Abstract
Sulfonolipids (SoLs) are a unique class of sphingolipids, featuring a sulfonate group compared to other sphingolipids. However, the biological functions and biosynthesis of SoLs in human microbiota have been poorly understood. Here we report the discovery and isolation of SoLs from a human opportunistic pathogen Chryseobacterium gleum DSM16776. We show for the first time the pro-inflammatory activity of SoLs with mice primary macrophages. Furthermore, we used both in vivo heterologous expression and in vitro biochemical reconstitution to characterize two enzymes, cysteate synthase and cysteate fatty acyltransferase, that are specifically involved in the biosynthesis of SoLs rather than other sphingolipids. Based on these two SoL-specific enzymes, our bioinformatics analysis showed a wider distribution of SoLs biosynthetic genes in microbes that had not been reported as SoLs producers. We selected four of these strains and verified their cysteate synthase and cysteate fatty acyltransferase activities in SoLs biosynthesis. Considering this wider distribution of SoL-specific biosynthetic enzymes in the context of SoLs’ activity in mediating inflammation, a common and fundamental biological process, it may suggest a more comprehensive function of SoLs at play.
Graphical Abstract
INTRODUCTION
Trillions of microbes colonize in all parts of the human body, including the skin, reproductive organs, oral cavity, and the intestines.1 Some of these microbial tribes are pathogenic and can contribute to a variety of disease conditions such as inflammation.2 Like other microorganisms that rely heavily on their small molecule metabolites to communicate with the environment, human symbiotic bacteria can also interact with the human host through such metabolites.3,4 However, the structures, functions, and biosynthesis of these molecules remain poorly understood.
Chryseobacterium bacteria are emerging causative pathogens involved in human infectious diseases.5,6 Among them, C. gleum species are resistant to many antibiotics, especially to β-lactam antibiotics by encoding metallo-β-lactamases.7,8 Recently, C. gleum has been frequently detected in neonatal respiratory tract infection,9 sepsis,10 urinary tract infection,11 and other inflammatory conditions. In our search for functional metabolites of human microbiota that may mediate microbe-host interactions, we observed that the butanone extract of C. gleum DSM16776, originally isolated from the female high vaginal swab, can cause a strong induction of inflammatory cytokines. This observation suggests that the infection caused by C. gleum might be partially mediated by the small molecule metabolites.
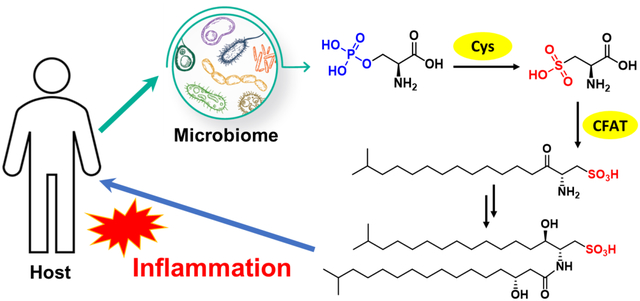
Here we describe the discovery of an unusual class of sphingolipids, sulfonolipids (SoLs), from the pathogenic bacterium C. gleum DSM16776, using bioassay-guided isolation in combination with mass spectrometry-based metabolomic analysis. While SoLs have been reported to possess cytotoxicity,12 antagonistic activity on Willebrand factor receptors,13 inhibitory effect on DNA polymerase,14 and inducing multicellular colonies of choanoflagellates,15–18 this is the first report on the pro-inflammatory activity of SoLs produced by a human pathogen, implying a potential role of this class of metabolites in mediating pathogenicity. Furthermore, we characterized two unique enzymes involved in the biosynthesis of SoLs but not other sphingolipids. We showed that these two SoL-specific enzymes catalyzed the conversion of L-phosphoserine to L-cysteate (named cysteate synthase; Cys) and the condensation between cysteate and fatty acyl-CoA (named cysteate fatty acyltransferase; CFAT). Interestingly, CFAT showed strict substrate specificity for cysteate rather than serine, with the latter being involved in the biosynthesis of other sphingolipids. Subsequently, our bioinformatic analysis revealed that homologs of Cys and CFAT are widely distributed in other bacteria that had not been reported as SoLs producers. The wide distribution of these SoL-specific biosynthetic enzymes, combined with SoLs’ activity in inducing inflammation, one of the most common and fundamental biological processes, may suggest a more profound role of SoLs in mediating microbe-host interactions than previously realized.
RESULTS AND DISCUSSION
Discovery of SoLs as A Pro-inflammatory Factor from C. gleum DSM16776.
In our search for pro-inflammatory functional metabolites of human microbiota, we observed the activity of the butanone extract of C. gleum DSM16776 from a female high vaginal swab in inducing inflammatory cytokines such as IL-1 and IL-6. After fractionation using silica gel chromatography, five major fractions were obtained and fraction F03 eluted by DCM:MeOH 10:1 exhibited the highest activity in inducing IL-1α (12-fold increase), IL-1ß (8-fold increase), and IL-6 (2-fold increase). We next implemented a bioassay guided fractionation combining UPLC-HR-QTOF MS-based metabolic analysis to identify active metabolites from F03. MS2 analysis of this fraction showed peaks with typical fragment ion at m/z 79.9576, corresponding to SO3−• group in negative ion mode. Then, we used a fragment ion scanning strategy to recognize peaks featuring sulfonic residues from the fraction, and observed many targeted molecules (Figure 1A, Supporting Information Figures S1–S6). Molecular networking analysis can cluster molecules with similar fragmentation patterns and visualize their tandem MS data as a relational spectral network. Therefore, we deconvoluted and filtered the raw UPLC-MS2 data of the fraction using MZmine program to get entries with abundant intensity (> 5.0 E3). Next, product ions with diagnostic m/z 79.95 were further filtered. Subsequently, the refined dataset was submitted to the online platform Global Natural Products Social Molecular Networking (GNPS) to generate a molecular network associated with sulfonic residue-containing metabolites (Figure 1B). Due to the limitation of data capacity in public databases, no hits of the targeted cluster were annotated. Compound Structure Identification (CSI): FingerID is a computational approach to searching molecular structure which combines MS/MS fragmentation tree computation and machine learning.19 Sirus 4 is a powerful tool that integrates CSI:FingerID for searching in molecular structure databases.20 Thus, we submitted the MS2 data of two typical sulfonic acid-containing metabolites (m/z 618.48 and 574.44) to Sirus 4.9.12 software, which were identified as sulfobacins A and B (SoLs A and B), respectively. Then we enlarged cultivation of C. gleum DSM16776 and isolated SoL A as the major SoL compound from the extract using various column chromatograph and preparative HPLC methods. The structure of SoL A was further confirmed by 1D and 2D NMR spectroscopy (Figure 1C, Supporting Information Figures S7–S13 and Table S3).21
Figure 1.
