Through analysis of preclinical models from endocrine receptor (ER)−/HER2+ and triple-negative (TN) inflammatory breast cancers (BCs) and of clinical BC samples, Villodre et al. (1) suggest that NDRG1 critically contributes to tumor growth and metastasis in aggressive ER-negative BCs. NDRG1 depletion reduces colony formation, migration, and invasion; number of tumor-initiating cells; and mTOR/AKT signaling in vitro and primary tumor growth and brain metastasis in vivo. NDRG1 expression is associated with aggressiveness features of clinical samples (ER−, TNBC, HER2+, high grade, metastatic samples) and shorter overall survival in multivariate analysis, including pathological stage and ER status. Because previous literature describes NDRG1 as a metastasis suppressor in less aggressive ER+ BC cell lines, the authors suggest that “NDRG1 has a context-dependent function in BC.” In fact, the clinical relevance of NDRG1 expression in BC has never been assessed in series large enough to allow analysis by molecular subtype. To fill this gap, we retrospectively examined NDRG1 mRNA expression in 8982 primary BC samples pooled from 36 public datasets (Supplementary Table 1, available online) (2), including notably 5929 ER+/HER2− and 1936 TN cases. The methods used are detailed in the Supplementary Methods (available online).
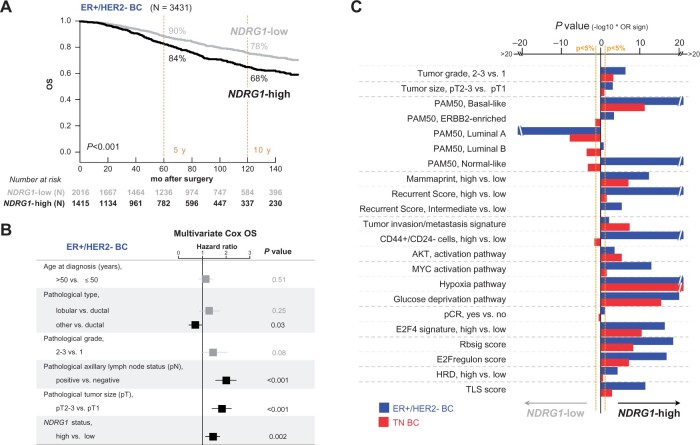
We confirmed the independent unfavorable prognostic value of high NDRG1 expression (cutoff = median) for overall survival in the whole population in multivariate analysis, including pathological stage, but also, in contrast with Villodre et al. (1), other major prognostic variables such as pathological tumor grade, type, and molecular subtypes (hazard ratio [HR] = 1.34, 95% confidence interval [CI] = 1.13 to 1.60). In analysis per molecular subtype, we found independent poor-prognosis value in the aggressive TN subtype (HR = 1.59, 95% CI = 1.01 to 2.49) but also in the less aggressive ER+/HER2− subtype (HR = 1.40, 95% CI = 1.14 to 1.73; Figure 1, A and B; Supplementary Table 2, available online). We extended NDRG1 association analyses to the ER+/HER2− samples (Figure 1, C): high expression was associated with aggressiveness features (high tumor grade and size, basal and ERBB2-enriched PAM50 subtypes, high relapse risk according to Mammaprint and Recurrence Score) and with signatures of “tumor invasion/metastasis” (3), CD44+/CD24− tumor-initiating cells (4), and AKT activation (5). As expected, NDRG1 expression was associated with signatures of MYC activation, hypoxia, and glucose deprivation (5). Combined with Villodre et al.’s results, the similarity of these correlations with those observed in aggressive TNBCs (Figure 1, C) suggests that NDRG1 may also serve as a therapeutic and/or prognostic target in the less aggressive ER+/HER2− subtype.
Figure 1.
Association of NDRG1 expression with overall survival (OS) and clinicopathological and molecular features. A) Kaplan-Meier postoperative OS curves according to NDRG1 status in endocrine receptor (ER)+/HER2− breast cancers (BCs). P values were calculated using a log-rank test. B) Multivariate Cox regression analyses for OS in ER+/HER2− BCs. P values were calculated using a Wald test. C) Association of NDRG1 mRNA status with clinicopathological and molecular features in ER+/HER2− BCs and triple-negative (TN) BCs. The statistical significance was assessed using a logistic regression (Logit link function). The bar plots represent the P values for association of each feature with NDRG1 in both ER+/HER2− and TN subtypes. The P values were −log10 transformed and pondered by the regression coefficient sign (positive when higher in NDRG1-high and negative when lower); the longer the bar, the lower the P value. All statistical tests were 2-sided. HRD = homologous recombination deficiency; pCR = pathological complete response; TLS = tertiary lymphoid structures.
