1 mM was used as an electron transfer intermediate.
Identification of reaction product.
The reaction product of l-proline dehydrogenation with the enzyme was identified by thin-layer chromatography. The reaction product was postulated to be Δ1-pyrroline-5-carboxylate or Δ1-pyrroline-2-carboxylate. Δ1-Pyrroline-5-carboxylate and Δ1-pyrroline-2-carboxylate were converted into glutamate and 4-aminobutyrate, respectively, by H2O2 treatment, and the two amino acids were separated and identified by thin-layer chromatography. As the result of the enzyme reaction, a ninhydrin spot corresponding to glutamate was detected on the plate in the thin-layer chromatography. This shows that the reaction product is Δ1-pyrroline-5-carboxylate but not Δ1-pyrroline-2-carboxylate.
Steady-state kinetics.
Steady-state kinetic analysis for l-proline dehydrogenation with Cl2Ind as the electron acceptor was carried out. Initial velocity experiments were done by varying the concentration of one substrate at a fixed concentration of other substrates as previously described (13). Double reciprocal plots of the initial velocity and the concentrations of substrates (l-proline and Cl2Ind) showed a series of parallel lines. The Km values were calculated from the secondary plot of the intercepts versus the reciprocal of the substrate concentration. From the plots of intercepts against l-proline, the Km value for l-proline was determined to be 2.05 mM. The Km value for Cl2Ind was similarly calculated to be 0.073 mM.
N-terminal amino acid sequences.
The N-terminal amino acid sequences of the two subunits were analyzed by an automated Edman degradation protein sequencer. The N-terminal amino acid sequences of the large α-subunit (23-amino-acid residues) and the small β-subunit (25-amino-acid residues) were determined to be MRLTEHPILDFSERRGRKVTIHF and XRSEAKTVIIGGGIIGLSIAYNLAK, respectively.
Absorption spectra and prosthetic group.
The purified enzyme did not lose its activity by dialysis against 10 mM potassium phosphate buffer (pH 7.0) containing 1 mM EDTA and 10% glycerol. The addition of 50 μM FAD or FMN to the enzyme solution had no effect on the activity.
The absorption spectrum of the purified T. profundus dye-l-proDH was detected. The enzyme showed two pronounced absorption peaks at ca. 380 and 450 nm in addition to that seen at 280 nm (data not shown). This spectrum shows that the enzyme may be a typical flavoprotein. The flavin compound in the extract of the purified enzyme with 1% parchloric acid was analyzed by HPLC. The flavin compound in the enzyme extract was identified to be FAD and not FMN.
Spectrophotometric determination of l-proline with dye-l-proDH.
The high specificity of dye-l-proDH for l-proline may be advantageous for use as an l-proline biosensor. Enzymatic determination of l-proline with dye-l-proDH using an electron transport system consisting of PMS and INT was examined. An increase in the absorbance at 490 nm after the incubation for 5 min at 50°C was measured. A linear relationship was obtained between the increase and the concentration of l-proline in the concentration range of 0.10 to 2.5 mM.
DISCUSSION
In this study, we found the occurrence of dye-l-proDH in several anaerobic hyperthermophilic archaea and first purified it to homogeneity from T. profundus DSM 9503, which exhibited the highest activity in the crude extract. In general, many kinds of dye-DHs such as succinate dehydrogenase and NADH dehydrogenase are localized on the surface of the cytoplasmic membrane and play an important role in incorporating electrons from the substrate into the electron transfer system (15, 20). Thus, the dye-l-proDH may function by electron incorporation from l-proline into the electron transfer system of the anaerobic hyperthermophilic archaea in Thermococcales such as Thermococcus and Pyrococcus species.
In the procedure of enzyme purification, the T. profundus dye-l-proDH appeared in the soluble fraction without a special solubilization procedure after the cell disruption by sonication. This suggests that the enzyme may be distributed on the surface of cytoplasmic membrane and may be released easily from the membrane similar to the case of the Mycobacterium phlei dye-linked l-malate dehydrogenase (7). The solubilization of many dye-DHs requires tedious procedures, such as surfactant extraction and ultracentrifugation, and often may result in a substantial loss of activity. Therefore, the easy solubilization of the T. profundus dye-l-proDH is advantageous for its large-scale preparation. In addition, the high stability of the enzyme is favorable for purification and application to bioprocesses.