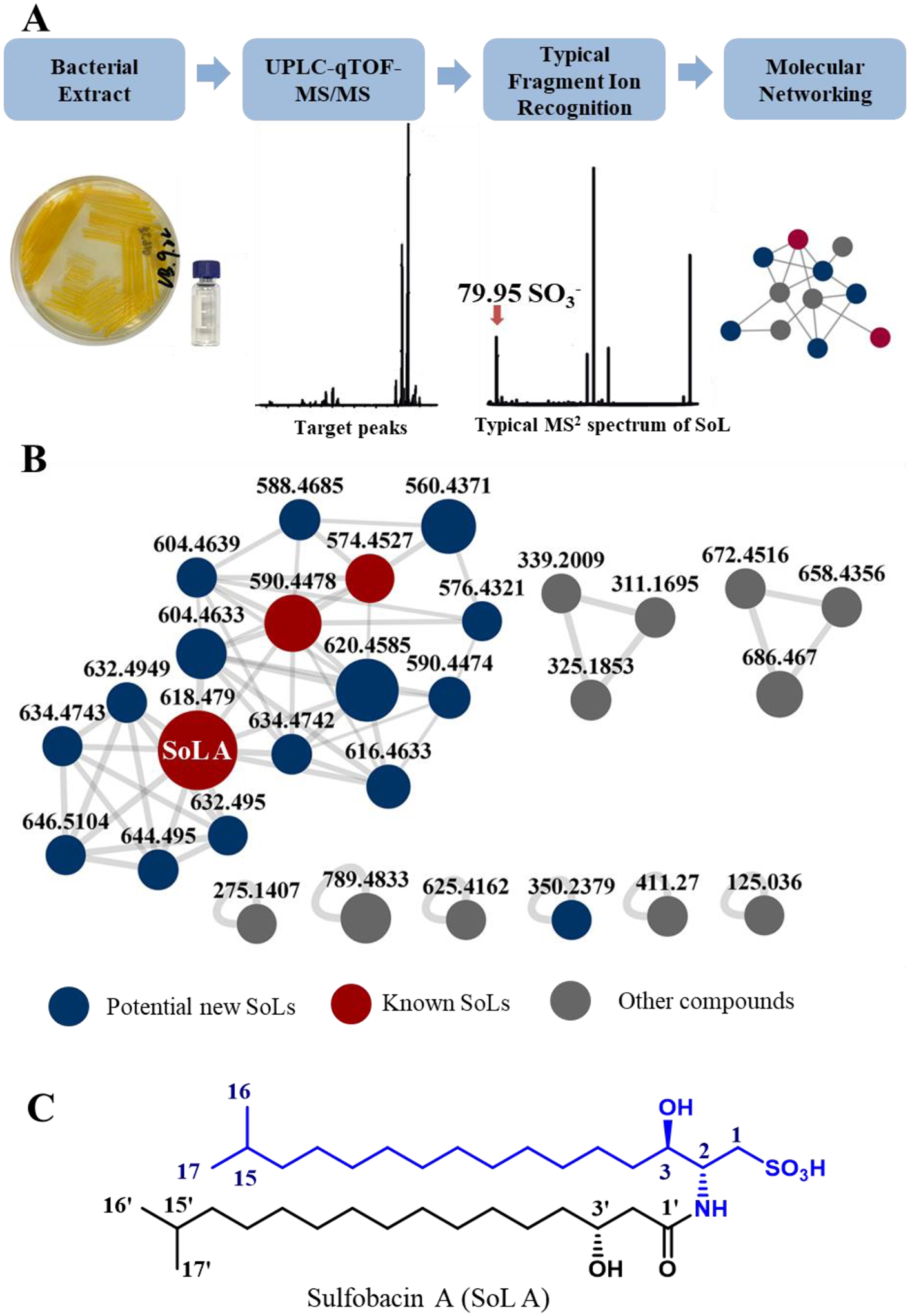
(A) Diagrammatic workflow for SoLs discovery from human opportunistic pathogen C. gleum DSM16776. (B) Negative ion mode feature-based molecular networking (FBMN) of C. gleum extracts. (C) Chemical structure of a major SoL compound, SoL A, isolated from C. gleum (capnine moiety in blue).
We next evaluated the pro-inflammatory activity of pure SoL A as a representative SoL member. We first measured the expression of inflammatory genes by RT-PCR analysis as this approach can test multiple genes simultaneously. The expression levels of several pro-inflammatory cytokines including IL-1α, IL-1ß, IL-6, and TNFα were significantly increased when the peritoneal macrophages isolated from wild type male C57Bl/6 mice were treated with 10 μM of SoL A (Figure 2A). Since the induction of these cytokines by SoL A showed a similar tread to that induced by lipopolysaccharide (LPS), a well-known compound that induces inflammation,22,23 we hypothesized that SoL A and LPS may share a similar mechanism in inducing pro-inflammatory cytokines. Thus, we tested the cytokine inducing effect of a mixture of SoL A and LPS. Intriguingly, the mixture of SoL A (10 μM) and LPS (100 ng/mL) showed less potency than LPS (100 ng/mL) alone in inducing IL-6 and IL-1ß (Figure 2B), suggesting that SoL A and LPS may have different pro-inflammatory mechanisms. This observation may also explain a previously observed anti-inflammatory effect of SoL B in a mouse model of LPS-induced acute inflammation, which showed a suppression of TNFα by SoL B.14 Thus, the seemingly bifunctional inflammatory mediation of SoLs may involve interesting mechanisms and is worth of further investigations. We also evaluated the pro-inflammatory activity of SoL A at the protein level using an ELISA assay (Figure 3) and observed the similar trend to that observed at the mRNA level.
Figure 2.
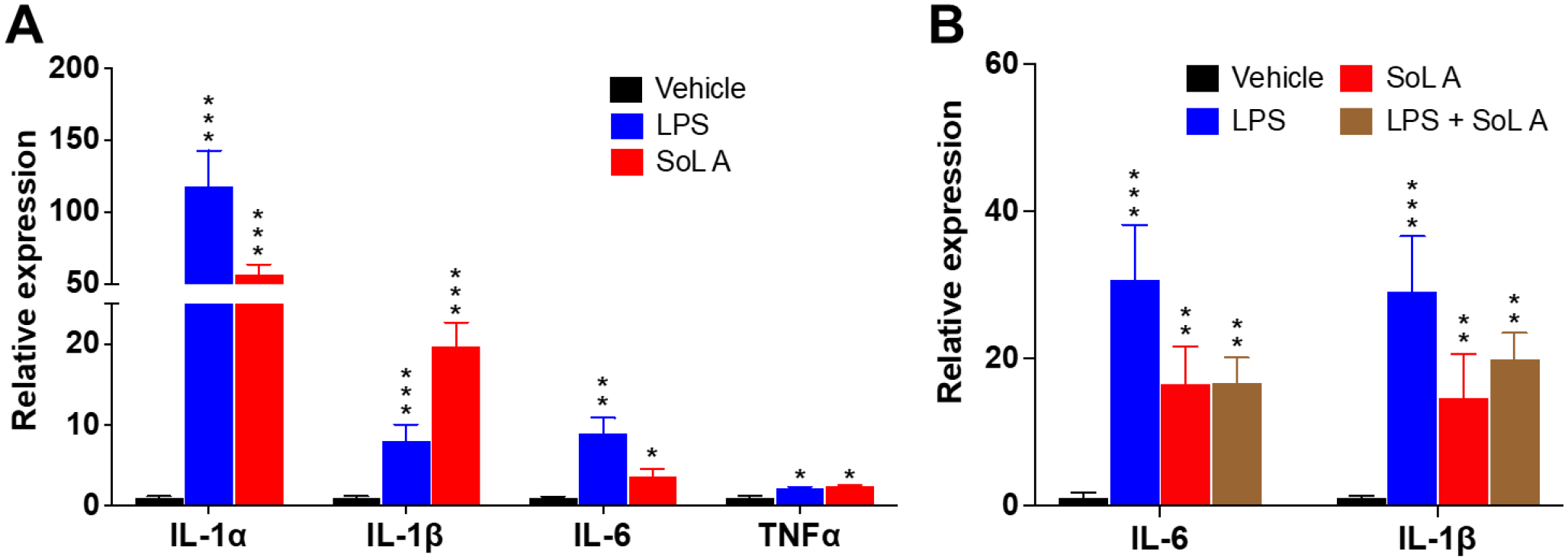
SoL A induced inflammatory response in primary macrophages isolated from wild type male C57Bl/6 mice. (A). Increased mRNA levels of pro-inflammatory cytokines induced by 10 μM of SoL A. LPS (100 ng/mL) used as a control; (B) Increased mRNA levels of IL-6 and IL-1ß induced by SoL A (10 μM) and a mixture of SoL A (10 μM) + LPS (100 ng/mL). LPS (100 ng/mL) used as a control. For each treatment, n=3. *, p<0.05; **, p< 0.01; ***, p< 0.001, versus vehicle.
Figure 3.
