ABSTRACT
This study aimed to identify differentially expressed genes (DEGs) and molecular pathways in eutopic endometrial stromal cells (EuESCs) from adenomyosis (AM) patients and to provide a new insight into the disease mechanisms. The gene expression profiles in adenomyotic EuESCs (A-EuESCs) and normal ESCs (N-ESCs) were analyzed by RNA-sequencing (RNA-Seq) and validated by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment analyses were performed to obtain insights into the functions of DEGs. The protein-protein interaction (PPI) network was constructed using the STRING database and visualized by Cytoscape software, and their hub genes were identified. A total of 458 up-/363 down-regulated genes were identified in A-EuESCs versus N-ESCs. The GO enrichment analysis showed that these genes were significantly enriched in calcium-dependent cell-cell adhesion. The most significant term of the KEGG pathway analysis was cytokine-cytokine receptor interaction. There were 145 nodes in the PPI network of the 157 DEGs, which were identified in significant enrichment pathway by the KEGG pathway analysis in N-ESCs and A-EuESCs. The PPI network revealed that IL-6 was a central hub gene. Besides, IL-6 was found as a central hub gene in the pro-inflammatory/chemotactic subnetwork, and EGF was noted as a central hub gene in the angiogenesis subnetwork. Our study indicated the alterations of transcriptomic profiles in A-EuESCs and provided new insights into the pathogenesis of AM. The A-EuESCs in women with AM have fundamental abnormalities that may predispose to pro-invasion/migration and angiogenesis.
KEYWORDS: Adenomyosis, transcriptome, RNA sequencing, eutopic endometrial stromal cells, inflammatory cytokines, chemokines, angiogenesis
Graphical abstract

Highlights
458 upregulated/363 downregulated genes were identified in A-EuESCs versus N-ESCs.
IL-6 was found as a central hub gene in the pro-inflammatory/chemotactic subnetwork.
EGF was noted as a central hub gene in the angiogenesis subnetwork.
A-EuESCs may predispose to pro-invasion/migration and angiogenesis.
Introduction
Adenomyosis (AM) is a common gynecological condition causing uterine enlargement, pelvic pain, menorrhagia and/or dysmenorrhea, in which and the condition is mainly refractory to drug treatments, and hysterectomy is sometimes essential for the complete alleviation of clinical symptoms [1,2]. Although several studies have concentrated on AM, but the etiology and pathogenesis of the AM have still remained elusive, which are worthy of further assessment [3,4].
As there is often a visualized histologic continuity between the ectopic and the basal endometrium in AM [5], it is widely accepted that AM results from the invagination of basalis endometrium into the myometrium through an altered or interrupted junctional zone, representing a highly specialized hormone-responsive structure located in the inner third of the myometrium [6–8]. Recent studies have suggested that a heritable or acquired alteration in the eutopic endometrium may play an essential role in the occurrence of AM [9–12]. Eutopic endometrium of the women with AM showed abnormal biological processes, including decreased apoptosis [8,13], increased proliferation [10] and angiogenesis [14], and impaired cytokine expression and local production of estrogens, which involved the pathogenesis of AM by enhancing the infiltration of the endometrium to the junctional zone and the growth of ectopic tissue [15]. Numerous AM-based studies have concentrated on the eutopic endometrium. Initially, gene expression profiles of endometrium from women with AM and age-matched healthy controls (HCs) had been explored with microarray platforms [16]. Subsequently, one study used RNA sequencing (RNA-Seq) to analyze differentially expressed genes (DEGs) that were in the eutopic endometrium of women with AM and HCs [17]. Recently, single-cell RNA sequencing (scRNA-seq) has been applied to identify the changes in gene expression patterns among ectopic lesions, eutopic endometrium, and normal endometrium at the single-cell level and to explore a potential novel pathogenesis of AM [18]. However, these studies were carried out without separating the endometrial stromal cells (ESCs) from glandular epithelial cells in eutopic endometrial tissue.
Invasion of abnormal adenomyotic eutopic endometrial stromal cells (A-EuESCs) has been reported in the etiology of AM [19,20]. The stromal cells may play a primary pathogenetic role in accelerating epithelial downgrowth [6]. In addition, both exogenous and endogenous interleukin-22 (IL-22) have enhanced the invasiveness of A-EuESCs in vitro [21]. Besides, A-EuESCs were proliferated more rapidly than normal endometrial stromal cells (N-ESCs), whether they were treated with or without estradiol (E2), medroxyprogesterone acetate (MPA), interleukin 6 (IL-6), lipopolysaccharide (LPS) and interferon γ (IFN-γ) [10,14]. A-EuECSs also expressed a higher level of AMP-activated protein kinase (AMPK) than N-ESCs [22]. As described above, studies on A-EuESCs were limited to the evaluation of expression levels of one or several particular genes [13,23,24], while few systematic studies have concentrated on the DEGs or the major pathways involved in AM from the perspective of A-EuESCs [25]. Hence, identifying distinct gene expression profiles in A-EuESCs and N-ESCs is of great significance to better understand the pathogenesis of AM.
As mentioned earlier, A-EuESCs exhibited dysregulation of pathways that globally predispose toward the development, invasion/migration, and survival of ectopic endometrial implants beyond the myometrial interface. It is reasonable to postulate the existence of intrinsic abnormalities in A-EuESCs from women with AM. In the present study, we performed the transcriptome analysis of A-EuESCs from women with or without AM by RNA-Seq, and aimed to identify DEGs and molecular pathways/networks in A-EuESCs and to provide new insights into underlying mechanisms of AM.
Materials and methods
Patients and tissue sample collection
Female patients with AM and women without AM as age-matched HCs were enrolled in the present study. All patients had not received hormone therapy or used intrauterine contraceptive device for at least 6 months prior to surgery. Eutopic endometrial tissues were collected by hysterectomy from symptomatic women with pathologically confirmed diffuse AM, and also from female age-matched HCs by hysterectomy who had no endometrial disorders (e.g., intramural myomas) and pathologically confirmed to be free of AM or endometriosis.
All eutopic endometrial tissues were collected on the days of 20–23 of the menstrual cycle (middle-secretory phase) for isolation and culture of primary ESCs [26]. The study was approved by the Ethic Committee of Integrated Traditional Chinese and Western Medicine Hospital of Jiangsu Province (Nanjing, China; Approval No. 2019LWKYS-001). Informed consent was obtained from all participants prior to enrollment.