We then compared NDRG1-high with NDRG1-low ER+/HER2− tumors regarding vulnerability or response to current and future systemic therapies of ER+/HER2− BC (2). The pathological complete response rate to neoadjuvant chemotherapy was not different. Differences existed for other therapies: NDRG1-high tumors showed lower probability of response to hormo ne therapy (E2F4-activation signature) and CDK4/6-inhibitors (RBsig and E2F-regulon scores), but higher probability of response to PARP-inhibitors (Homologous Recombination Deficiency signature) and immune checkpoint-inhibitors (TLS score). Analysis of ER+/HER2− BC cells using publicly available drug sensitivity data (6) and proteomics data (7) showed positive correlations between NDRG1_pT346 expression and sensitivity to MK-2206 AKT-inhibitor (P = 1.60E-2) and alpelisib PI3K-inhibitor (P = 6.99E-02) without correlation with sensitivity to AZD8055 dual mTOR-inhibitor.
Our observations not only reinforce the results of Villodre et al. (1) in a larger clinical series of aggressive BCs but also suggest potential prognostic, predictive, and/or therapeutic relevance for NDRG1 in less aggressive ER+/HER2− BCs, which deserves further investigation.
Funding
This work has been supported by grants from the Ligue Nationale Contre Le Cancer (EL2019.LNCC/FB, EL2022/FB), and Association Ruban Rose (FB).
Notes
Role of the funder: The study sponsors had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
Disclosures: The authors have no conflict of interest to disclose.
Author contributions: Conceptualization of the research: AN, PF, and FB; Supervision: FB; Writing-original draft: AN and FB; Writing-review and editing: AN, PF, EM, and FB.
Data Availability
All data sets of primary breast cancer were collected from and are available in public databases listed in Supplementary Table 1 (available online). Cell lines drug sensitivity data and proteomics data are publicly available and were derived from the Genomics of Drug Sensitivity in Cancer dataset (6) and protein array dataset from the Cancer Cell Line Encyclopedia (7) using the online DepMap web tool (https://depmap.org, version 21Q4).
Supplementary Material
Contributor Information
Alexandre de Nonneville, Laboratory of Predictive Oncology, Equipe labellisée Ligue Nationale Contre Le Cancer, Centre de Recherche en Cancérologie de Marseille, Institut Paoli-Calmettes, INSERM UMR1068, CNRS UMR725, Marseille, France.
Pascal Finetti, Laboratory of Predictive Oncology, Equipe labellisée Ligue Nationale Contre Le Cancer, Centre de Recherche en Cancérologie de Marseille, Institut Paoli-Calmettes, INSERM UMR1068, CNRS UMR725, Marseille, France.
Emilie Mamessier, Laboratory of Predictive Oncology, Equipe labellisée Ligue Nationale Contre Le Cancer, Centre de Recherche en Cancérologie de Marseille, Institut Paoli-Calmettes, INSERM UMR1068, CNRS UMR725, Marseille, France.
François Bertucci, Laboratory of Predictive Oncology, Equipe labellisée Ligue Nationale Contre Le Cancer, Centre de Recherche en Cancérologie de Marseille, Institut Paoli-Calmettes, INSERM UMR1068, CNRS UMR725, Marseille, France.
References
- 1. Villodre ES, Hu X, Eckhardt BL, et al. NDRG1 in aggressive breast cancer progression and brain metastasis [published online ahead of print]. J Natl Cancer Inst. 2021;djab222. doi: 10.1093/jnci/djab222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bertucci F, Finetti P, Goncalves A, Birnbaum D. The therapeutic response of ER+/HER2− breast cancers differs according to the molecular basal or luminal subtype. NPJ Breast Cancer. 2020;6(1):1–7. doi: 10.1038/s41523-020-0151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Desmedt C, Haibe-Kains B, Wirapati P, et al. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res. 2008;14(16):5158–5165. doi: 10.1158/1078-0432.CCR-07-4756. [DOI] [PubMed] [Google Scholar]
- 4. Creighton CJ, Li X, Landis M, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA. 2009;106(33):13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gatza ML, Lucas JE, Barry WT, et al. A pathway-based classification of human breast cancer. Proc Natl Acad Sci USA. 2010;107(15):6994–6999. doi: 10.1073/pnas.0912708107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Picco G, Chen ED, Alonso LG, et al. Functional linkage of gene fusions to cancer cell fitness assessed by pharmacological and CRISPR-Cas9 screening. Nat Commun. 2019;10(1):2198. doi: 10.1038/s41467-019-09940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ghandi M, Huang FW, Jané-Valbuena J, et al. Next-generation characterization of the cancer cell line encyclopedia. Nature. 2019;569(7757):503–508. doi: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data sets of primary breast cancer were collected from and are available in public databases listed in Supplementary Table 1 (available online). Cell lines drug sensitivity data and proteomics data are publicly available and were derived from the Genomics of Drug Sensitivity in Cancer dataset (6) and protein array dataset from the Cancer Cell Line Encyclopedia (7) using the online DepMap web tool (https://depmap.org, version 21Q4).