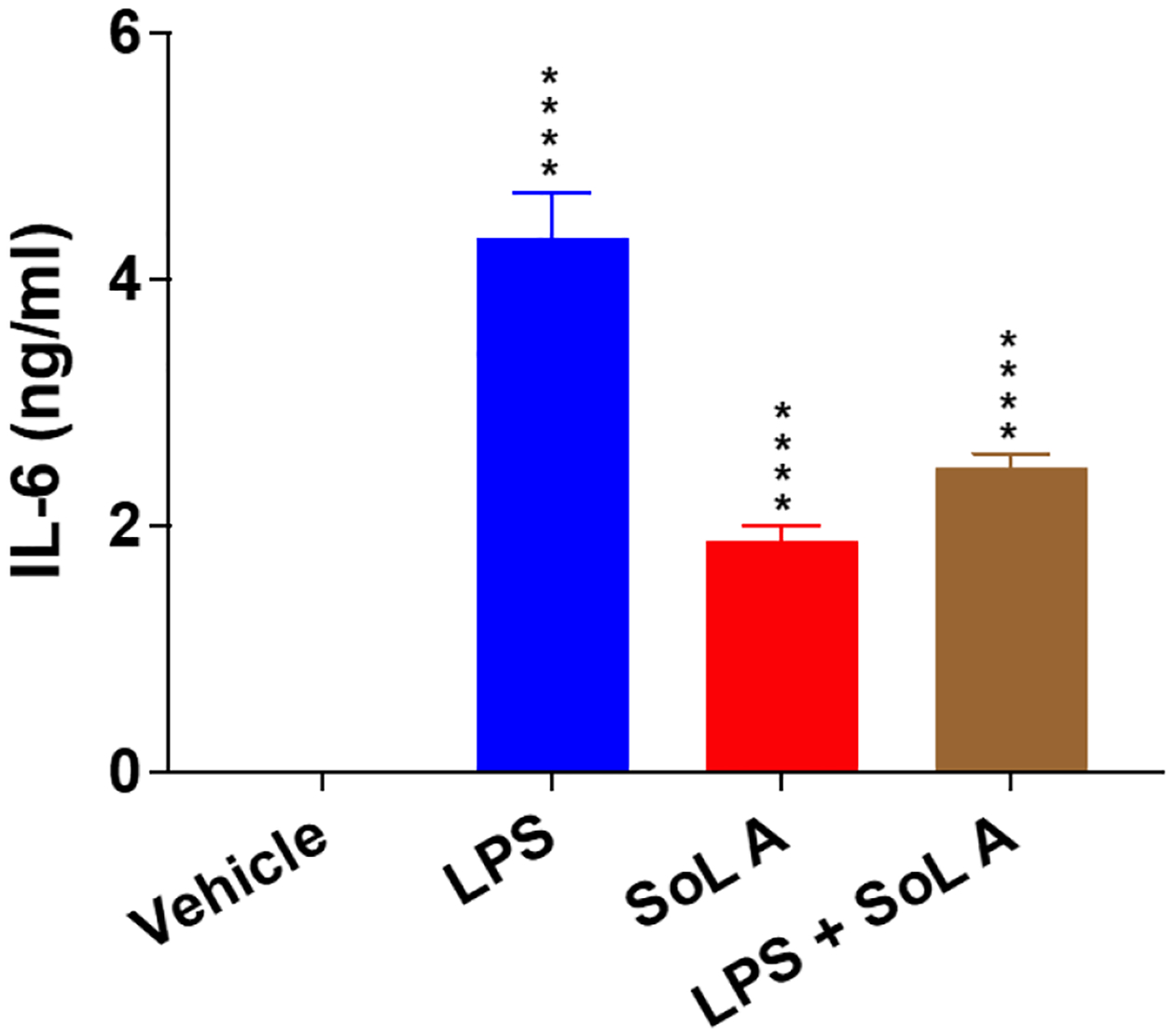
SoL A induced IL-6 protein production and secretion in macrophages. LPS (100 ng/mL) used as a control. For each treatment, n=3. ****, p< 0.0001, versus vehicle.
Two Key Enzymes, Cysteate Synthase (Cys) and Cysteate Fatty Acyltransferase (CFAT), Involved in SoL A Biosynthesis
SoLs are an unusual class of sphingolipid, in which the hydroxyl group of sphingosine is substituted by a sulfonate.24 Compared with sphingosine, capnine is the precursor generated from cysteic acid and fatty acyl-CoA (Figure 1C).25 According to previous research,26,27 we proposed that there are four enzymes involved in the biosynthesis of SoLs (Supporting Information Figure S14). The first enzyme, Cys, is responsible for the synthesis of L-cysteate that subsequently serves as the substrate for the second enzyme, CFAT, to create the capnine core. Then, following a 3-keto reduction and attachment of an amide linked fatty acid, different SoLs are created.24 Among these enzymes, Cys and CFAT are unique for SoL biosynthesis.
Characterization of Cys-catalyzed Conversions of L-phosphoserine to L-cysteate.
The PLP (pyridoxal 5’-phosphate)-dependent enzyme MA3297, identified from Methanosarcina acetivorans, was previously confirmed to convert L-phosphoserine and sulfite to L-cysteate and phosphate.28 The OrfA in Streptomyces lincolnensis, a homolog of MA3297, can also convert L-phosphoserine to L-cysteate that is a moiety of cysteoamide.29 Bioinformatic search using OrfA as a query in C. gleum DSM16776 genome revealed that there are three candidate homologs, Cys1, Cys2, and Cys3, which showed sequence identity of 12%, 14%, and 17%, respectively (Table 1). The low sequence identity indicates that these three homologs may have different functions or substrate specificity. To test their activity, we first cloned the cys1–3 genes from the genomic DNA of C. gleum DSM16776 and expressed these genes in E. coli BL21 (DE3). Subsequently, we performed an in vitro biochemical assay following the purification of Cys1–3 (Supporting Information Figure S15). This assay was performed by adding enzyme Cys and substrates including L-phosphoserine and sodium sulfite (Figure 4). The reaction without enzyme Cys or sodium sulfite were designed as the negative control. To facilitate the analysis of the final products, Marfey’s method producing L-FDAA derivatives was used (Supporting Information Figure S16) and the L-FDAA derivatives were detected by high-performance liquid chromatography (HPLC). By comparing the retention times and mass data of the L-FDAA derivatives with those derivatized from standard amino acids, we observed that Cys1 and Cys2 converted L-phosphoserine and sodium sulfite to L-cysteate and phosphate, and the catalytic efficiency of Cys1 was better than Cys2 (Figures 4A and 4B), while Cys3 did not exhibit this activity (Figure 4C). Thus, Cys1 and Cys2 were the candidate enzymes involved in the biosynthesis of SoLs. Cys1 and Cys2 may be isozymes and may contribute to increasing SoLs yields and chemical diversity, which may play a role in the biological process of the producing strains.
Table 1.
Homologs of OrfA and SpSpt in C. gleum DSM16776
| Query protein | Accession number | Target protein | Size (aa) | Accession number | Identity/Similarity (%) | Proposed function |
|---|---|---|---|---|---|---|
| OrfA | ANS62969 | Cysl | 346 | EFK34534.1 | 12/27 | pyridoxal-phosphate dependent protein |
| Cys2 | 331 | EFK33362.1 | 14/28 | putative cystathionine beta-synthase | ||
| Cys3 | 304 | EFK34612.1 | 17/27 | cysteine synthase A | ||
| SpSpt | Q93UV0 | CFAT1 | 425 | EFK34533.1 | 30/48 | aminotransferase, class I/II |
| CFAT2 | 399 | EFK35504.1 | 31/52 | glycine C-acetyltransferase | ||
| CFAT3 | 419 | EFK35137.1 | 33/51 | aminotransferase, class I/II |
Figure 4.