Primary cell culture
The isolation and culture of A-EuESCs and N-ESCs were carried out based on previously described procedures with slight modification [27]. Briefly, endometrial samples obtained by surgery were immediately placed in ice-cold sterile phosphate-buffered saline (PBS) and transferred to the laboratory. Tissues were thrice washed with sterile PBS and minced into small pieces, and then incubated with 0.1% (w/v) collagenase type II (Sigma-Aldrich, St. Louis, MO, USA) in a shaking water bath for 0.5 h at 37°C. The cell suspension was sequentially filtered through a 100-μm filter, and then, through a 40-μm cell strainer (BD Falcon, Bedford, MA, USA), followed by removing the debris and epithelial cells, respectively. The cell suspension was collected and centrifuged at 200 × g for 5 min to obtain ESCs. The pellet was re-suspended in a Dulbecco’s modified Eagle’s medium (DMEM)/F12 (1:1) (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS) (Gibico, New York, NY, USA) and 1% penicillin/streptomycin (Invitrogen), 10 nmol/L 17-estradiol (Sigma-Aldrich), and 1 umol/L medroxyprogesterone acetate (MPA) (Sigma-Aldrich) [28]. Cells were seeded at a density of 2 × 105 cells per T25 flask and incubated in a 5% CO2 atmosphere at 37°C. The cultured ESCs were identified by immunocytochemical staining for vimentin and cytokeratin 8 as previously described (data were not shown) [29].
RNA-seq and data analysis
To preserve the biological properties of the ESCs, RNA-Seq was performed using primary cells (all passage 1) when the cell confluence reached 80% at 6–7 days after culture. Total RNA was extracted from cultured cells using TRIzol® reagent (Invitrogen) according to the manufacturer’s instructions. RNA-Seq transcriptome library was prepared through a TruSeqTM RNA sample preparation kit (Illumina Inc., San Diego, CA, USA) using 1 μg of total RNA [30,31]. Then, mRNA sequencing was performed on an Illumina NovaSeq 6000 platform (Illumina Inc.) by Shanghai Majorbio Bio-pharm Technology Co.,Ltd (Shanghai, China). To identify DEGs in two different groups, the expression level of each transcript was calculated according to transcripts per million (TPM) reads. RSEM software was used to quantify gene abundances. DESeq2 R package (ver. 1.22.2) was utilized for annotation and differential expression analysis. DEGs were identified with adjusted P < 0.05 and absolute Log2 fold-change (FC) ≥ 1. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment analyses were performed to identify significant biological processes and pathways [32,33], using Goatools and KOBAS, respectively, with P-value < 0.05.
In order to explore the relationship between key genes and GO terms/KEGG pathways at a clearer glance, chord plots of GO terms and KEGG pathways were also drawn. The data were analyzed using Majorbio cloud platform (www.majorbio.com). The data discussed in the present study were deposited in the Gene Expression Omnibus (GEO) database (Accession No. GSE157718) [34].
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted from the ESCs (all passage 2) using TRIzol® reagent as described previously. The cDNA was synthesized using the a HiScript®III Reverse Transcriptase kit (Vazyme Biotech Co., Ltd., Nanjing, China) for qRT-PCR. The thermocycling conditions of the reverse transcription were as follows: Removing genomic DNA by gDNA wiper at 42 °C for 2 min; synthesizing first- strand cDNA at 25 °C for 5 min, 37 °C for 45 sec, and at 85 °C for 5 sec. qPCR was subsequently performed using an AceQ qPCR SYBR Green Master Mix system (Vazyme Biotech Co., Ltd.) according to the manufacturer’s instructions. Experiments were performed by a QuantStudio™ 5 Real-Time PCR system (Applied Biosystems, Waltham, MA, USA). The thermocycling conditions of the qPCR were as follows: pre-denaturation at 95 °C for 5 min; 40 cycles at 95 °C for 10 sec, and at 60 °C for 30 sec; and the final dissociation stage (at 95 °C for 15 sec, at 60 °C for 60 sec and at 95 °C for 15 sec) was performed at the end of the amplification procedure. The relative mRNA expression levels were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH), as a
reference gene, and calculated using the 2‑ΔΔCT method [35,36]. The qRT-PCR reactions were performed in triplicate in 96-well optical reaction plates. The GraphPad Prism 9.0 software package (GraphPad Software Inc., San Diego, CA, USA) was used to draw figures. The primers used for RT-qPCR are listed in Supplementary Table 1.
Construction of protein-protein interaction (PPI) network
The Search Tool for the Retrieval of Interacting Genes (STRING) database (ver. 11.0; https://string-db.org/) was used to elucidate the interactive relationships of the DEGs identified in significantly enriched pathways (adjusted P < 0.05) by the KEGG pathway analysis [37]. The interacting pairs with a confidence score greater than 0.4 were considered significant and were retained. Subsequently, Cytoscape software (ver. 3.8.1) was used to establish the PPI network [38]. The network topology property indicators, including degree centrality, betweenness centrality, and closeness centrality were analyzed using CytoNCA in Cytoscape software. A node with a higher score of network topology property indicators indicated a more important role in that node in the PPI network, which was considered as a hub node [39].
Cytokine assay
N-ESCs (n = 15) and A-EuESCs (n = 15) at passage 2 were cultured at a density of 5 × 105 cells per a 60-mm dish. The conditioned medium from each dish was collected after 48 h of inoculation, then, centrifuged at 2,500 rpm for 5 min at room temperature, and the culture supernatants were filtered through a 0.22-μm pore-sized filter and stored at −80°C. The levels of interleukin-6 (IL-6) and epidermal growth factor (EGF) in the supernatants were determined using enzyme-linked immunosorbent assay (ELISA) kits (Elabscience Biotechnology Co., Ltd., Wuhan, China). The measurements were performed according to the manufacturer’s instructions [40]. Each experiment was carried out in triplicate and repeated three times.
