ABSTRACT
Pulmonary arterial hypertension (PAH) is a cardiovascular disease that has high incidence and causes massive deaths. miR-155-5p/PYGL pathway was revealed to play a crucial role in PAH by weighted gene co-expression network analysis (WGCNA). The potential mechanism of miR-155-5p in regulating hypoxia-induced pulmonary artery smooth muscle cell (PASMC) function was analyzed through in vitro experiments. Hypoxia treatment stimulated the proliferation of PASMCs and increased the expression of vascular endothelial growth factor (VEGF) and hypoxia-inducible factor-1α (HIF-1α). At the same time, revealed by qRT-PCR and western blot, the level of miR-155-5p was raised, and the level of PYGL was decreased in hypoxia-induced PASMCs. Through CCK-8 assay, transwell assay and flow cytometry, it was revealed that miR-155-5p inhibitor remarkably inhibited the cell proliferation and migration and decreased the proportion of hypoxia-stimulated PASMCs in S and G2/M phases. Dual-luciferase reporter system was subsequently applied to validate the straight regulation of miR-155-5p on PYGL based on the analysis of online database. Furthermore, siPYGL was revealed to reverse the influence of miR-155-5p inhibitor on hypoxia-induced PASMCs. These outcomes indicate that the increased level of miR-155-5p in hypoxia-stimulated PASMCs could enhance the cell proliferation, cell migration, and cell cycle progression by targeting PYGL directly. This study may supply novel treatment strategies for PAH.
Abbreviations: PH, pulmonary hypertension; PAH, pulmonary arterial hypertension; WGCNA, weighted gene co-expression network analysis; PASMCs, pulmonary artery smooth muscle cells; VEGF, vascular endothelial growth factor; HIF-1α, hypoxia-inducible factor-1α; SMCs, smooth muscle cells; DEGs, differentially expressed genes; GEO, Gene Expression Omnibus; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; FBS, fetal bovine serum; OD, optical density; BCA, bicinchoninic acid; PVDF, polyvinylidene fluoride; PBS, phosphate-buffered saline; BP, biological process; MF, molecular function; CC, cell component.
KEYWORDS: miR-155-5p, hypoxia‐induced pulmonary arterial hypertension, PYGL, WGCNA
GRAPHICAL ABSTRACT
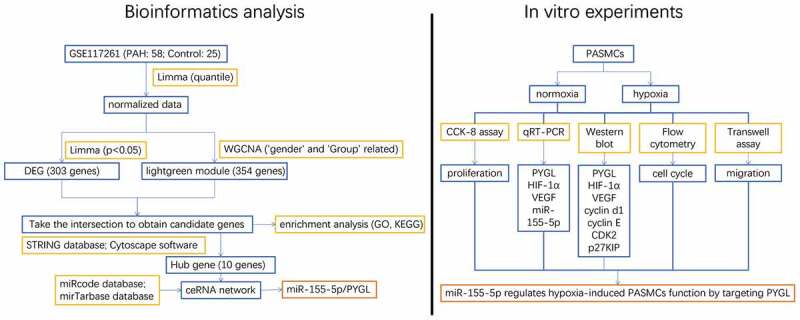
Highlights
It was found that miR-155-5p / PYGL pathway plays an important role in PAH by weighted gene co-expression network analysis (WGCNA).
Hypoxia promoted PASMCs proliferation, increased miR-155-5p level and decreased PYGL expression.
miR-155-5p regulates PASMCs proliferation, migration, and cell cycle.
PYGL targets miR-155-5p directly.
PYGL knockdown reversed the beneficial effect of miR-155-5p inhibitor on hypoxia-induced PASMCs.
1. Introduction
Pulmonary hypertension (PH) is one of the most common cardiovascular diseases that has high incidence and causes massive deaths. Pulmonary vascular resistance is usually persistently elevated during the progression of PH. Pathological studies have found that vascular contracture of pulmonary arterioles, intima hyperplasia and remodeling, and microthrombosis are all the main features of PH. Right-heart failure will be eventually induced by PH [1,2]. As a category of PH, pulmonary arterial hypertension (PAH) is not necessarily a single illness, but probably a complication or syndrome related to other diseases[3]. The proliferation of pulmonary artery smooth muscle cells (PASMCs) is a major symbol of pulmonary vascular remodeling and is the main cause of the occurrence and progression of PAH [4,5].