Similar dye-l-proDH activity was found in a PutA protein in bacteria such as E. coli and serovar Typhimurium, which can use l-proline as a carbon and nitrogen source (9, 16). The PutA protein is known to be a bifunctional dehydrogenase with both dye-l-proDH and NAD-dependent Δ1-pyrroline-5-carboxylate dehydrogenase activities that catalyze the oxidation of l-proline to l-glutamate via Δ1-pyrroline-5-carboxylate (11, 12). Of the two dehydrogenases, the dye-l-proDH is a FAD-dependent dehydrogenase that interacts with the cytoplasmic membrane-associated respiratory chain (1). The E. coli dye-l-proDH catalyzes oxidation of l-hydroxyproline at a rate of 3% that found for l-proline, whereas d-proline and 17 common l-amino acids do not function as substrates (18). The enzyme from serovar Typhimurium is highly specific for l-proline and does not act on l-hydroxyproline or d-proline (12). The Km values for l-proline have been determined to be 60 and 83 mM for the E. coli and serovar Typhimurium enzymes respectively (12, 18). In substrate specificity, the T. profundus dye-l-proDH is similar to those enzymes and showed high specificity for l-proline. However, an extremely low Km value for l-proline (2.05 mM) in the T. profundus enzyme was recognized. The dye-l-proDH solubilized from the E. coli membrane consists of two subunits with a molecular mass of 124 kDa (the native molecular mass is 200 to 260 kDa) (18), and the primary structure has been determined by gene analysis (22). On the other hand, the molecular mass of the T. profundus dye-l-proDH is about 160 kDa with two different kinds of subunits (α2β2 structure) as shown in this study. Thus, the subunit structure of the T. profundus enzyme is largely different from that of the E. coli one. In addition, the N-terminal amino acid sequences in the two subunits of T. profundus enzyme exhibited low homology with that of the E. coli enzyme. In contrast, the N-terminal amino acid sequences of the larger subunit (α) of the T. profundus enzyme showed high homology (ca. 60%) with those of the P. abyssi Sox (sarcosine oxidase) A gene product and P. horikoshii d-nopaline dehydrogenase, which were postulated from the genome sequences (Fig. 5). The smaller subunit (β) of the T. profundus enzyme exhibited a high sequence homology (ca. 62%) with those of the P. horikoshii SoxB and rat liver dimethylglycine dehydrogenase precursor. We examined the activities of sarcosine oxidase, d-nopaline dehydrogenase, and dimethylglycine dehydrogenase with the T. profundus enzyme. The enzyme exhibited no activity for the three kinds of reactions. These observations strongly suggest that the T. profundus l-proDH may be a novel type of the FAD-dependent l-proline oxidoreductase. The complete sequencing of the two peptide of T. profundus enzyme is expected to afford more detailed information about their catalytically functional and molecular structural characteristics, and the gene cloning of the enzyme is now under investigation.
FIG. 5.
Alignment of N-terminal amino acid sequence. (A) N-terminal amino acid sequences of the α-subunit of T. profundus dye-l-proDH [T. pro dye-L-proDH(α)] and those of the putative SoxA gene product (P. aby SoxA; NCB accession no. C75144) from P. abyssi and putative d-nopaline dehydrogenase (P. hori D-nopDH; NCB accession no. H71183) from P. horikoshii. (B) N-terminal amino acid sequences of the β-subunit of T. profundus dye-l-proDH [T. pro dye-L-proDH(β)] and those of the putative SoxB gene product (P. hori SoxB; NCB accession no. B71184) from P. horikoshii and rat liver dimethylglycine dehydrogenase precursor (rat liver dmgDH; GenBank accession no. X55995). Asterisks represent conserved residues among the enzymes.
We showed here that T. profundus dye-l-proDH did not lose the activity at up to 70°C in the presence of KCl and NaCl. This shows that the enzyme is an extremely thermostable dye-DHs like Archaeoglobus fulgidus d-lactate dehydrogenase (17). In addition, the T. profundus dye-l-proDH exhibits high stability over a wide range of pH (pH 4 to 10) and can be stored for a long period of more than 3 months (data not shown) at a low temperature such as 4°C. This high stability is very useful for the simple purification of the enzyme and its application. We showed here that the enzyme could be applicable to the determination of l-proline. In particular, the high specificity of the enzyme for l-proline may be advantageous for the application to an l-proline biosensor. Development of the practical method for l-proline determination using the T. profundus dye-l-proDH is in progress.