Statistical analysis
The statistical analysis was performed using SPSS 22.0 software (IBM Corp., Armonk, NY, USA). Continuous variables were presented as mean ± standard deviation (SD). Categorical data were expressed as number (percentage). The Chi-square and test or the Fisher’s exact test, whatever appropriate, were used for data analysis. The independent two-sample t-test or the Mann-Whitney-U test, whatever appropriate, were utilized to compare the continuous variables between the two groups. P < 0.05 was considered statistically significant.
Results
Clinical characteristics in the AM and control groups
A total of 18 female patients with AM and 18 age-matched female HCs were enrolled in our study. All patients’ endometrial tissues were collected by hysterectomy during the secretory phase. Besides, 18 patients with AM were multipara who aged 32 to 53 (median age, 41.6) years old; 18 age-matched female HCs were multipara who aged 34 to 55 (median age, 42.3) years old. None of the age-matched female HCs suffered from AM. The clinical data of participants are presented in Table 1.
Table 1.
Patients characteristics and symptoms of the study population
| Variable | Control (n = 18) | AM (n = 18) | P-value |
|---|---|---|---|
| Age, years | |||
| mean ± SD | 41.6 ± 3.78 | 42.3 ± 4.16 | 0.271 |
| Body mass index (BMI), kg/m2 | |||
| mean ± SD | 20.9 ± 2.63 | 21.4 ± 3.19 | 0.347 |
| Gravidity, n (%) | |||
| 0 | 1 (5.6%) | 2 (11.1%) | 0.546 |
| ≥1 | 17 (94.4%) | 16 (88.9%) | |
| Parity, n (%) | |||
| 0 | 2 (11.1%) | 3 (16.7%) | 0.630 |
| ≥1 | 16 (88.9%) | 15 (83.3%) | |
| Menorrhagia, n (%) | |||
| Yes | 0 (0.0%) | 12 (66.7%) | 0.000 |
| No | 18 (100.0%) | 6 (33.3%) | |
| Dysmenorrhea, n (%) | |||
| Yes | 0 (0.0%) | 15 (83.3%) | 0.000 |
| No | 18 (100.0%) | 3 (16.7%) | |
| Previous miscarriages, n (%) | |||
| 0 | 14 (77.8%) | 13 (72.2%) | 0.700 |
| ≥1 | 4 (22.2%) | 5 (27.8%) | |
RNA-Seq analysis and identification of DEGs
To better understand the molecular mechanism of AM, we conducted a comparative transcriptomic analysis on ESCs from 3 AM and 3 age-matched female HCs. Totally, 29655 annotated mRNAs were identified, of which 821 mRNAs (458 upregulated and 363 downregulated) were significantly deregulated (Figure 1(a,b)). The top 10 genes that were significantly upregulated or downregulated are listed in Table 2. The principle component analysis (PCA) showed that the A-EuESCs exhibited distinct gene expression profiles compared with N-ESCs (Figure 1(c)). Hierarchical cluster analysis of the DEGs in ESCs from AM and control groups indicated that the gene expression patterns were clustered separately after unsupervised clustering (Figure 1(d)). The above-mentioned results suggested that the gene expression profiles in A-EuESCs in AM group were significantly altered compared with N-ESCs in control group.
Figure 1.

Differential expression analysis in N-ESCs and A-EuESCs (a) Volcano plot shows DEGs in N-ESCs and A-EuESCs. (b) Bar chart showed the number of upregulated and downregulated DEGs in A-EuESCs compared with N-ESCs. (c) Unsupervised principal component analysis (PCA) of different cell lines (3 N-ESCs and 3 A-EuESCs). (d) Hierarchical clustering analysis of the DEGs in N-ESCs group (3 N-ESCs are abbreviated as N1, N2, and N3, respectively) and A-EuESCs group (3 A-EuESCs are abbreviated as A1, A2, and A3, respectively). Yellow and blue bar charts represent significantly upregulated and downregulated genes, respectively.
Table 2.
The DEGs in the A-EuESCs compared to N-ESCs
| Gene Symbol | Gene ID | P-value | P-adjust | log2FC |
|---|---|---|---|---|
| Up-regulation genes (Top 10) | ||||
| HOXC8 | 3224 | 1.53E-39 | 0.00094 | 8.97 |
| IL33 | 90865 | 2.59E-32 | 4.27E-11 | 8.37 |
| MYH1 | 4619 | 1.44E-31 | 0.036 | 7.67 |
| TCF21 | 6943 | 6.44E-24 | 2.98E-11 | 7.48 |
| ADAMTSL1 | 92949 | 8.43E-24 | 5.95E-06 | 7.43 |
| MYH2 | 4620 | 1.4E-23 | 0.00054 | 7.29 |
| CCL7 | 6354 | 1.36E-22 | 0.0017 | 7.23 |
| CCL11 | 6356 | 2.27E-22 | 0.0045 | 7.21 |
| ACAN | 176 | 3.61E-22 | 0.0037 | 6.92 |
| CACNG7 | 59284 | 1.49E-20 | 8.66E-05 | 6.90 |
| Down-regulation genes (Top 10) | ||||
| SLC6A2 | 6530 | 4.59E-12 | 1.5E-09 | −9.34 |
| MYCN | 4613 | 2.75E-07 | 2.8E-05 | −7.22 |
| KCTD8 | 386617 | 3.39E-10 | 7.34E-08 | −6.53 |
| NPBWR1 | 2831 | 5.81E-05 | 0.0025 | −6.26 |
| NKD2 | 85409 | 0.000112 | 0.0042 | −6.07 |
| F13A1 | 2162 | 0.000267 | 0.0083 | −5.91 |
| MMP7 | 4316 | 0.002097 | 0.041 | −5.90 |
| LPL | 4023 | 0.001222 | 0.026 | −5.80 |
| COCH | 1690 | 5.33E-66 | 9.95E-62 | −5.75 |
| DTX1 | 1840 | 0.0015 | 0.030 | −5.71 |
FC: fold change
Verification of the mRNA expression levels of DEGs
To confirm the accuracy of RNA-Seq, the top 10 upregulated and top 10 downregulated genes in N-ESCs (n = 15) and A-EuESCs (n = 15) were selected for further validation by qRT-PCR in an additional set of endometrial tissues. It was revealed that HOXC8 (P < 0.05), IL-33 (P < 0.01), MYH1 (P < 0.01), TCF21 (P < 0.01), ADAMTSL1 (P < 0.01), MYH2 (P < 0.01), CCL7 (P < 0.001), CCL11 (P < 0.05) and CACNG7 (P < 0.01) genes were significantly up-regulated in A-EuESCs compared with N-ESCs (Figure 2(a)), and the expression level of ACAN (P = 0.25) did not significantly differ between N-ESCs and A-EuESCs. Moreover, SLC6A2 (P < 0.001), MYCN (P < 0.001), KCTD8 (P < 0.01), NPBWR1 (P < 0.05), NKD2 (P < 0.01), COCH (P < 0.05), and DTX1 (P < 0.01) genes were significantly down-regulated in A-EuESCs compared with N-ESCs (Figure 2(b)), and the expression levels of F13A1 (P = 0.90) and LPL (P = 0.17) did not significantly differ between N-ESCs and A-EuESCs. Meanwhile, the other upregulated genes including ADAM12, SERPINE1, ADGRA2, ANGPTL4, IL6, NTF3, CCL2, TLR3, GATA6, HGF, and KDR were also selected for validation, which are shown in Supplementary Figure 1, and the validation results achieved by qRT-PCR were almost identical to the RNA-Seq results.