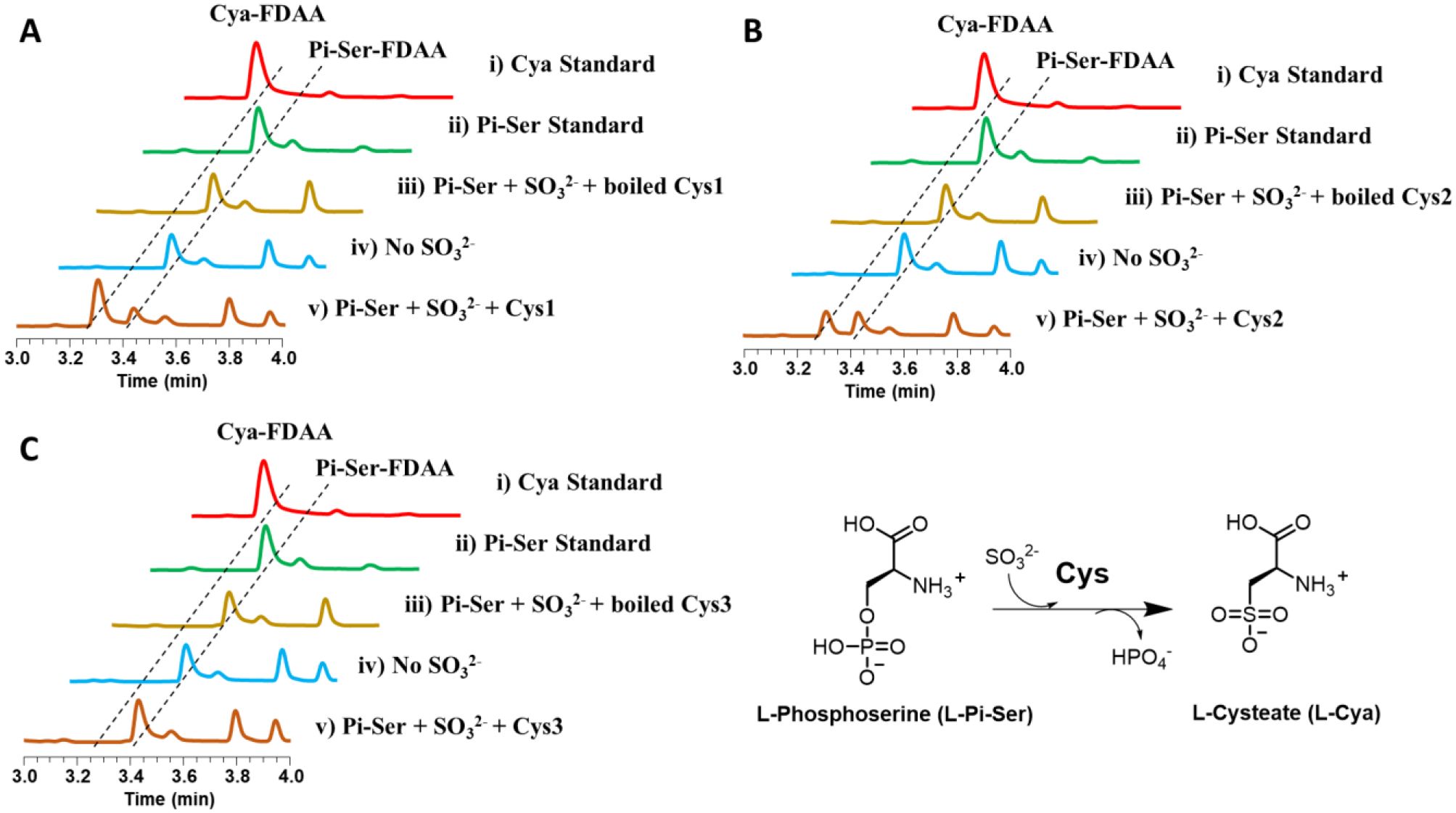
HPLC analysis of in vitro Cys assays followed by derivatization with L-FDAA. (A) Cys1, (B) Cys2, (C) Cys3. Reactions were performed as follows: (i) L-cysteate standard, (ii) L-phosphoserine standard, (iii) L-phosphoserine + Na2SO3 + boiled Cys, (iv) L-phosphoserine + Cys, and (v) L-phosphoserine + Na2SO3 + Cys. Cya: cysteate; Cya-FDAA: cysteate-FDAA derivative; Pi-Ser: phosphoserine; Pi-Ser-FDAA: phosphoserine-FDAA derivative.
Characterization of CFAT-catalyzed Condensation of Cysteate and Fatty Acyl-CoA.
Serine palmitoyltransferase (Spt), a PLP dependent enzyme, has been well reported in many eukaryotes to catalyze the first step in the de novo sphingolipid biosynthesis pathway. Spt has been characterized for catalyzing the condensation of serine and palmitoyl-CoA to form 3-ketodihydrosphingosine (3-KDS), the sphingoid base which is the starting point for all sphingolipids. Spt is ubiquitous in eukaryotes and is also found in certain prokaryotes. The first purification of a bacterial Spt, SpSpt from Sphingomonas paucimobilis, was reported in 200130 and it catalyzed the condensation of L-serine and palmitoyl-CoA to produce 3-KDS by heterologous expression in E. coli. We speculated that Spt homolog(s) in C. gleum DSM16776 may catalyze the condensation of 13-methylmyristoyl-CoA and cysteate to form 2-amino-3-keto-15-methylhexadecane-1-sulfonic acid (3-keto capnine). Due to taking cysteate, instead of serine, as the substrate, cysteate fatty acyltransferase (CFAT) is used to stand for such Spt homologs in this paper. After bioinformatic search using SpSpt as a query in C. gleum DSM16776, three candidate homologs, CFAT1, CFAT2, and CFAT3, showed sequence identity of 30%, 31%, and 33%, respectively (Table 1). Sequence alignment of Q93UV0 (SpSpt) with CFAT1–3 revealed that CFAT1–3 contain the conserved lysine residue required for PLP binding (Supporting Information Figure S17). Interestingly, we found that cfat1 gene was adjacent to cys1 in the genome of C. gleum DSM16776 (Supporting Information Figure S18). We cloned the cfat1-3 genes from the genome of this strain and transferred the individual recombinant plasmids to E. coli BL21 (DE3) (Supporting Information Table S1). To investigate the activity of cfat1-3, in vivo feeding assay was performed. After feeding 2 mM cysteate in E. coli BL21 (DE3) harboring cfat1, cfat2, or cfat3, the LC-MS analysis showed that only E. coli BL21 (DE3) containing cfat1 produced new peaks 1–4, the masses of which were consistent with our predicted capnine-like analogs (Figure 5A; Supporting Information Figures S19–S21). This suggests that cfat1 is an essential enzyme responsible for the biosynthesis of SoLs. Next, we confirmed the function of CFAT1 by in vitro biochemical assay with purified CFAT1 (Supporting Information Figure S15C) and substrates including cysteate and palmitoyl-CoA. As shown in Figure 5A, CFAT1 catalyzed the condensation of cysteate and palmitoyl-CoA to form compound 3, 2-amino-3-keto-octadecane-1-sulfonic acid, one capnine-like analog as confirmed by MSMS spectrum. The C-S bond can be dissociated in a homolytic manner or a heterolytic manner and two prominent ions, SO3–• and HSO3−, were observed. The abundance of the 79.9574 and 80.9652 ions raised the possibility that they could be used as diagnostic product ions for SoLs (Supporting Information Figures S22 and S23).31,32 Predicted compounds 1, 2 and 4 belong to capnine-like analogs and were also confirmed by MSMS spectrum (Supporting Information Figures S24–S26). We further explored the substrate specificity of CFAT1 toward different substrates including palmitoyl acid, phosphoserine, serine, and cysteine, with which CFAT1 failed to form any products (Supporting Information Figure S27). These results demonstrate that CFAT1 selectively recognizes cysteate and is specific to SoL biosynthesis in contrast to biosynthetic pathways of other sphingolipids that take serine.
Figure 5. Comparative LC-ESI analysis of production of key enzymatic conversions.