As small molecules regulating gene expression, miRNAs have been described that they are involved in the adjustment of tumor [6], autoimmune diseases [7] and various cardiovascular diseases, including atherosclerosis [8], hypertension [9], and PH [10,11]. miRNA can induce vascular endothelial cell injury, smooth muscle cells (SMCs) proliferation, migration, and abnormal deposition of extracellular matrix by adjusting the expression levels of related genes and participate in the initiation and progression of PAH [12]. Some miRNAs are of great significance in vascular function regulations, such as miRNA-126, and they can regulate the process of thrombosis, cell proliferation, and apoptosis [13]. As an upstream signal molecule, miRNAs can regulate the biological functions of cells and are decisive in the remodeling of pulmonary vasculature. Accumulating evidence has revealed that miRNA may potentially become a novel target to effectively diagnose and cure PAH [14].
Weighted gene co-expression network analysis (WGCNA) is a data package in the R language [15], which is a method for joint expression network analysis of gene expression data. It is used by many researchers in mining gene expression data [16,17]. WGCNA makes it easier to find key genes and their possible functions, greatly improving the accuracy and speed of research [17]. It has achieved a number of biologically meaningful results in gene expression data analysis of yeast, mice, humans, and other species [18,19]. WGCNA is also used to study the relationship between genes. If a group of genes have identical functions or occur in the same biological pathways, they can be regarded as a module, and then the relationship with external information at the gene module level can be studied, which will greatly reduce the complexity of the problem [20,21].
This study was designed to explore the role of miR-155-5p/PYGL in PAH. First, WGCNA and differentially expressed genes (DEGs) analysis were conducted to analyze PAH-related genes based on data from the Gene Expression Omnibus (GEO) database. Combined with the clinical data, gene modules significantly associated with clinical features were selected. miR-155-5p/PYGL was confirmed to play a crucial role in the progress of PAH. Meanwhile, we explored the role of miR-155-5p/PYGL in PAH through in vitro experiments. The major findings of the current study may help develop a novel therapeutic target for PAH.
2. Materials and methods
2.1. Bioinformatics analysis
The GSE117261 dataset (containing lung samples from 58 PAH patients and 25 healthy controls) was obtained from the GEO database (https://www.ncbi.nlm.nih.gov/geo/) and analyzed accordingly [22]. The expression matrix was annotated with GPL6244, Affymetrix human gene 1.0 ST array (transcription/gene), and normalized using the limma software package and the ‘quantify’ method [23]. As shown in Table 1, data of ‘gender’ (male or female) and ‘Group’ (PAH patients or healthy controls) were collected and analyzed for clinical characteristics [15]. A network was built by the ‘blockwise modules’ function of the WGCNA software, and minModuleSize was set to 80. The genes significantly associated with clinical features (P < 0.05) were identified and chosen for further analysis. Then, considering the complex mechanism of PAH, we defined DEGs with parameter P < 0.05. The DEGs and the genes in important PAH-related modules were summarized and integrated to acquire candidate genes.
Table 1.
Clinical data of patients
| Group | Healthy controls (n = 25) |
PAH patients (n = 58) |
|
|---|---|---|---|
| Age (range) | 1–64 (30.1%) | 7–79 (69.9%) | |
| Gender | Male | 18 (54.5%) | 15 (45.5%) |
| Female | 7 (14%) | 43 (86%) | |
Functional enrichment analysis of the candidate genes was carried out, and the candidate genes were enriched by DAVID (https://david.ncifcrf.gov/) [24]. The highly enriched terms of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were collected. Then, the online tool STRING (https://string-db.org/) was used to research the protein–protein interaction (PPI) of the candidate genes overlapped by DEGs and WGCNA [25]. The interaction between proteins was calculated and sequenced by the ‘Degree’ method to obtain the key genes.
The miRcode database was employed to predict the lncRNA–miRNA interaction. The miRNA-mRNA interaction was predicted by mirTarbase [26]. Hypergeometric experiments were conducted with a threshold (P < 0.01) to evaluate the abundance of miRNAs shared by lncRNA and mRNA [27]. The ceRNA network was visualized via the software of Cytoscape.
2.2. Culture of human PASMCs
Primary human PASMCs were gained from Shanghai Fenghui Biological Research Co., Ltd. (Shanghai, China) and cultured in SMC medium consist of 10% fetal bovine serum (FBS). PASMCs were cultured in an incubator (gas composition: 3% O2 and 5% CO2) for 24 hours to establish the hypoxia model [28]. Normoxia (gas composition: 21% O2 and 5% CO2) was set as a control group.
