Abstract
Despite an increasing interest in horizontal gene transfer in bacteria, the role of generalized transduction in this process has not been well investigated yet. Certainly one of the reasons is that only a small fraction of general transducing bacteriophages have been characterized, because many bacterial hosts needed for propagation and identification are not culturable or are simply unknown. A method for host-independent detection of transducing bacteriophages was developed. Phage-encapsulated DNA was used as a template for PCR amplification of 16S ribosomal DNA using primers specific for the 16S rRNA genes of most eubacteria. Sequencing of the cloned amplification products permits the identification of the host bacteria. The Salmonella phage P22 was used as an example. Applying this method to a sample of the supernatant of the mixed liquor in the aeration tank of an activated sludge treatment works revealed the presence of transducing phages infecting several bacterial species for which such phages have not yet been described. This method is suitable for estimating the contribution of generalized transduction to horizontal gene transfer in different habitats.
Horizontal gene transfer in bacteria is an important factor in evolution. The interest in gene flux in the environment has also been stimulated by the discussion of the risks associated with the release of genetically modified microorganisms. In recent years, increased attempts have been made to assess the contribution of the various gene transfer mechanisms. Most effort has been put into plasmid-mediated conjugation and to a lesser extent into transformation, the uptake of free DNA. Phage-mediated transduction, however, has been widely neglected. One of the reasons may be that only a few generalized transducing phages from well-established laboratory strains are known.
In a previous study, we have shown that at least 95% of the natural strains of Salmonella enterica serovar Typhimurium harbor prophages (6). About 62% of the released phage strains could be assayed for their ability to transduce host genetic markers. With only two exceptions, all of them appeared to be generalized transducing phages. The detection of released phage particles from a lysogenic culture depends on the availability of suitable and sensitive bacterial indicator strains. The test for the transducing ability of the phages detected depends also on the availability of suitable mutant strains as recipients. Therefore, only 62% of the phages could be assayed.
Obviously, it would be rather unproductive to conduct a similar study for bacterial species that are not as well characterized and as well established in laboratories as serovar Typhimurium and its available mutant collections. If one asks for the overall occurrence of generalized transducing phages in natural habitats, still another problem interferes: 90 to 99.9% of all bacteria are considered unculturable (2). All these species would escape such an investigation. Therefore, it was desirable to develop a method for host-independent detection of transducing phage particles.
MATERIALS AND METHODS
Concentration of phage particles and destruction of free DNA.
Lysates of phage P22H5 (c2 mutant) and of phage λcI857 Sam7 were sterilized with membrane filters (pore size, 0.45 μm; Schleicher & Schuell, Dassel, Germany) and concentrated by ultracentrifugation (38.5-ml tubes; 2 h; 22,000 rpm; 5°C; Kontron TGA-50; rotor TST 28.38). The sediment was resuspended in 500 μl of DNase buffer (50 mM Tris-HCl [pH 7.5], 10 mM MgCl2) and spiked with 1 μg of the control plasmid pHpaC (pUC19H with an amplifiable insert of phage ES18 DNA). To degrade nucleic acids, 100 μl of DNase I, 30 μl of RNase, and 30 μl of lysozyme were added (enzymes were from Boehringer, Mannheim, Germany) (stock solutions were at 10 mg ml−1) and shaken overnight at 6°C. The sample was extracted with an equal volume of chloroform and centrifuged for 30 min at 3,000 rpm at room temperature in a Heraeus Minifuge. The supernatant was incubated with 100 μl of lysozyme for 1 h at 37°C. After a further chloroform treatment and subsequent centrifugation, 100 μl of DNase, 30 μl of RNase, and 30 μl of lysozyme were added and incubated overnight at 6°C. Then, the phages were assumed to be ready for phenol extraction.
Environmental sample.
From a sample of the supernatant of the mixed liquor in the aeration tank of an activated sludge treatment works (treatment plants Munich II, Munich, Germany) (samples are referred to as sewage water), coarse particles were removed by centrifugation (SS34, 5,000 rpm, 30 min; Sorvall) and filtration (Folded Filters; Schleicher & Schuell). Subsequently, the material was treated like phage lysates. Combined phage sediments of 1 liter of sewage water were resuspended in 3 ml of DNase buffer, and other volumes were adjusted accordingly.
Extraction of bacteriophage-encapsulated DNA.
Phage-encapsulated DNA was extracted with the phenol-chloroform method (all components from Roth, Karlsruhe, Germany) according to the method of Maniatis et al. (4). DNA was precipitated at −80°C overnight by addition of 0.4 volumes of 5 M ammonium acetate and 2 volumes of 99% (vol/vol) ethanol. After centrifugation, DNA was washed with 70% (vol/vol) ethanol, dried under vacuum, and resolved overnight in double-distilled H2O by shaking at 6°C. The DNA concentration was determined spectrophotometrically at 260 nm (Gene Quant; Pharmacia, Cambridge, United Kingdom). DNA quality and size were controlled electrophoretically.