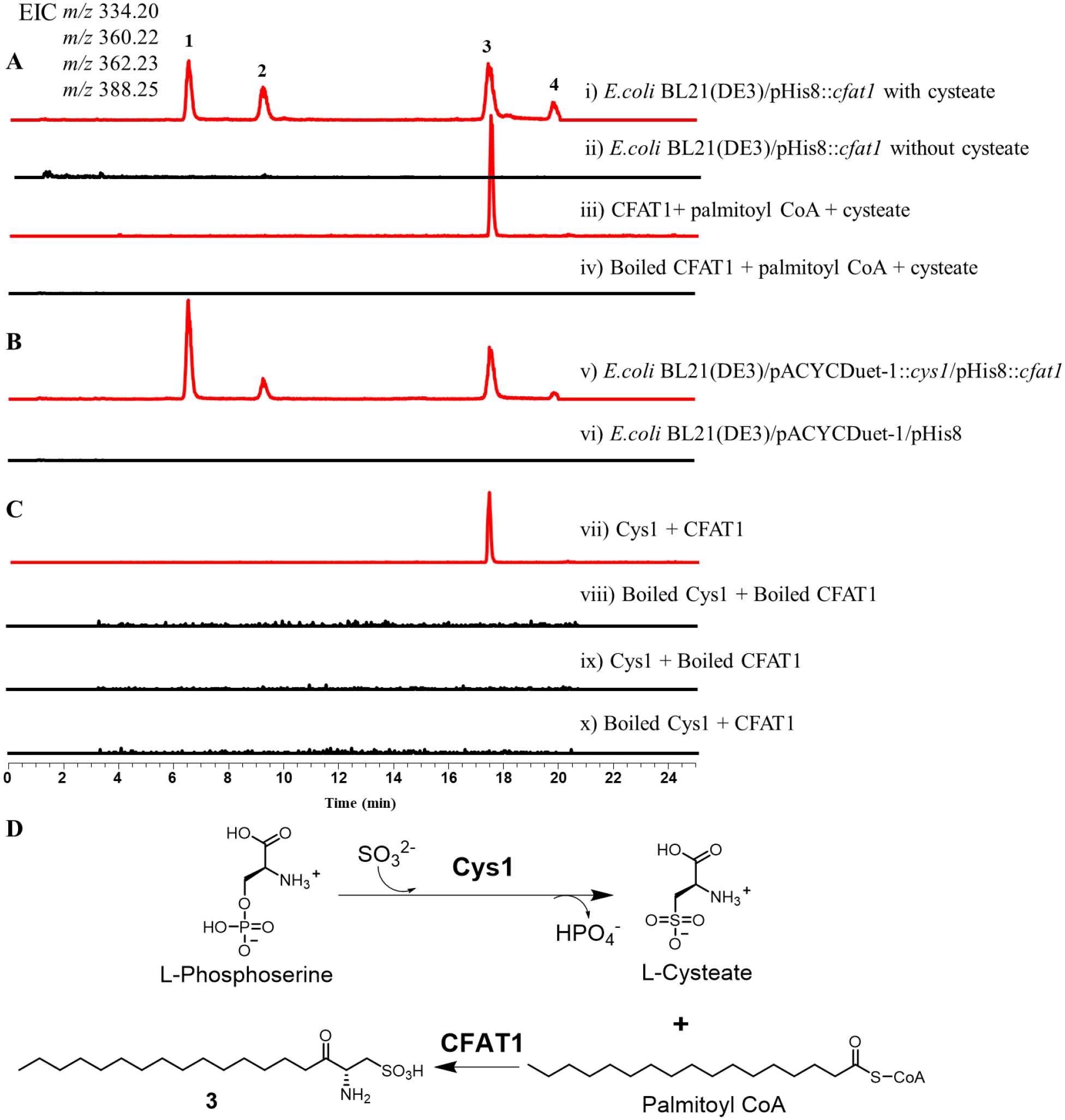
The extracted ion chromatograms (EICs) were shown at m/z 334.20 [M - H]− for 1, m/z 360.22 [M - H]− for 2, m/z 362.23 [M - H]− for 3, and m/z 388.25 [M - H]− for 4. (A) Production of E. coli BL21(DE3)/pHis8::cfat1 fed with cysteate or not (i and ii) and in the in vitro CFAT1 reaction (iii and iv). (B) Production of E. coli BL21(DE3)/pACYC::cys1/pHis8::cfat1 (v) and E. coli BL21(DE3)/pACYC/pHis8 (vi). (C) One-pot assay of Cys1 and CFAT1(vii, viii, ix and x). (D) The biosynthetic pathway of 3.
In vivo Co-expression Assay and in vitro One-pot Reaction
Since cys1 and cfat1 are clustered in the genome of C. gleum DSM16776 (Supporting Information Figure S18) and the catalytic efficiency of Cys1 was higher than Cys2 (Figures 4A and 4B), it suggested that cys1 is more likely involved in SoLs biosynthesis than cys2. To further elucidate the function of cys1 and cfat1 in C. gleum DSM16776, we performed co-expression of both genes in E. coli BL21 (DE3) as well as one-pot in vitro assay. Indeed, co-expression produced capnine-like analogs 1–4 (Figure 5B), the same compounds as produced by cfat1-harboring E. coli BL21 (DE3) when fed with cysteate (Figure 5A). This result also indicated that cys1 can produce cysteate in engineered E. coli. Next, we confirmed the function of Cys1 and CFAT1 by an in vitro one-pot biochemical assay. Upon incubation of L-phosphoserine, sodium sulfite, palmitoyl-CoA in the presence of Cys1 and CFAT1, LC-MS analysis revealed that Cys1 and CFAT1 coupled catalysis produced capnine-like compound 3 as expected (Figures 5C and 5D).
Distribution of SoL-specific Biosynthetic Genes
Sphingolipids are essential molecules and play important roles in both eukaryotic and prokaryotic organisms. As an unusual class of sphingolipids, SoLs containing a sulfonate group have been identified from certain bacteria.15,18,25 In 2017, a metabolomic analysis of the mice gut bacteria identified eighteen SoLs analogs via high-resolution mass spectrometry.25 Given that CFAT is a conserved gene specifically involved in SoLs biosynthesis in contrast to biosynthesis of other sphingolipids, we analyzed the distribution of Spt homologs including CFAT. We performed a sequence similarity network (SSN) analysis of 2,917 Spt homologs from the nonredundant protein sequence database. CFATs responsible for biosynthesis of SoLs fell into one cluster that is separate from other Spt homologs likely involved in the biosynthesis of other sphingolipids (Figure 6). From this CFATs cluster, we selected four different strains other than C. gleum DSM16776 in an attempt to produce SoLs and verify the wide distribution of SoL biosynthetic genes. Flavobacterium johnsoniae DSM2064 isolated from soil, Alistipes timonensis DSM25383 isolated from human fecal sample, as well as two animal associated strains, Chryseobacterium scophthalmum JUb44 associated with Caenorhabditis elegans and Algoriphagus machipongonensis DSM24695 associated with Choanoflagellates were selected. Using our characterized Cys1 and CFAT from C. gleum DSM16776 as hooks, we selected one Cys and one CFAT as candidates from each of these four strains. Then we purified these proteins (Supporting Information Figure S28) and confirmed their functions involved in SoL production by both in vitro reconstitution and co-expression assay using the same methods described above. All the selected Cys enzymes could convert L-phosphoserine to L-cysteate, while all the selected CFAT enzymes could catalyze the condensation of cysteate and palmitoyl-CoA to form 3 (Supporting Information Figures S29–S33). During our preparation of this manuscript, another CFAT from Flavobacterium johnsoniae DSM2064 required for capnine biosynthesis was reported and named Fjoh_241933 that was the same as CFAT-DSM2064 in this paper. Their expression of gene fjoh_2419 in E. coli led to an accumulation of capnine-like molecules, but without characterizing the function of this enzyme by in vitro assay. Thus, mining of CFAT homologs revealed their wide distribution across various bacteria and might imply a broader involvement of SoLs in mediating host inflammatory process.
Figure 6.
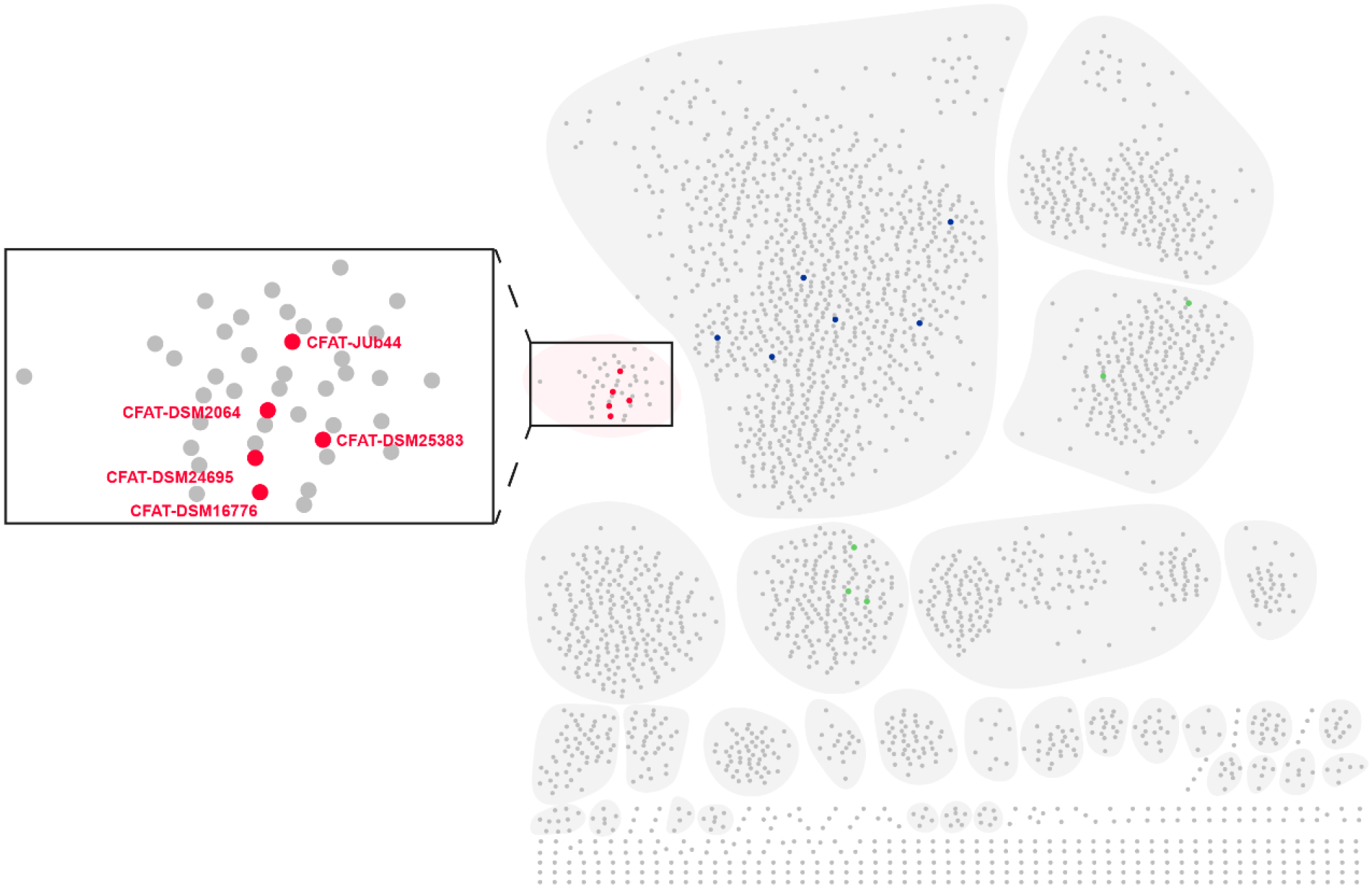
Distribution of Spt homologs in different organisms. SSN analysis of the Spt homologs in diverse organisms and visualized by Cytoscape.34 CFATs responsible for SoLs biosynthesis fell into a separate cluster as highlighted in the box, five of which (shown in red) were characterized in this study. Some reported prokaryote-derived Spt responsible for condensation of serine and fatty acyl-CoA are shown in green while some reported eukaryote-derived Spt responsible for condensation of serine and fatty acyl-CoA are shown in blue.