Figure 2.

Verification of the expression levels of DEGs by qRT-PCR qRT-PCR of top 10 upregulated (a) or downregulated (b) DEGs by RNA-Seq in A-EuESCs (n = 15) compared with N-ESCs (n = 15). GAPDH was used as an internal control for normalization. At least three independent experiments were performed for statistical evaluation. Error bars represent standard deviation (SD). *P < 0.05, *P < 0.01, *P < 0.001, A-EuESCs vs. N-ESCs; ns, not statistically significant.
Functional annotation analysis of DEGs
GO annotation was performed on 821 DEGs in N-ESCs and A-EuESCs to obtain insights into their biological functions. Binding (GO:0005488), catalytic activity (GO:0003824), and molecular function regulator (GO:0098772) were the most enriched molecular functions. Cell part (GO:0044464), organelle (GO:0043226), and membrane (GO:0016020) were the most enriched cellular components. Cellular process (GO:0009987), biological regulation (GO:0065007), and developmental process (GO:0032502) were the most enriched biological processes (Figure 3(a)). KEGG annotation was performed on 821 DEGs in N-ESCs and A-EuESCs to obtain insights into their pathways involved. The first three pathways with the highest number of DEGs were signal transduction (n = 121), cancers: overview (n = 74) and endocrine system (n = 57) (Figure 3(b)).
Figure 3.

Functional annotation analysis of DEGs (a) GO annotation was performed using 821 DEGs in N-ESCs and A-EuESCs to obtain insights into their biological functions. The DEGs were classified into three functional groups, including biological process, cellular component and molecular function. (b) KEGG annotation was performed using 821 DEGs in N-ESCs and A-EuESCs to obtain insights into their pathways involved.
Functional enrichment analysis of DEGs
To define the biological functions of the 821 DEGs, GO and KEGG pathway enrichment analyses were performed. The GO term enrichment analysis showed that the up-regulated DEGs were significantly enriched in regulation of angiogenesis (GO:0045765), tissue morphogenesis (GO:0048729) and regulation of vasculature development (GO:1901342) (Figure 4(a)). Meanwhile, the down-regulated DEGs were significantly enriched in regulation of chemotaxis (GO:0050920), cell-cell signaling (GO:0007267), and synaptic signaling (GO:0099536) (Figure 4(b)). The functions of all DEGs were significantly enriched in calcium-dependent cell-cell adhesion (GO:0016339), proteoglycan metabolic process (GO:0006029), and negative regulation of chemotaxis (GO:0050922) (Figure 4(c)).
Figure 4.

GO and KEGG pathway enrichment analyses of the significant DEGs. (a) The top 25 enriched GO biological processes of the upregulated DEGs. (b) The top 25 enriched GO biological processes of the downregulated DEGs. (c) The top 25 enriched GO biological processes of all DEGs. (d) The top 25 enriched KEGG pathways of the upregulated DEGs. (e) The top 25 enriched KEGG pathways of the downregulated DEGs. (f) The top 25 enriched KEGG pathways of all DEGs.
The KEGG pathway analysis revealed that the up-regulated DEGs were significantly enriched in cytokine-cytokine receptor interaction (map04060), TNF signaling pathway (map04668) and vascular smooth muscle contraction (map04270) (Figure 4(d)), while the down-regulated DEGs were significantly enriched in prostate cancer (map05215), Ras signaling pathway (map04014) and microRNAs in cancer (map05206) (Figure 4(e)). The KEGG enrichment analysis showed that all the DEGs were mostly significantly enriched in cytokine-cytokine receptor interaction (map04060), MAPK signaling pathway (map04010), and axon guidance (map04360) (Figure 4(f)).
Analysis of chord plot
The four designated GO terms, including calcium-dependent cell-cell adhesion, regulation of angiogenesis, extracellular matrix organization, and regulation of chemotaxis, belong to biological process (BP) subontology. The presence of IL-6, LPAR1, NTF3, CCL26, and PTK2B was directly related to the regulation of chemotaxis (Figure 5(a)). Importantly, the upregulated transcripts of forkhead box protein C2 (FOXC2) and hypoxia-inducible factor 1A (HIF1A) were involved in the regulation of angiogenesis (Figure 5(a)). Several MMP family genes (MMP3, MMP7, MMP10, MMP12 and MMP27), with a particularly pronounced down-regulation in A-EuESCs, were found to be associated with extracellular matrix organization (Figure 5(a)). Furthermore, we observed a significant downregulation of protocadherin-beta (PCDHB) family genes (PCDHB3, PCDHB4, PCDHB5, PCDHB9, PCDHB13, and PCDHB16) in A-EuESCs, which were associated with calcium-dependent cell-cell adhesion (Figure 5(a)). The KEGG pathway analysis revealed that a significantly upregulated chemokine family member genes (CCL2, CCL7, CCL11, CCL26, and CXCL12) were associated with chemokine signaling pathway (map04062) (Figure 5(b)). In addition to the above-mentioned chemokines, several pronounced upregulated proinflammatory cytokines (IL-33, LIF, IL-6 and IL-15) in A-EuESCs were involved in cytokine-cytokine receptor interaction (map04060) (Figure 5(b)).