PCR amplification.
The amplification reaction mixture contained 10 ng of DNA, 5 μl of 10× PCR buffer, 1.5 mM MgCl2 (for 16S ribosomal DNA [rDNA]) or 2.5 mM MgCl2 (for plasmid control amplification), 2 U of native Taq polymerase (all components were from MBI Fermentas, St. Leon-Rot, Germany), 10 mM (each) deoxynucleotide triphosphates (PCR Nucleotide Mix; Boehringer) and 0.3 mM (each) primers. The final reactions/mixture volume was 50 μl. The mixture was incubated in a thermal cycler (RoboCycler; Stratagene). The cycling program was the following: initial denaturation at 95°C for 5 min; 30 cycles of 95°C for 1 min, primer annealing at 50°C for 1 min, and DNA synthesis at 72°C for 2 min; and a final extension step at 72°C for 7 min. All primers used in this study (Table 1) were synthesized by MWG Biotech (Ebersberg, Germany).
TABLE 1.
PCR and sequencing primers used in this studya
| Primer | Sequence (5′ to 3′) |
|---|---|
| 28F* | AAG AGT TTG ATC CTG GCT CAG A |
| 525F | AAC TCC GTG CCA GCA GC |
| 1492R* | TAC GGC TAC CTT GTT ACG ACT T |
| M13 reverse | CAG GAA ACA GCT ATG AC |
| M13 (−20) forward | GTA AAA CGA CGG CCA GT |
Cloning and sequencing.
Amplification products were analyzed by electrophoresis in a 0.8% agarose gel, cloned into the plasmid pCR 2.1-TOPO by TA cloning (Invitrogen, Groningen, The Netherlands), and transformed into Escherichia coli strain TOP10F′ (Invitrogen). Transformants were selected on Luria broth agar plates containing 100 μg of ampicillin ml−1, 50 μg of 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid ml−1, and 200 mM isopropyl-β-d-thiogalactopyranoside. Putative positive recombinant plasmids were isolated by boiling the plasmid preparations (3) and verified by EcoRI digestion (New England Biolabs, Beverly, Mass.). The plasmids selected for sequencing were purified using Jet Quick Clean-Up (Genomed, Bad Oyenhausen, Germany) following the manufacturer's recommendations. The plasmid DNA concentration was determined spectrophotometrically at 260 nm (Gene Quant; Pharmacia). Plasmid DNA (1.2 μg) was used for each sequencing reaction. Cloned 16S rDNAs were completely sequenced by M13 reverse, M13 (−20) forward, and 525F primers (sequences of primers are shown in Table 1) using the DyeTerminator Cycle-Sequencing Kit AmpliTaq FS and the automatic sequence analyzer ABI PRISM 377 DNA Sequencer (Perkin-Elmer).
Nucleotide sequence accession numbers.
Sequences have been submitted to GenBank. Accession numbers from AF324530 to AF324539 were assigned.
RESULTS
The concept.
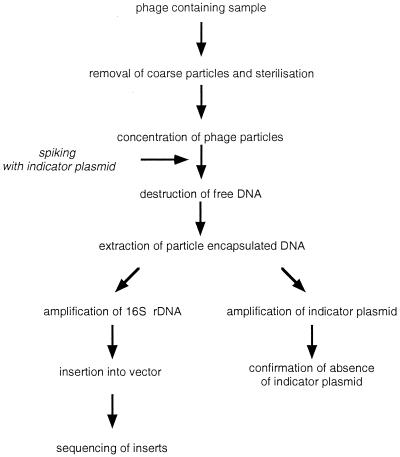
A lysate of a generalized transducing phage contains the entire genome of its propagating host strain as headful-sized fragments, thus representing a diluted complete gene library of the (killed) cells, including also the gene(s) for 16S rRNA. These 16S rRNA genes consist of highly conserved and variable sections, which are characteristic for the respective species or genus, like fingerprints. Therefore, we intended to extract phage-encapsulated DNA after removal of cells and complete destruction of all free nucleic acids, to amplify previously phage-encapsulated bacterial 16S rDNA by PCR. Primers used for amplification correspond with slight modifications to the primers fD1 and rP2, which were described elsewhere and allow amplification of almost full-length 16S rDNA of most eubacteria (7). After cloning of the amplification products, a collection of clones was expected to carry the 16S rDNA of those bacteria in the sample which have released general transducing phages. After the inserts are sequenced, the variable sections should yield information on which bacteria the transducing phages were released.