CONCLUSIONS
In search of human microbial functional metabolites, we discovered unique SoLs from the human opportunistic pathogen C. gleum DSM16776, using integrated bioassay-guided isolation and mass spectrometry-based metabolic and cheminformatics analysis. These SoLs showed strong activities in inducing mouse macrophage inflammatory cytokines. While inflammation is a fundamental biological process, pro-inflammatory activity had not been reported for SoLs. Thus, this study expands the potential role of SoLs in mediating microbe-host interactions. Using both in vivo heterologous expression and in vitro reconstitution, two SoLs biosynthetic enzymes, Cys and CFAT, were characterized and their functions in SoLs biosynthesis were established. Cys catalyzes a conversion of L-phosphoserine to L-cysteate while CFAT subsequently catalyzes a condensation between cysteate and fatty acyl-CoA. This is the first report that characterized these two steps as a coupled reaction for SoLs biosynthesis. Although CFAT shares relatively high homology with Spt, it showed a strict substrate specificity for cysteate over serine. Thus, the coupled reaction of Cys and CFAT is specific for biosynthesis of SoLs but not other sphingolipids. Finally, our bioinformatics analysis of SoL-specific CFATs revealed a wider distribution of this enzyme in microbes than previously observed and we verified the Cys and CFAT activities in four of these strains, suggesting that the production of SoLs and their pro-inflammatory activities may have a broader impact on microbe-host interactions than previously realized.
METHODS
Cultivation and Fractionation of C. gleum DSM16776.
The strain was cultured in 2.5 L ultra-yield flasks containing 800 mL of 5 g/L peptone media in incubator shakers at 30 °C and 220 rpm shaking. After 5 days, the culture broth was extracted with an equal volume methyl ethyl ketone (MEK) and the resulting organic phase was concentrated under vacuum by rotary evaporation. Cell pellets were disrupted using a high-pressure homogenizer NanoGenizer (Genizer LLC, Irvine, CA, USA), and subsequently extracted by MeOH and concentrated under vacuum by rotary evaporation. Dried organic extracts from culture broth and cell pellets were combined, redissolved in MeOH, and applied to silica gel for open column chromatography. Elution with a stepwise gradient of DCM:MeOH mixtures (15:1, 10:1, 7:1, 3:1, 1:1) afforded 5 major fractions. Fractions identified as active in the inflammation cytokine induction assays were further fractionated on Sephadex LH-20 column, using the same inflammation cytokine induction assay.
Isolation of SoL A.
The active subfraction from Sephadex LH-20 column was purified by semi-preparative HPLC using a Phenomenex Kinetex EVO-C18 Column (100 Å, 5μm, 10×250 mm) and eluted at 3.8 ml/min in a gradient of solvents A (0.1% NH4OH in water) and B (methanol): 80% B for 20 min to afford SoL A (1.63 mg). The concentration of SoL A from the cultivation was estimated as ~9.0 mg/L (14.5 μM) by a comparative LC-MS analysis between the crude extract and isolated pure SoL A.
Isolation and Treatment of Mouse Peritoneal Macrophages.
Primary mouse macrophages were collected by introducing 3 mL of 3% thioglycolate into the mice through intraperitoneal injection. After 3 days, 10 mL of PBS were introduced intraperitoneally to flush out cells. These cells were seeded in culture dishes containing DMEM with 10% FBS for 1 hour before being washed with serum-free DMEM two times to remove unattached cells. To treat the macrophages, cells were incubated for 6 to 24 hours in DMEM without FBS and an addition of LPS (E. coli O111:B4; Sigma-Aldrich, CAS# 93572-42-0) or SoL A or semi-pure fraction at the indicated concentration. Cells were finally washed with Dulbecco’s phosphate buffered saline two times before being lysed for total RNA or protein extraction.
Quantitative Real-time PCR.
TRIzol (Invitrogen) along with the RNeasy™ Mini Kit (Qiagen) were used to extract and purify total RNA from lysed macrophage cells prior to reverse transcription using a First-strand cDNA Synthesis System (Marligen Bioscience, MD). Fast Start Universal SYBR Green Master (Rox) (Roche Applied Science) was used to perform quantitative real-time PCR on an Eppendorf Realplex2 Mastercycler (Eppendorf, Hamburg, Germany). The following primers were used: Mouse 18S rRNA (internal control), 5’-CGCGGTTCTATTTTGTTGGT-3’ (forward) and 5’-AGTCGGCATCGTTTATGGTC-3’ (reverse); Mouse TNFα, 5’-CGTCAGCCGATTTGCTATCT-3’ (forward) and 5’-CGGACTCCGCAAAGTCTAAG-3’ (reverse); Mouse IL-6, 5’-AGTTGCCTTCTTGGGACTGA-3’ (forward) and 5’-TCCACGATTTCCCAGAGAAC-3’ (reverse); Mouse IL-1α, 5’-ATGGCCAAAGTTCCTGACTTG-3’ (forward) and 5’-TGACGTTTCAGAGGTTCTCAG-3’ (reverse); Mouse IL-1ß, 5’-GCCCATCCTCTGTGACTCAT-3’ (forward) and 5’-AGGCCACAGGTATTTTGTCG-3’ (reverse). The amplification program was 95 °C for 10 min followed by 40 cycles of 95 °C for 10 s, 60 °C for 15 s, and 68 °C for 20 s. Finally, a melting curve analysis from 60 °C to 95 °C every 0.2 °C was performed. Gene abundances were calculated by relative quantification and normalized to the internal control, 18S rRNA.
Enzyme-linked Immunosorbent Assay (ELISA).
ELISA kits for mouse IL-6 were purchased from eBioscience (San Diego, CA) and used following the manufacturer’s instructions. For the analysis, the macrophage-conditioned culture media was diluted 30–180 folds in 96 well microplates. These microplates were then read using a SpectraMax M5 microplate reader.
Statistical Analysis.
The experiments were carried out in three biological replicates. All data were presented as mean ± S.D. Statistical significance was calculated by ordinary one-way ANOVA using the GraphPad Prism statistical program (GraphPad Software Inc., San Diego, CA). p≤0.05 was considered statistically significant.
In-vivo Assay of cfat Activity.