2.3. Cell Counting Kit-8 (CCK-8) assay
PASMCs were seeded in 96-well plates and treated in light of diverse experimental protocols. Then, cells were further incubated for 2 h after CCK-8 solution was added to each well. Finally, the absorbance at 450 nm was measured to calculate the relative cell viability [29].
2.4. Quantitative reverse transcription PCR (qRT-PCR)
The whole RNA was obtained with Trizol (Invitrogen, USA) [30]. The concentration and purity of RNA were quantified through optical density (OD) at 260 nm and 280 nm. TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster, CA, USA) and PrimeScriptTM RT reagent Kit (Takara, Japan) were used to make miRNA and total RNA reversely transcribed into cDNA, respectively. Then, SYBR Green reagent (Takara) was used for quantification. U6 and GAPDH were made as the internal control for miR-155-5p and other genes. The related primers are presented in Table 2.
Table 2.
Primers for qRT-PCR
| Forward-primer | Reverse-primer | |
|---|---|---|
| PYGL | TATAAGTGAGCTGGCCCAAG | TCTGGACTCATGCTCTGACA |
| HIF-1α | GAACGTCGAAAAGAAAAGTCTCG | CCTTATCAAGATGCGAACTCACA |
| VEGF | CCCTGATGAGATCGAGTACA | AGGAAGCTCATCTCTCCTAT |
| β-actin | TGAGAGGGAAATCGTGCGTGAC | AAGAAGGAAGGCTGGAAAAGAG |
| miR-155-5p | CGCGTTAATGCTAATCGTGATA | AGTGCAGGGTCCGAGGTATT |
| RT-primer | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAACCCC | |
| U6 | CTCGCTTCGGCAGCACA | AACGCTT CACGAATTTGCGT |
2.5. Western blot
The PASMCs were lysed with RIPA buffer (Beyotime, Shanghai, China), and the concentration of protein was quantified using bicinchoninic acid (BCA) protein analysis kit (Beyotime) [31]. Subsequently, 25 μg protein was added in 10% SDS-polyacrylamide gel electrophoresis and moved onto a polyvinylidene fluoride (PVDF) membrane. After being sealed with skim milk, the PVDF membrane was incubated with the primary antibody at 4°C overnight. They were cleaned with phosphate-buffered saline (PBS) three times, with 10 minutes each time, and then incubated with secondary antibodies for 2 hours. ECL chemiluminescence reagent (Beijing Kangwei century Biotechnology Co., Ltd., Beijing, China) was prepared. The PVDF membrane was incubated, and the reaction protein was observed using an enhanced chemiluminescence detection system (BioRad, Hercules, USA).
The primary antibody: PYGL (1:1000, Santa Cruz, USA), HIF-1α (1:1000, Cell Signaling Technology, Inc.), VEGF (1:1000, Cell Signaling Technology, Inc.), cyclin d1 (1:1,000; cat. no. 2978; Cell Signaling Technology, Inc.), cyclin E (1:1,000; cat. no. 20808; Cell Signaling Technology, Inc.), CDK2 (1:1,000; cat. no. 2546; Cell Signaling Technology, Inc.), p27KIP (1:1,000; cat. no. 3686; Cell Signaling Technology, Inc), GAPDH (1:1000, Santa Cruz, USA).
2.6. PASMC cell cycle detected by flow cytometry
Based on the manufacturer’s protocol (BD Biosciences, USA), the PASMCs were fixed with 70% ethanol overnight. Then, cells were dyed in the dark at 37°C for 30 minutes after the addition of 500 µL PI staining solution. The distribution of PASMCs in three phases (G0/G1, S, and G2/M phases) was calculated by a BD FACSCanto II flow cytometry (BD Biosciences, USA) [32].
2.7. Transfection
miRNA-NC/miR-155-5p mimics/miR-155-5p inhibitor was transfected into PASMCs using Lipofectamine RNAimax (Thermo Fisher, USA). For a specific transfection scheme, please refer to the manual provided by the kit manufacturer.
2.8. Transwell assay
Transwell incubator was put in a 24-well plate. Then, the lower chamber was added with 500 µL DMEM medium. The PASMCs from diverse treatments were resuspended in a serum-free DMEM medium and then inoculated into the upper chamber. 1 × 105 PASMCs were inoculated into each well. After incubation at a constant temperature for 24 hours, a cotton swab was used to gently wipe the upper layer. Afterward, PASMCs were fixed with 4% paraformaldehyde and stained with 1% crystal violet for 15 minutes. At last, 10 views were casually chosen, and the quantity of cells was tallied up with 200 magnification [33].