Figure 5.

Chord plot analysis of GO terms and KEGG pathways. (a) GO chord plot of 4 designated GO terms belonging to the Biological Process (BP) sub-ontology for A-EuESCs. (b) KEGG chord plot of 2 designated KEGG pathways for A-EuESCs. The genes are linked to their assigned terms or pathways via colored ribbons. Genes are ordered according to the observed log2 fold-change (log2FC), which can be shown in descending intensity of red squares next to the selected genes.
PPI networks of the DEGs in N-ESCs and A-EuESCs
PPI networks of the 157 DEGs identified in significant enrichment pathway (adjusted P < 0.05) by the KEGG pathway analysis in N-ESCs and A-EuESCs were constructed using the STRING database and visualized by the Cytoscape software. In the PPI networks of 157 DEGs, there were 145 nodes and 793 edges (Figure 6(a)). According to the ranking of network topology property indicator of degree centrality, the top 10 nodes were separately identified as hub genes, and IL-6 was a central hub gene in the network, with the maximum number of degree (n = 61) (see Table 3). Besides, PPI subnetwork of the DEGs related to inflammatory cytokines and chemokines were constructed as described above. In these PPI subnetworks, there were 71 nodes and 177 edges (Figure 6(b)). As mentioned earlier, the top 10 nodes were separately identified as hub genes (Table 4), and IL-6 was also a central hub gene in the subnetworks (n = 61) (Table 4). Moreover, another subnetwork of the DEGs related to the regulation of angiogenesis was constructed, including 82 nodes and 240 edges (Figure 6(c)). Similarly, the top 10 nodes were also identified as hub genes (Table 5), and EGF was a central hub gene in this subnetwork (n = 55).
Figure 6.

The constructed PPI networks and subnetworks of the DEGs in N-ESCs and A-EuESCs. (a) PPI network of the 157 DEGs identified in significant enrichment pathway (adjusted P < 0.05) by the KEGG pathway analysis in N-ESCs and A-EuESCs. (b) PPI subnetwork of the DEGs related to inflammatory cytokines and chemokines. (c) PPI subnetwork of the DEGs related to the regulation of angiogenesis. The red circular nodes represent the upregulated DEGs and the blue circular nodes represent the downregulated DEGs; black edges indicate the interaction between two proteins. The larger the circular nodes, the greater the centrality in the PPI network.
Table 3.
The hub nodes in the PPI network according to the score of network topology property indicators
| Rank | Node | Locus | Gene description | Degree |
|---|---|---|---|---|
| 1 | IL6 | 7p15.3 | interleukin 6 | 61 |
| 2 | EGF | 4q25 | epidermal growth factor | 55 |
| 3 | BDNF | 11p14.1 | brain derived neurotrophic factor | 43 |
| 4 | HGF | 7q21.11 | hepatocyte growth factor | 42 |
| 5 | KDR | 4q12 | kinase insert domain receptor | 41 |
| 6 | FGF2 | 4q28.1 | fibroblast growth factor 2 | 40 |
| 7 | CXCL12 | 10q11.21 | C-X-C motif chemokine ligand 12 | 38 |
| 8 | NGF | 1p13.2 | nerve growth factor | 38 |
| 9 | ERBB2 | 17q12 | erb-b2 receptor tyrosine kinase 2 | 34 |
| 10 | CCL2 | 17q12 | C-C motif chemokine ligand 2 | 33 |
Table 4.
The hub nodes in the PPI subnetwork constructed by the DEGs related to inflammatory cytokines and C-C motif chemokines
| Rank | Node | Locus | Gene description | Degree |
|---|---|---|---|---|
| 1 | IL6 | 7p15.3 | interleukin 6 | 61 |
| 2 | CXCL12 | 10q11.21 | C-X-C motif chemokine ligand 12 | 38 |
| 3 | CCL2 | 17q12 | C-C motif chemokine ligand 2 | 33 |
| 4 | IL15 | 4q31.21 | interleukin 15 | 23 |
| 5 | CCL11 | 17q12 | C-C motif chemokine ligand 11 | 13 |
| 6 | IL33 | 9p24.1 | interleukin 33 | 12 |
| 7 | CCL7 | 17q12 | C-C motif chemokine ligand 7 | 11 |
| 8 | CD40 | 20q13.12 | CD40 molecule | 7 |
| 9 | CSF1 | 1p13.3 | colony stimulating factor 1 | 7 |
| 10 | EGF | 4q25 | epidermal growth factor | 7 |
Table 5.
The hub nodes in the PPI subnetwork constructed by the DEGs related to the regulation of angiogenesis
| Rank | Node | Locus | Gene description | Degree |
|---|---|---|---|---|
| 1 | EGF | 4q25 | epidermal growth factor | 55 |
| 2 | HGF | 7q21.11 | hepatocyte growth factor | 42 |
| 3 | FGF2 | 4q28.1 | fibroblast growth factor 2 | 40 |
| 4 | ERBB2 | 17q12 | erb-b2 receptor tyrosine kinase 2 | 34 |
| 5 | CCL2 | 17q12 | C-C motif chemokine ligand 2 | 33 |
| 6 | SERPINE1 | 7q22.1 | serpin family E member 1 | 24 |
| 7 | MMP3 | 11q22.2 | matrix metallopeptidase 3 | 21 |
| 8 | ANGPT2 | 8p23.1 | angiopoietin 2 | 16 |
| 9 | CXCL12 | 10q11.21 | C-X-C motif chemokine ligand 12 | 8 |
| 10 | IL6 | 7p15.3 | interleukin 6 | 8 |
Quantitative analysis of ESCs-secreted protein levels of IL-6 and EGF
To further detect ESCs-secreted protein levels of IL-6 and EGF in the cell culture supernatants, two ELISA kits were used. The protein levels of IL-6 and EGF in supernatants were significantly higher in A-EuESCs than those in N-ESCs (Figure 7(a,b)). These results further confirmed that IL-6 and EGF were key hub genes in the subnetworks of the DEGs related to the regulation of inflammatory cytokines/chemokines and angiogenesis.
Figure 7.