The plasmid pLLH105 containing cfat1 (Table S1) was transformed into E. coli BL21 (DE3). A single colony was inoculated into a 10 mL culture of LB supplemented with kanamycin at 37 °C overnight. 500 ul culture broth was transferred to 50 ml LB with kanamycin and grew at 37 °C to OD600 ~0.6, then cooled to 20 °C before IPTG was added to a final concentration of 0.2 mM and cysteate was added to a final concentration of 2 mM. The culture was incubated at 20 °C for an additional 16 h. Cell pellets harvested and resuspended in 5 mL of deionized water was lysed by sonication. 5 ml butanone was added to extract the product. The butanone phase was dried and dissolves in 100 ul MeOH for analysis by HPLC-DAD-ESIMS (Kinetex C18 HPLC column, 4.6 × 100 mm, 2.4 μm, 100 Å), with gradient elution of solvent A (H2O + 0.1% NH4OH) and solvent B (MeCN + 0.1% NH4OH) at 0.5 ml/min over a 20 min period from 30% to 90% B (negative ionization mode). Likewise, the constructed plasmids harboring cfat2, cfat3, cfat-DSM2064, cfat-DSM25383, cfat-JUb44, or cfat-DSM24695 were transformed into E. coli BL21 (DE3). The productions were processed and analyzed using the method as mentioned above.
Characterization of Cys-Catalyzed Conversions in Vitro.
The in vitro Cys activity assay was guided by a previously described method.29 The assays were carried out in a 50 μL reaction mixture containing 50 mM Tris-HCl (pH 7.5), 2 mM L-phosphoserine, 2 mM Na2SO3, 0.2 mM PLP, and 20 μM Cys. The reactions proceeding in the absence of the enzyme or Na2SO3 were utilized as the negative controls. After incubation at 30 °C for 20 min, the reactions were quenched by adding an equal volume of acetonitrile. After centrifugation (12000 rpm, 10 min), 20 μL of the supernatant was added to 1 M NaHCO3 (15 μL) and 1% of L-FDAA solution in acetone (5 μL). The solutions were heated at 50 °C for 30 min and cooled down to rt, and the reaction was quenched by the addition of 2 N HCl (20 μL). Standard amino acids (L-phosphoserine and L-cysteate) were reacted with L-FDAA in the same way. The sample was analyzed by HPLC-DAD-ESIMS (Kinetex C18 HPLC column, 4.6 × 100 mm, 2.4 μm, 100 Å,0.5 mL/min gradient elution from 10% to 100% H2O/MeCN over 10 min with constant 0.1% formic acid; mAU at 340nm).
In-vitro Assay of Recombinant CFAT Activity.
CFAT activity was tested in vitro according to the methods used for this class of enzymes.35 CFAT was first converted to the holo-form by dialysis against freshly prepared 20 mM potassium phosphate (pH 7.5) containing 150 mM NaCl and 25 uM PLP for 1 h at 4 °C. The activity of CFAT was measured by detecting the formation of 3. A final concentration of 10 uM CFAT in 20mM potassium phosphate buffer (pH 7.5, 150 mM NaCl) was incubated with 20 mM L-phosphoserine and 1.6 mM palmitoyl-CoA in a final volume of 125 ul. The reaction was incubated at 37 °C for 20 min and was quenched and extracted with an equal volume of CHCl3/CH3OH (2:1, v/v). The sample was centrifuged at 13,300 rpm for 30 min, and the organic phase was allowed to dry. The residue was dissolved in 100 ul of CH3OH and analyzed by HPLC-DAD-ESIMS (Kinetex C18 HPLC column, 4.6 × 100 mm, 2.4 μm, 100 Å), using the same gradient elution as described above for in-vivo assay of cfat activity.
Co-expression of cys and cfat Genes in E. coli.
The constructed plasmids, pLLH105 containing cfat1 and pLLH112 containing cys1, were transformed to form E. coli BL21 (DE3)/pLLH105+ pLLH112. A single colony was selected to inoculate a 10 mL culture of LB supplemented with chloramphenicol and kanamycin at 37 °C overnight. 500 ul culture broth was transferred to 50 ml LB with chloramphenicol and kanamycin and grown at 37 °C to OD600 ~0.6, then cooled to 20 °C before IPTG was added to a final concentration of 0.2 mM. The culture was incubated at 20 °C for another 16 h. Cells were harvested by centrifugation at 5,000 ×g for 10 min at 4 °C. Cell pellets were resuspended in 5 mL of deionized water and sonicated for 5 min to lyse the cells. 5 ml butanone was added to extract the product. The butanone phase was separated, dried, and dissolved in 100 ul MeOH for analysis by HPLC-DAD-ESIMS in negative ionization mode, using the same HPLC column and gradient elution as described above. The co-expression of cys and cfat genes from four other strains, F. johnsoniae DSM2064, A. timonensis DSM25383, C. scophthalmum JUb44 and A. machipongonensis DSM24695, as well as their co-production were processed and analyzed using the same method described above.
One-pot Assay of Cys and CFAT.
The assays were carried out in a 125 μL reaction mixture containing 50 mM Tris−HCl (pH 7.5), 150 mM NaCl, 2 mM L-phosphoserine, 2 mM Na2SO3, 0.2 mM PLP, 1.6 mM palmitoyl-CoA, 20 μM Cys and 10 μM Spt. The reaction was incubated at either 30 °C or 37 °C for 20 min, and then the reaction was quenched and extracted with an equal volume of CHCl3/CH3OH (2:1, v/v). The sample was centrifuged at 13,300 rpm for 30 min, and the organic phase was allowed to dry. The residue was dissolved in 100 ul of CH3OH and analyzed by HPLC-DAD-ESIMS in negative ionization mode, using the same HPLC column and gradient elution as described above.
Bioinformatic Analysis.
Cys and Spt (including CFAT) were searched and the proposed functions were accomplished by using Blast programs (http://blast.ncbi.nlm.nih.gov/Blast.cgi).36 Spt protein sequences were searched against NCBI nonredundant protein database (May 2020) using hmmscan (v3.1b2)37 with the parameter “-E 1E-10 --domE 1E-10”. The matched Spt homologs were filtered to remove sequences <400 bp or >600 bp with a custom script, and clustered using CD-HIT (v4.8.1)38 with the parameter “-c 0.6”. Finally, we obtained 2,917 sequences. The protein similarity network was calculated using the EFI web tool,39 and visualized using the Cytoscape software (v3.8.2).40
Supplementary Material
ACKNOWLEDGMENTS
J. Li. acknowledges a partial support from National Institutes of Health (NIH) grants P20GM103641 and P20GM109091, and a National Science Foundation EPSCoR Program OIA-1655740. This work was also partially supported by fundings from Shenzhen Bay Laboratory (21230051 to X. Tang and SZBL2021080601004 to H. Tian). We thank P. J. Pellechia and T. Johnson from UofSC NMR Facility for help with acquiring NMR data.
Footnotes
Supporting Information
The Supporting Information is available free of charge via the Internet at http://pubs.acs.org/.
LC-MS based metabolomics analysis procedure, information of primers, plasmids, and strains, 1H, 13C, and 2D NMR as well as HR-ESIMS spectra of SoL A, and in vivo and in vitro analysis of SoLs biosynthetic enzymes.
The authors declare no competing financial interest.