2.9. Dual-luciferase reporter assay
293 T was inoculated onto a 24-well plate, and then pGL3-PYGL-3-UTR (WT)/pGL3-PYGL mut-3-UTR (MT) plasmid and miRNA-NC/miR-155-5p mimics were co-transfected at 37°C for 8 hours. The relative luciferase activities were quantified with the dual-luciferase reporter Kit (Promega, USA) in accordance with the producer’s instructions [34].
2.10. Statistical analysis
The statistics were analyzed using GraphPad Prism Software 7.0 (Los Angeles, USA). The data was displayed as ‘mean ± SD.’ Bilateral t-test was applied to test the individual differences between the two groups. When P < 0.05, the difference was considered to be statistically significant.
3. Results
For the sake of explore the function of miR-155-5p/PYGL in PAH, this study first revealed the significant role of miR-155-5p/PYGL in PAH by bioinformatics analysis. The possible mechanism of miR-155-5p regulating the function of hypoxia-induced PASMCs was investigated through in vitro experiments. Specifically, the function of miR-155-5p/PYGL in the cell proliferation, cell migration, and cell cycle of hypoxia-induced PASMCs was studied by qRT-PCR, western blot, CCK-8, transwell assay, and flow cytometry. The relationship between miR-155-5p and PYGL was identified using a dual-luciferase reporter system.
3.1. Modules and genes related to PAH
Phylogenetic tree analysis showed that there were two outliers (GSM3290090 (PAH) and GSM3290146 (control group)) in the sample population. We marked the samples with a red line and deleted the samples upon the red line (Figure 1(a)). To ensure that there were no outliers, a phylogenetic tree was drawn again (Figure 1(b)). On account of two criteria: (1) the lowest power with a scale-free topology fitting index of 0.90 and (2) a relatively high average connectivity, 7 was picked as the appropriate soft threshold power for analysis (Figure 1(c,d)). We obtained 23 co-expression modules in total, with the quantity of genes ranging from 176 to 2627 (Figure 2(a,b)). There were two modules, midnight blue (cor = −0.6, P = 5e-09) and light green (cor = 0.74, P = 2e-15), significantly correlated with clinical features (Figure 2(c)). The ‘light green’ module, which contained 354 genes and scored the highest (0.74, Figure 2(c)), was eventually selected for further analysis. We used the ‘verboseScatterplot’ function to visualize the correlation between the members in the ‘light green’ module and the importance of genes, and further determined its significant correlation with clinical features (cor = 0.73 and P = 3.8e-60) (Figure 2(d)). According to the network heat map, little correlation was shown between different modules (Figure 2(e)).
Figure 1.
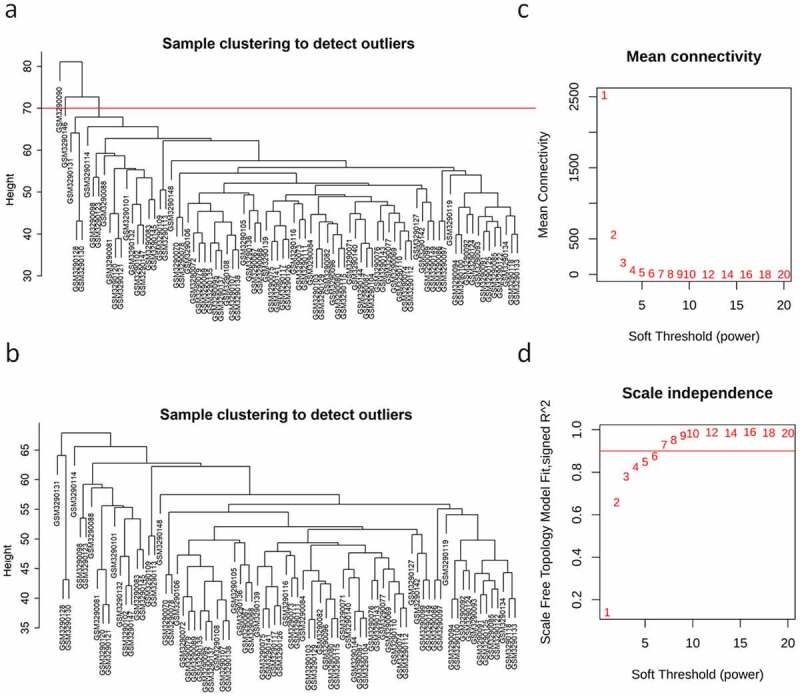
Identification of outliers and definition of soft threshold. (a, b) The clustering tree before and after the filter; the red line indicates the cutting position. (c, d) Determination of the value of power; (c) the average connectivity (Y-axis) decreases with the increase of soft threshold power (X-axis); (d) the scale-free fitting index (Y-axis) at different power (X-axis). The red line in (d) indicates that the correlation coefficient is equal to 0.9.