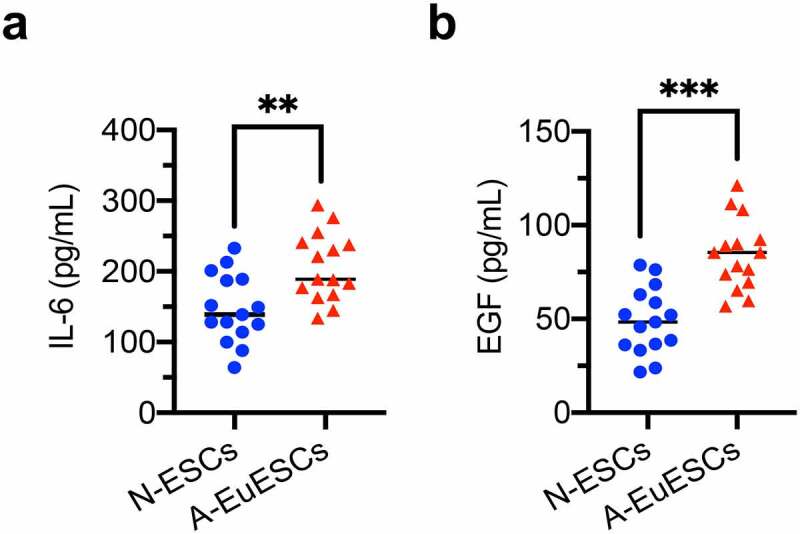
ESCs-secreted protein levels of hub genes in the cell culture supernatants ESCs-secreted protein levels in the cell culture supernatants from N-ESCs (n = 15) and A-EuESCs (n = 15) were determined using ELISA kits. (a) The concentration of IL-6 in the cell culture supernatants. (b) The concentration of EGF in the cell culture supernatants. *P < 0.05, *P < 0.01, *P < 0.001, A-EuESCs vs. N-ESCs.
Discussion
To date, no study has directly investigated the transcriptomic profile of EuESCs from women with clinically significant AM. Given the cyclical changes in the uterine endometrium [41,42], the A-EuESCs and N-ESCs were collected in the mid-secretory phase of the menstrual cycle, as confirmed by histopathological examination, for RNA-Seq. Hence, our study presented the first genome-wide view of the gene expression profiles in secretory EuESCs from female patients with AM.
In the present study, we compared the transcriptomic profiles between A-EuESCs and N-ESCs. The potential functions of dysregulated genes in A-EuESCs and N-ESCs, were predicted through the GO and KEGG pathway enrichment analyses. The GO enrichment analysis revealed that the DEGs in N-EuESCs and A-EuESCs were mainly enriched in cell-cell adhesion, angiogenesis, extracellular matrix organization, chemotaxis, etc. Meanwhile, the KEGG pathway enrichment analysis revealed that the abovementioned DEGs were mainly enriched in cytokine-cytokine receptor interaction, MAPK signaling pathway, PI3K-AKT signaling pathway, and chemokine signaling pathway.
The three tandem-arrayed protocadherin (PCDH) gene clusters, including Pcdh-α, Pcdh-β, and Pcdh-γ, play fundamental roles in the development of the vertebrate central nervous system [43]. A growing body of evidence suggested that PCDHs are widely involved in the pathogenesis and progression of multiple types of cancers by enhancing invasion and metastasis [44,45]. According to the results of chord plot analysis, several PCDH-β family member genes (PCDHB3, PCDHB4, PCDHB5, PCDHB9, PCDHB13 and PCDHB16), with a particularly pronounced down-regulation in A-EuESCs were directly related to calcium-dependent cell-cell adhesion. Downregulation of PCDH-β genes may affect invasion and migration of A-EuESCs by regulating intercellular adhesion and cell spreading. Meanwhile, 2 known genes, including FOXC2 and SERPINE1, with a particularly pronounced up-regulation in A-EuESCs were directly related to in extracellular matrix organization. Up-regulation of these genes may promote cell invasion and migration by degrading extracellular matrix. In addition, we found that the chemotactic genes, including IL-6, LPAR1, NTF3, CCL26, PTK2B, ADGRA2, ANGPTL4, and SERPINE1 were expressed through up-regulation in A-EuESCs, suggesting that A-EuESCs may have more movement tendency than N-EuESCs.
Previous studies have shown that inflammation was accumulated in the eutopic endometrium compared with in the control endometrium [46,47], which was supported by the results of the comparison between the A-EuESCs and N-ESCs in the present study. The PPI subnetworks showed that IL-6 acted as the most significant hub gene and interacted with several important inflammatory cytokines and chemokines. Recently, Xiang et al. has reported the increased mRNA expression of SERPINE1 in both eutopic and ectopic endometrium compared with that in controls during proliferative and secretory phase, while the altered expression of SERPINE1 in cellular components of endometrial tissues remained elusive [17]. Remarkably, our study further demonstrated that SERPINE1 was significantly upregulated in A-EuESCs compared with N-ESCs.
It has also been shown that angiogenesis participates in the pathophysiology of abnormal uterine bleeding and subfertility in AM [48]. Wang et al. demonstrated that A-EuESCs treated with β-estradiol presented stronger pro-angiogenetic capacities, accompanied by the increased expression levels of VEGFB and ANGPTL4 proteins [14]. Furthermore, our chord plot analysis revealed that upregulated genes of FOXC2, TLR3, GATA6, HGF, IL-6, PTK2B, ANGPTL4, and HIF1A were involved in the regulation of angiogenesis (Figure 5). Importantly, the PPI subnetworks showed that EGF acted as most significant hub gene and interacted with other angiogenic factors (HGF, ERBB2, FGF2 and SERPINE1) (Figure 6). These results demonstrated that A-EuESCs have the characteristics of pro-angiogenic activity compared with N-ESCs. Besides, the recent scRNA-seq analysis has shown that the cell motility, cell proliferation, angiogenesis, and inflammation terms were enriched in eutopic endometrium versus normal endometrium. The DEGs were mainly functioned in angiogenesis and cell mobility-related cytoskeleton regulation and chemotaxis. However, there is no information related to stromal cell subpopulation [18]. Hence, the overlapping of our findings with the previous studies may reflect the universal mechanisms underlying the AM and the reliability of our data.