Reference
- 1.Koppel N; Balskus EP Exploring and understanding the biochemical diversity of the human microbiota. Cell Chem. Biol 2016, 23, 18–30. [DOI] [PubMed] [Google Scholar]
- 2.Donia MS; Cimermancic P; Schulze CJ; Brown LCW; Martin J; Mitreva M; Clardy J; Linington RG; Fischbach MA A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 2014, 158, 1402–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen LJ; Esterhazy D; Kim SH; Lemetre C; Aguilar RR; Gordon EA; Pickard AJ; Cross JR; Emiliano AB; Han SM; et al. Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature 2017, 549, 48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen LJ; Kang HS; Chu J; Huang YH; Gordon EA; Reddy BVB; Ternei MA; Craig JW; Brady SF Functional metagenomic discovery of bacterial effectors in the human microbiome and isolation of commendamide, a GPCR G2A/132 agonist. Proc. Natl. Acad. Sci. U.S.A 2015, 112, E4825–E4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yadav VS; Das BK; Mohapatra S; Ahmed MN; Gautam H; Kapil A; Sood S; Dhawan B; Chaudhry R Clinical correlation and antimicrobial susceptibility pattern of Chryseobacterium spp.: A three year prospective study. Intractable Rare Dis. Res 2021, 10, 37–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin J-N; Teng S-H; Lai C-H; Yang C-H; Huang EY-H; Lin H-F; Lin H-H Comparison of the Vitek MS and Bruker Matrix-assisted laser desorption ionization-time of flight mass spectrometry systems for identification of Chryseobacterium isolates from clinical specimens and report of uncommon Chryseobacterium infections in humans. J. Clin. Microbiol 2018, 56, e00712–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsouvalas CP; Mousa G; Lee AH; Philip JA; Levine D Chryseobacteriurn gleum isolation from respiratory culture following community-acquired pneumonia. Am. J. Med. Case Rep 2020, 21, e921172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellais S; Naas T; Nordmann P Molecular and biochemical characterization of ambler class A extended-spectrum beta-lactamase CGA-1 from Chryseobacterium gleum. Antimicrob. Agents Chemother 2002, 46, 966–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Virok DP; Abrok M; Szel B; Tajti Z; Mader K; Urban E; Talosi G Chryseobacterium gleum - a novel bacterium species detected in neonatal respiratory tract infections. J. Matern. Fetal Neonatal Med 2014, 27, 1926–1929. [DOI] [PubMed] [Google Scholar]
- 10.Singhal L; Gupta V; Mehta V; Singla N; Janmeja AK; Chander J Sepsis due to Chryseobacterium gleum in a diabetic patient with chronic obstructive pulmonary disease: a case report and mini review. Jpn. J. Infect. Dis 2017, 70, 687–688. [DOI] [PubMed] [Google Scholar]
- 11.Rajendran P; Muthusamy S; Balaji VK; Rakesh GJ; Easow JM Urinary tract infection due to Chryseobacterium gleum, an uncommon pathogen. Indian J. Pathol. Microbiol 2016, 59, 551–553. [DOI] [PubMed] [Google Scholar]
- 12.Chaudhari PN; Wani KS; Chaudhari BL; Chincholkar SB Characteristics of sulfobacin A from a soil isolate Chryseobacterium gleum. Appl. Biochem. Biotechnol 2009, 158, 231–241. [DOI] [PubMed] [Google Scholar]
- 13.Kamiyama T; Umino T; Satoh T; Sawairi S; Shirane M; Ohshima S; Yokose K Sulfobacins A and B, novel von Willebrand factor receptor antagonists. I. Production, isolation, characterization and biological activities. J. Antibiot 1995, 48, 924–928. [DOI] [PubMed] [Google Scholar]
- 14.Maeda J; Nishida M; Takikawa H; Yoshida H; Azuma T; Yoshida M; Mizushina Y Inhibitory effects of sulfobacin B on DNA polymerase and inflammation. Int. J. Mol. Med 2010, 26, 751–758. [DOI] [PubMed] [Google Scholar]
- 15.Alegado RA; Brown LW; Cao SG; Dermenjian RK; Zuzow R; Fairclough SR; Clardy J; King N A bacterial sulfonolipid triggers multicellular development in the closest living relatives of animals. Elife 2012, 1, e00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beemelmanns C; Woznica A; Alegado RA; Cantley AM; King N; Clardy J Synthesis of the rosette-inducing factor RIF-1 and analogs. J. Am. Chem. Soc 2014, 136, 10210–10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cantley AM; Woznica A; Beemelmanns C; King N; Clardy J Isolation and synthesis of a bacterially produced inhibitor of rosette development in choanoflagellates. J. Am. Chem. Soc 2016, 138, 4326–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woznica A; Cantley AM; Beemelmanns C; Freinkman E; Clardy J; King N Bacterial lipids activate, synergize, and inhibit a developmental switch in choanoflagellates. Proc. Natl. Acad. Sci. U.S.A 2016, 113, 7894–7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duhrkop K; Shen H; Meusel M; Rousu J; Bocker S Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc. Natl. Acad. Sci. U.S.A 2015, 112, 12580–12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duhrkop K; Fleischauer M; Ludwig M; Aksenov AA; Melnik AV; Meusel M; Dorrestein PC; Rousu J; Bocker S SIRIUS 4: a rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 2019, 16, 299–302. [DOI] [PubMed] [Google Scholar]
- 21.Kamiyama T; Umino T; Itezono Y; Nakamura Y; Satoh T; Yokose K Sulfobacins A and B, novel von willebrand factor receptor antagonists. II. Structural elucidation. J. Antibiot 1995, 48, 929–936. [DOI] [PubMed] [Google Scholar]
- 22.Dumitru CD; Ceci JD; Tsatsanis C; Kontoyiannis D; Stamatakis K; Lin JH; Patriotis C; Jenkins NA; Copeland NG; Kollias G; et al. TNF-α induction by LPS is regulated posttranscriptionally via a Tpl2/ERK-dependent pathway. Cell 2000, 103, 1071–1083. [DOI] [PubMed] [Google Scholar]
- 23.Meng F; Lowell CA Lipopolysaccharide (LPS)-induced macrophage activation and signal transduction in the absence of Src-family kinases Hck, Fgr, and Lyn. J. Exp. Med 1997, 185, 1661–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrison PJ; Dunn TM; Campopiano DJ Sphingolipid biosynthesis in man and microbes. Nat. Prod. Rep 2018, 35, 921–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker A; Pfitzner B; Harir M; Schaubeck M; Calasan J; Heinzmann SS; Turaev D; Rattei T; Endesfelder D; zu Castell W; et al. Sulfonolipids as novel metabolite markers of Alistipes and Odoribacter affected by high-fat diets. Sci. Rep 2017, 7, 13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White RH Biosynthesis of the sulfonolipid 2-amino-3-hydroxy-15-methylhexadecane-1-sulfonic acid in the gliding bacterium Cytophaga johnsonae. J. Bacteriol 1984, 159, 42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abbanat DR; Godchaux W; Polychroniou G; Leadbetter ER Biosynthesis of a sulfonolipid in gliding bacteria. Biochem. Biophys. Res. Commun 1985, 130, 873–878. [DOI] [PubMed] [Google Scholar]
- 28.Graham DE; Taylor SM; Wolf RZ; Namboori SC Convergent evolution of coenzyme M biosynthesis in the Methanosarcinales: cysteate synthase evolved from an ancestral threonine synthase. Biochem. J 2009, 424, 467–478. [DOI] [PubMed] [Google Scholar]
- 29.Wang M; Chen D; Zhao Q; Liu W Isolation, structure elucidation, and biosynthesis of a cysteate-containing nonribosomal peptide in Streptomyces lincolnensis. J. Org. Chem 2018, 83, 7102–7108. [DOI] [PubMed] [Google Scholar]
- 30.Ikushiro H; Hayashi H; Kagamiyama H A water-soluble homodimeric serine palmitoyltransferase from Sphingomonas paucimobilis EY2395T strain - Purification, characterization, cloning, and overproduction. J. Biol. Chem 2001, 276, 18249–18256. [DOI] [PubMed] [Google Scholar]
- 31.Williams BJ; Barlow CK; Kmiec KL; Russell WK; Russell DH Negative ion fragmentation of cysteic acid containing peptides: cysteic acid as a fixed negative charge. J. Am. Soc. Mass. Spectr 2011, 22, 1622–1630. [DOI] [PubMed] [Google Scholar]
- 32.Corcelli A; Lattanzio VMT; Mascolo G; Babudri F; Oren A; Kates M Novel sulfonolipid in the extremely halophilic bacterium Salinibacter ruber. Appl. Environ. Microb 2004, 70, 6678–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vences-Guzman MA; Pena-Miller R; Hidalgo-Aguilar NA; Vences-Guzman ML; Guan ZQ; Sohlenkamp C Identification of the Flavobacterium johnsoniae cysteate-fatty acyl transferase required for capnine synthesis and for efficient gliding motility. Environ. Microbiol 2021, 23, 2448–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shannon P; Markiel A; Ozier O; Baliga NS; Wang JT; Ramage D; Amin N; Schwikowski B; Ideker T Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 2003, 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raman MCC; Johnson KA; Yard BA; Lowther J; Carter LG; Naismith JH; Campopiano DJ The external aldimine form of serine palmitoyltransferase: structural, kinetic, and spectroscopic analysis of the wild-type enzyme and HSAN1 mutant mimics. J. Biol. Chem 2009, 284, 17328–17339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altschul SF; Gish W; Miller W; Myers EW; Lipman DJ Basic local alignment search tool. J. Mol. Biol 1990, 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 37.Finn RD; Clements J; Eddy SR HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 2011, 39, W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu L; Niu B; Zhu Z; Wu S; Li W CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zallot R; Oberg N; Gerlt JA The EFI web resource for genomic enzymology tools: leveraging protein, genome, and metagenome databases to discover novel enzymes and metabolic pathways. Biochemistry 2019, 58, 4169–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su G; Morris JH; Demchak B; Bader GD Biological network exploration with Cytoscape 3. Curr. Protoc. Bioinform 2014, 47, 8.13.1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