Removal of free DNA.
The method was developed and conditions were optimized using a lysate of the generalized transducing Salmonella phage P22 as a test system.
The most critical step in the procedure is the complete removal of free DNA which would contribute 16S rDNA of the lysed host cells. To control the efficacy of DNA destruction, we used two approaches.
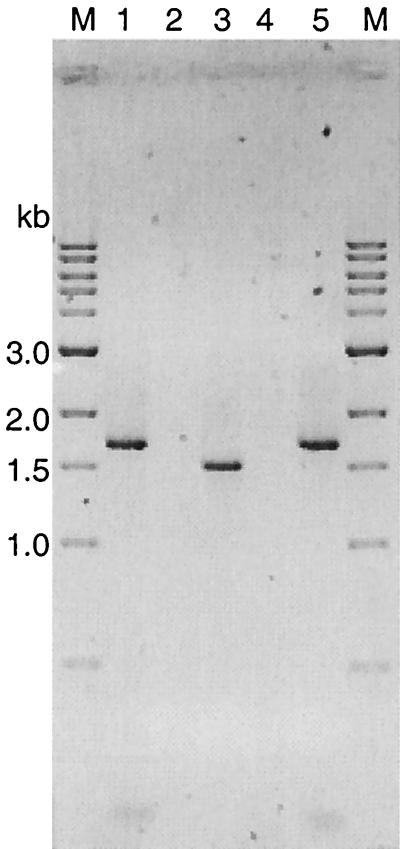
(i) We spiked the P22 lysate before adding the DNase I with a high concentration of plasmid pHpaC having an insert of length similar to that of 16S rDNA, 1.6 kb (Fig. 1). The plasmid was linearized by NdeI treatment to simulate free bacterial DNA. PCR amplification with standard universal and reverse primers [M13 (−20) forward and M13 reverse, respectively] yielded the expected product of 1.6 kb. Two additional bands of about 900 (visible only at high loading) and 200 bp were visible, which possibly derive from secondary binding sites (Fig. 2, lanes 1 and 5). When this external DNA was degraded so that it could not be detected by PCR amplification, it was assumed that all free bacterial DNA, present in a sample to a smaller amount, would also be destroyed. Figure 2 shows the results with a spiked lysate of phage P22. Lane 2 demonstrates that no control plasmid could be amplified, whereas lane 3 shows the amplification result of the same P22 DNA preparation when using the 16S rDNA-specific primers: a clear band appears at about 1.5 kb, the expected size of the Salmonella 16S rDNA amplification product.
FIG. 1.
Protocol for preparation and amplification of phage-encapsulated DNA.
FIG. 2.
Agarose gel analysis of PCR amplification products. We performed 16S rDNA amplification with primers 28F and 1492R and control insert amplification with standard primers M13 reverse and M13 (−20) forward. Lane M, 1-kb ladder (New England Biolabs); lanes 1 and 5, amplification product of the control insert of plasmid pHpaC; lanes 2 to 4, test for the presence of the control insert of plasmid pHpaC or 16S rDNA, respectively, in phage lysates. All lysates were spiked with pHpaC, free DNA was degraded, and encapsulated DNA subsequently was extracted. Lane 2, control insert amplification of a P22 lysate; lane 3, 16S rDNA amplification of a P22 lysate; lane 4, 16S rDNA amplification of a lambda lysate. The 900-bp pHpaC amplification product is invisible at the applicated DNA amounts in lanes 1 and 5.
(ii) The same steps were performed with the nontransducing E. coli phage lambda. As Fig. 2 (lane 4) shows, no amplification product could be found using the 16S rDNA primers, indicating that no free 16S rDNA of lysed host cells contaminated the sample and that the positive result with P22 was due to the phage-encapsulated 16S rDNA of the host.
Initially, we had considerable problems. The DNase treatment was optimized in such a way that the control plasmid could not be detected by PCR. Nevertheless, 16S rDNA was amplified even in the negative lambda control. A systematic search revealed that 16S rDNA traces were present in the Taq polymerase used for PCR amplification and therefore came into the sample after DNase treatment. A comparison of various Taq polymerases showed that all tested enzymes produced by genetically modified E. coli strains contained traces of 16S rDNA. The only tested product which did not contribute contaminating DNA was native Taq polymerase from MBI-Fermentas. This was consistent using different batches of enzyme from the same supplier. This could be a consequence of the higher purity of the enzyme. But, it is more probable that our primers did not bind to Thermus aquaticus 16S rDNA. The available T. aquaticus sequence, which may not be complete, has no homology with the primer 28F and shows two mismatches with the primer 1492R. Native Taq polymerases from other suppliers were not tested.