In summary, these findings, for the first time, revealed the overall characteristics of A-EuESCs from the perspective of transcriptomic profiles. Besides, there are several limitations in our study. First, endometrial samples analyzed herein were from the mid-secretory phase of menstrual cycle, and study of gene dysregulation during proliferative phase may increase the understanding of abnormalities of A-EuESCs in the pathogenesis of AM. Second, the sample size was small, and further study should be conducted with a larger sample size. Third, the dysregulated genes were only validated through qRT-PCR and ELISA in ESCs, and no validation was carried out by in situ biospecimens. Last but not least, the biological functions of several DEGs, including HOXC8, TCF21, CACNG7, FOXC2, SERPINE1, ADGRA2, NTF3, TLR3, GATA6, KCTD8 and DTX1 in the pathophysiology of AM remained unclear and should be further explored by in vitro and in vivo studies.
Conclusions
The present study provided an important basis to prior focal studies that revealed fundamental abnormalities in A-EuESCs in women with AM. The findings of the DEGs lay a foundation for further investigation to elucidate the mechanisms of AM, and the altered pathways in A-EuESCs may predispose to pro-invasion/migration and angiogenesis, which may be involved in the development of AM. The DEGs and pathways may assist scholars in the development of further efficacious therapies for AM. However, specific roles and mechanisms of the DEGs in A-EuESCs should be investigated and confirmed in the future study.
Supplementary Material
Funding Statement
The Second Batch of Scientific Research Project of Jiangsu Provincial Bureau of Traditional Chinese Medicine (No. JD201814), Xiangcheng District Science and Technology Project of Suzhou (No. SYSD2020059), The Workstation Construction Project of Lanyuan Community Service Center (No. 18-G2S-G22-003), and Graduate Research and Practice Innovation Plan of Jiangsu Province (No. 021093002436).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Authors’ contributions
LG and YL conceived the study, executed the study, and wrote the manuscript. YC, MH, JC, MC and ZW contributed to patient recruitment, data collection, and data evaluation. TG and GW supervised the patient recruitment, manuscript editing, drafting, and the editing and approval of the final version of this paper for submission. All authors approved the final manuscript.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21655979.2022.2077614
References
- [1].Benetti-Pinto CL, Mira TAA, Yela DA, et al. Pharmacological treatment for symptomatic adenomyosis: a systematic review. Rev Bras Ginecol Obstet. 2019;41(9):564–574. [DOI] [PubMed] [Google Scholar]
- [2].Struble J, Reid S, Bedaiwy MA.. Adenomyosis: a clinical review of a challenging gynecologic condition. J Minim Invasive Gynecol. 2016;23(2):164–185. [DOI] [PubMed] [Google Scholar]
- [3].Guo SW. The pathogenesis of adenomyosis vis-a-vis endometriosis. J Clin Med. 2020;9(2):485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chapron C, Vannuccini S, Santulli P, et al. Diagnosing adenomyosis: an integrated clinical and imaging approach. Hum Reprod Update. 2020;26(3):392–411. [DOI] [PubMed] [Google Scholar]
- [5].Vannuccini S, Petraglia F, El Moshy S. Recent advances in understanding and managing adenomyosis. F1000Res. 2019;8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Vannuccini S, Tosti C, Carmona F, et al. Pathogenesis of adenomyosis: an update on molecular mechanisms. Reprod Biomed Online. 2017;35(5):592–601. [DOI] [PubMed] [Google Scholar]
- [7].Liu H, Lang JH. Is abnormal eutopic endometrium the cause of endometriosis? The role of eutopic endometrium in pathogenesis of endometriosis. Med Sci Monit. 2011;17(4):RA92–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chen L, Li C, Guo J, et al. Eutopic/ectopic endometrial apoptosis initiated by bilateral uterine artery occlusion: a new therapeutic mechanism for uterus-sparing surgery in adenomyosis. PLoS One. 2017;12(4):e0175511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Matsumoto Y, Iwasaka T, Yamasaki F, et al. Apoptosis and Ki-67 expression in adenomyotic lesions and in the corresponding eutopic endometrium. Obstet Gynecol. 1999;94(1):71–77. [DOI] [PubMed] [Google Scholar]
- [10].Yang JH, Wu MY, Chen CD, et al. Altered apoptosis and proliferation in endometrial stromal cells of women with adenomyosis. Hum Reprod. 2007;22(4):945–952. [DOI] [PubMed] [Google Scholar]
- [11].Goteri G, Lucarini G, Montik N, et al. Expression of vascular endothelial growth factor (VEGF), hypoxia inducible factor-1alpha (HIF-1alpha), and microvessel density in endometrial tissue in women with adenomyosis. Int J Gynecol Pathol. 2009;28(2):157–163. [DOI] [PubMed] [Google Scholar]
- [12].Nie J, Lu Y, Liu X, et al. Immunoreactivity of progesterone receptor isoform B, nuclear factor kappaB, and IkappaBalpha in adenomyosis. Fertil Steril. 2009;92(3):886–889. [DOI] [PubMed] [Google Scholar]
- [13].Li J, Yanyan M, Mu L, et al. The expression of Bcl-2 in adenomyosis and its effect on proliferation, migration, and apoptosis of endometrial stromal cells. Pathol Res Pract. 2019;215(8):152477. [DOI] [PubMed] [Google Scholar]
- [14].Wang YY, Duan H, Wang S, et al. Upregulated Talin1 synergistically boosts beta-estradiol-induced proliferation and pro-angiogenesis of eutopic and ectopic endometrial stromal cells in adenomyosis. Reprod Biol Endocrinol. 2021;19(1):70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Benagiano G, Brosens I. The endometrium in adenomyosis. Womens Health (Lond). 2012;8(3):301–312. [DOI] [PubMed] [Google Scholar]