REFERENCES
- 1.Abrahamson J L, Baker L G, Stephenson J T, Wood J M. Proline dehydrogenase from Escherichia coli K12: properties of the membrane-associated enzyme. Eur J Biochem. 1983;134:77–82. doi: 10.1111/j.1432-1033.1983.tb07533.x. [DOI] [PubMed] [Google Scholar]
- 2.Adams M W W, Kelly R M. Enzymes from microorganisms in extreme environments. Chem Eng News. 1995;December 18:32–42. [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Davis B J. Disc electrophoresis. 2. Method and application to human serum proteins. Ann N Y Acad Sci. 1975;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- 5.Dirmeier R, Keller M, Frey G, Huber H, Stetter K O. Purification and properties of an extremely thermostable membrane-bound sulfur-reducing complex from the hyperthermophilic Pyrodictium abyssi. Eur J Biochem. 1998;252:486–491. doi: 10.1046/j.1432-1327.1998.2520486.x. [DOI] [PubMed] [Google Scholar]
- 6.Frew J E, Hill H A. Electrochemical biosensors. Anal Chem. 1987;59:933A–944A. doi: 10.1021/ac00142a001. [DOI] [PubMed] [Google Scholar]
- 7.Imai T, Brodie A F. A phospholipid-requiring enzyme, malate-vitamin K reductase. J Biol Chem. 1973;248:7487–7494. [PubMed] [Google Scholar]
- 8.Laemmli U K. Cleavage of structural proteins during the assembly of the bacteriophase T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 9.Maloy S R. Cellular and molecular biology. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella typhimurium. Vol. 1. Washington, D.C.: American Society for Microbiology; 1987. pp. 1513–1519. [Google Scholar]
- 10.Meister A. The α-keto analogues of arginine, ornitine, and lysine. J Biol Chem. 1954;206:577–585. [PubMed] [Google Scholar]
- 11.Menzel R, Roth J. Purification of the putA gene product. A bifunctional membrane-bound protein from Salmonella typhimurium responsible for the two-step oxidation of proline to glutamate. J Biol Chem. 1981;256:9755–9761. [PubMed] [Google Scholar]
- 12.Menzel R, Roth J. Enzymatic properties of the purified PutA protein from Salmonella typhimurium. J Biol Chem. 1981;256:9762–9766. [PubMed] [Google Scholar]
- 13.Ohshima T, Tanaka S. Dye-linked l-malate dehydrogenase from thermophilic Bacillus species DSM 465. Purification and characterization. Eur J Biochem. 1993;214:37–42. doi: 10.1111/j.1432-1033.1993.tb17893.x. [DOI] [PubMed] [Google Scholar]
- 14.Ohshima T, Nishida N. Purification and characterization of extremely, thermostable glutamate dehydrogenase from a hyperthermophilic archaeon; Thermococcus litoralis. Biocatalysis. 1994;11:117–129. [Google Scholar]
- 15.Olsiewski P J, Kaczorowski G J, Walsh C. Purification and properties of d-amino acid dehydrogenase, an inducible membrane-bound iron-sulfur flavoenzyme from Escherichia coli B. J Biol Chem. 1980;255:4487–4494. [PubMed] [Google Scholar]
- 16.Ostrovsky de Spicer P, Maloy S. PutA protein, a membrane-associated flavin dehydrogenase, acts as a redox-dependent transcriptional regulator. Proc Natl Acad Sci USA. 1993;90:4295–4298. doi: 10.1073/pnas.90.9.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reed D W, Hartzell P L. The Archaeoglobus fulgidusd-lactate dehydrogenase is a Zn2+ flavoprotein. J Bacteriol. 1999;181:7580–7587. doi: 10.1128/jb.181.24.7580-7587.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scarpulla R C, Soffer R L. Membrane-bound proline dehydrogenase from Escherichia coli. Solubilization, purification, and characterization. J Biol Chem. 1978;253:5997–6001. [PubMed] [Google Scholar]
- 19.Slater G G. Stable pattern formation and determination of molecular size by pore-limit electrophoresis. Anal Chem. 1969;41:1039–1041. doi: 10.1021/ac60277a003. [DOI] [PubMed] [Google Scholar]
- 20.Thomson J W, Shapiro B M. The respiratory chain NADH dehydrogenase of Escherichia coli: isolation of an NADH:quinone oxidoreductase from membranes and comparison with the membrane-bound NADH:dichlorophenolindophenol oxidoreductase. J Biol Chem. 1981;256:3077–3084. [PubMed] [Google Scholar]
- 21.Willie A, Jorns M S. Discovery of a third coenzyme in sarcosine oxidase. Biochemistry. 1995;34:16703–16707. doi: 10.1021/bi00051a019. [DOI] [PubMed] [Google Scholar]
- 22.Xia M, Zhu Y, Cao X, You L, Chen Z. Cloning, sequencing and analysis of a gene encoding Escherichia coli proline dehydrogenase. FEMS Microbiol Lett. 1995;127:235–242. doi: 10.1111/j.1574-6968.1995.tb07479.x. [DOI] [PubMed] [Google Scholar]