Application of the method to a sample of sewage water.
Once the procedure was established with P22, we applied it to a sample of sewage water. To control reliability, spiking with the plasmid pHpaC was performed in parallel with P22 used as a positive control and phage lambda used as a negative control. Since both controls worked as expected and because no amplifiable pHpaC plasmid was left over after DNase treatment, all amplified material seen in an agarose gel was considered to derive from template DNA previously encapsulated in phage particles that were present in the sewage water.
These amplification products were cloned and the inserts of 10 positive clones were sequenced as described. A BLAST (1) search detected very high similarities (96 to 99%) with 16S rDNA from four different bacterial species in the databases (Table 2).
TABLE 2.
16S rDNAs from transducing phages in the carbon decomposition stage of a sewage water purification plant
| Accession no. of phage-derived 16S rDNAs | Species with highest degree of similarity (accession no.a) | % Similarities |
|---|---|---|
| AF324532–AF324536 | Aeromonas hydrophila (AF099022) | 98–99 |
| AF324537 | Acinetobacter johnsonii (X81663) | 99 |
| AF324538 | Acinetobacter johnsonii (X81663) | 96 |
| AF324530 | Aeromonas veronii (AF099024) | 99 |
| AF324531 | Aeromonas allosaccharophila (S39232) | 99 |
| AF324539 | Arcobacter cryaerophilus (U34387) | 99 |
GenBank and EMBL databases.
DISCUSSION
The main problem in establishing a method for detection of transducible 16S rDNA in aquatic samples was the need for complete elimination of free bacterial DNA. This problem was solved by optimizing degradation conditions and by choosing a suitable Taq polymerase that was free of amplifiable 16S rDNA. Complete destruction of free DNA was confirmed by spiking the sample with a well-defined plasmid.
The amplified 16S rDNA from such samples was thought to derive from phage-encapsulated DNA. We cannot exclude the possibility that gene transfer agent-like elements, which have been found in Rhodopseudomonas capsulata, may also be involved (8). The process performed by these gene transfer agents resembles generalized transduction. But, if these particles are different from phages, they may be rare, and to our knowledge they have never been found in other species.
We have shown that this approach allows detection of generalized transducing phage particles without isolation and cultivation of the releasing host cells. Therefore, it will be possible also to detect transducing phages released from nonculturable species. Already the first application detected phages from four different gram-negative bacterial species. Half of the sequenced amplification products were most closely related to Aeromonas hydrophila, although for the genus Aeromonas, transducing phages have not yet been described. We emphasize, however, that the frequency of identified sequences does not necessarily reflect the quantitative composition of the bacterial community of the habitat. There may be dominant bacterial species which release no or only few transducing phages, while rarely represented species may use transduction as the main mechanism for gene transfer.
This method, which also worked well with samples from other habitats (unpublished data), will elucidate the natural hosts of generalized transducing phages in nature and the contribution of these phages to horizontal gene transfer among bacterial communities.
By application of suitable primers, this method could also detect viruses of higher organisms that are able to contribute to horizontal gene transfer among their hosts. It is known that retroviruses occasionally pick up cellular proto-oncogenes by excision errors. This process reflects the formation of specialized transducing particles of bacteriophage lambda and was detected by deleterious effects in the new host cells. Since retroviruses integrate themselves into the host genome at random positions, they may transfer other host genes in a similar way, which has not yet been detected, since they are less detrimental to their hosts. It is also possible that viruses other than retroviruses may perform some kind of general transduction as a consequence of erroneous DNA packaging.
ACKNOWLEDGMENTS
This work was supported by a grant from the German Ministry for Education, Science, Research, and Technology (BEO 21/0311230).
We thank J. C. Fry, Cardiff University, Cardiff, United Kingdom, and T. Zilker, TU, Munich, Germany, for critically reading and correcting the manuscript.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holmes D S, Quigley M. A rapid boiling method for preparation of bacterial plasmids. Anal Biochem. 1981;114:193. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- 4.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 5.Noller H F. Structure of ribosomal RNA. Annu Rev Biochem. 1983;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- 6.Schicklmaier P, Schmieger H. Frequency of generalized transducing phages in natural isolates of the Salmonella typhimurium complex. Appl Environ Microbiol. 1994;61:1637–1640. doi: 10.1128/aem.61.4.1637-1640.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yen H C, Hu N T, Marrs B L. Characterization of the gene transfer agent made by an overproducer mutant of Rhodopseudomonas capsulata. J Mol Biol. 1979;131:157–168. doi: 10.1016/0022-2836(79)90071-8. [DOI] [PubMed] [Google Scholar]