- [16].Herndon CN, Aghajanova L, Balayan S, et al. Global transcriptome abnormalities of the eutopic endometrium from women with adenomyosis. Reprod Sci. 2016;23(10):1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Xiang Y, Sun Y, Yang B, et al. Transcriptome sequencing of adenomyosis eutopic endometrium: a new insight into its pathophysiology. J Cell Mol Med. 2019;23(12):8381–8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Liu Z, Sun Z, Liu H, et al. Single-cell transcriptomic analysis of eutopic endometrium and ectopic lesions of adenomyosis. Cell Biosci. 2021;11(1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Parrott E, Butterworth M, Green A, et al. Adenomyosis—A result of disordered stromal differentiation. Am J Pathol. 2001;159(2):623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Huang N, Xu L, Qiu Y, et al. Down-regulated miR-124-3p enhanced the migration and epithelial-stromal transformation of endometrial stromal cells extracted from eutopic endometrium in subjects with adenomyosis by up-regulating neuropilin 1. Tissue Cell. 2021;69:101474. [DOI] [PubMed] [Google Scholar]
- [21].Wang Q, Wang L, Shao J, et al. IL-22 enhances the invasiveness of endometrial stromal cells of adenomyosis in an autocrine manner. Int J Clin Exp Pathol. 2014;7(9):5762–5771. [PMC free article] [PubMed] [Google Scholar]
- [22].Xue J, Zhang H, Liu W, et al. Metformin inhibits growth of eutopic stromal cells from adenomyotic endometrium via AMPK activation and subsequent inhibition of AKT phosphorylation: a possible role in the treatment of adenomyosis. Reproduction. 2013;146(4):397–406. [DOI] [PubMed] [Google Scholar]
- [23].Liu Y, Wang X, Wan L, et al. TIPE2 inhibits the migration and invasion of endometrial cells by targeting beta-catenin to reverse epithelial-mesenchymal transition. Hum Reprod. 2020;35(6):1377–1390. [DOI] [PubMed] [Google Scholar]
- [24].Cheong ML, Lai TH, Wu WB. Connective tissue growth factor mediates transforming growth factor beta-induced collagen expression in human endometrial stromal cells. PLoS One. 2019;14(1):e0210765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhai J, Li S, Sen S, et al. m(6)A RNA methylation regulators contribute to eutopic endometrium and myometrium dysfunction in adenomyosis. Front Genet. 2020;11:716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Evron A, Goldman S, Shalev E. Effect of primary human endometrial stromal cells on epithelial cell receptivity and protein expression is dependent on menstrual cycle stage. Hum Reprod. 2011;26(1):176–190. [DOI] [PubMed] [Google Scholar]
- [27].Ryan IP, Schriock ED, Taylor RN. Isolation, characterization, and comparison of human endometrial and endometriosis cells in vitro. J Clin Endocrinol Metab. 1994;78(3):642–649. [DOI] [PubMed] [Google Scholar]
- [28].Lockwood CJY. Steroid-modulated stromal cell tissue factor expression: a model for the regulation of endometrial hemostasis and menstruation [J]. J Clin Endocrinol Metab. 1993;77:1014–1019. [DOI] [PubMed] [Google Scholar]
- [29].Bruse C, Guan Y, Carlberg M, et al. Basal release of urokinase plasminogen activator, plasminogen activator inhibitor-1, and soluble plasminogen activator receptor from separated and cultured endometriotic and endometrial stromal and epithelial cells. Fertil Steril. 2005;83(4):1155–1160. [DOI] [PubMed] [Google Scholar]
- [30].Mutz KO, Heilkenbrinker A, Lönne M, et al. Transcriptome analysis using next-generation sequencing. Curr Opin Biotechnol. 2013;24(1):22–30. [DOI] [PubMed] [Google Scholar]
- [31].Sun X, Liu Q, Huang J, et al. Transcriptome-based stemness indices analysis reveals platinum-based chemo-therapeutic response indicators in advanced-stage serous ovarian cancer. Bioengineered. 2021;12(1):3753–3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gene Ontology Consortium . The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34(D1):D322–D326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Clough E, Barrett T. The Gene Expression Omnibus database. Methods Mol Biol. 2016;1418:93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. [DOI] [PubMed] [Google Scholar]
- [36].Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–622. [DOI] [PubMed] [Google Scholar]
- [37].Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(D1):D808–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kohl M, Wiese S, Warscheid B. Cytoscape: software for visualization and analysis of biological networks. Methods Mol Biol. 2011;696:291–303. [DOI] [PubMed] [Google Scholar]
- [39].Tang Y, Li M, Wang J, et al. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems. 2015;127:67–72. [DOI] [PubMed] [Google Scholar]
- [40].Konstantinou GN. Enzyme-Linked Immunosorbent Assay (ELISA). Methods Mol Biol. 2017;1592:79–94. [DOI] [PubMed] [Google Scholar]
- [41].Page EW, Glendening MB, Parkinson D. Cyclic biochemical changes in the human endometrium, with special reference to the fibrinolytic enzyme. Am J Obstet Gynecol. 1951;62(5):1100–1105. [DOI] [PubMed] [Google Scholar]
- [42].Ueki M, Watanabe K, Kanki S, et al. Menstrual cyclic changes in a progesterone receptor in normal human endometrium. Bull Osaka Med Sch. 1978;24(1):16–22. [PubMed] [Google Scholar]
- [43].Han MH, Lin C, Meng S, et al. Proteomics analysis reveals overlapping functions of clustered protocadherins. Mol Cell Proteomics. 2010;9(1):71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].van Roy F. Beyond E-cadherin: roles of other cadherin superfamily members in cancer. Nat Rev Cancer. 2014;14(2):121–134. [DOI] [PubMed] [Google Scholar]
- [45].Lin Y, Ge X, Zhang X, et al. Protocadherin-8 promotes invasion and metastasis via laminin subunit gamma2 in gastric cancer. Cancer Sci. 2018;109(3):732–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Li C, Chen R, Jiang C, et al. Correlation of LOX5 and COX2 expression with inflammatory pathology and clinical features of adenomyosis. Mol Med Rep. 2019;19(1):727–733. [DOI] [PubMed] [Google Scholar]
- [47].Carrarelli P, Yen CF, Funghi L, et al. Expression of inflammatory and neurogenic mediators in adenomyosis. Reprod Sci. 2017;24(3):369–375. [DOI] [PubMed] [Google Scholar]
- [48].Harmsen MJ, Wong CFC, Mijatovic V, et al. Role of angiogenesis in adenomyosis-associated abnormal uterine bleeding and subfertility: a systematic review. Hum Reprod Update. 2019;25(5):647–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