Figure 2.
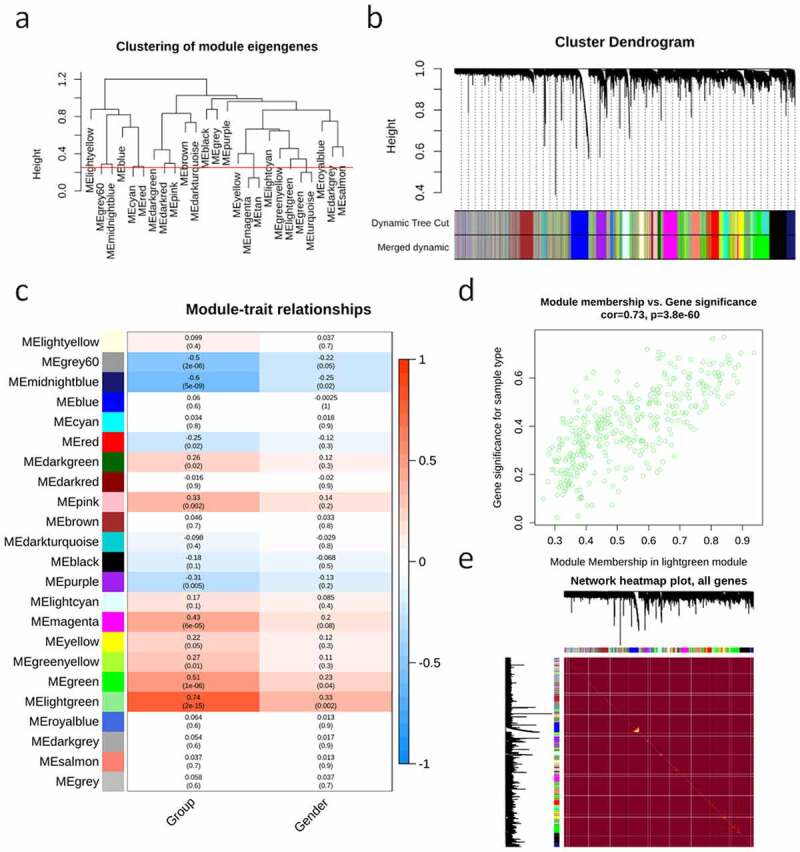
Network structure, gene importance and module members. (a) The clustering tree showing the characteristic genes of the module; the red line (0.25) indicates that the correlation coefficient is 0.75, and the modules under the red line will be merged. (b) Analysis on the cluster tree (top), main module (middle) and merging module (bottom) of genes. (c) Analysis of the correlation (top) and P value (bottom) of module characteristic genes (row) and clinical features (column). (d) MM and GS correlation diagram. (e) Network heat maps of all genes.
3.2. Functional enrichment analysis, PPI network analysis, and ceRNA network construction
We obtained 303 DEGs with a trend of differential expression (P < 0.05). The candidate genes were enriched in 63 GO terms significantly (P < 0.05), containing 38 biological processes (BP), 13 molecular functions (MF) ,and 12 cell components (CC). Twelve terms (the first four terms of each category) were described in a circular graph (Figure 3(a)). The KEGG enrichment pathways were visualized with Cytoscape (Figure 3(b)). Complement and coagulation cascade (hsa04610), osteoclast differentiation (hsa04380), and cytokine–cytokine receptor interaction (hsa04060) were significantly enriched (P < 0.05). A PPI network of candidate genes was built using the STRING database (Figure 4(a)). We obtained 10 hub genes (TLR4, S100A12, LILRB2, MNDA, MMP9, TLR1, S100A9, FPR1, APOB, and CLEC4D) (Figure 4(b)). At the same time, a ceRNA network containing three lncRNAs (LINC01270, AC026790.1, and GLIS3-AS1), nine miRNAs (hsa-miR-155-5p, hsa-miR-4735-3p, hsa-miR-93-5p, etc.), and three mRNAs (PYGL, SLC25A37, and ANKS1B) was acquired through miRcode database and mirTarbase (Figure 4(c)). miR-155-5p is closely related to a variety of diseases and may become a marker for the treatment and prognosis of PAH [35–38]. In addition, miR-155-5p could play a key role by targeting PYGL, and this was further verified by in vitro experiments.
Figure 3.
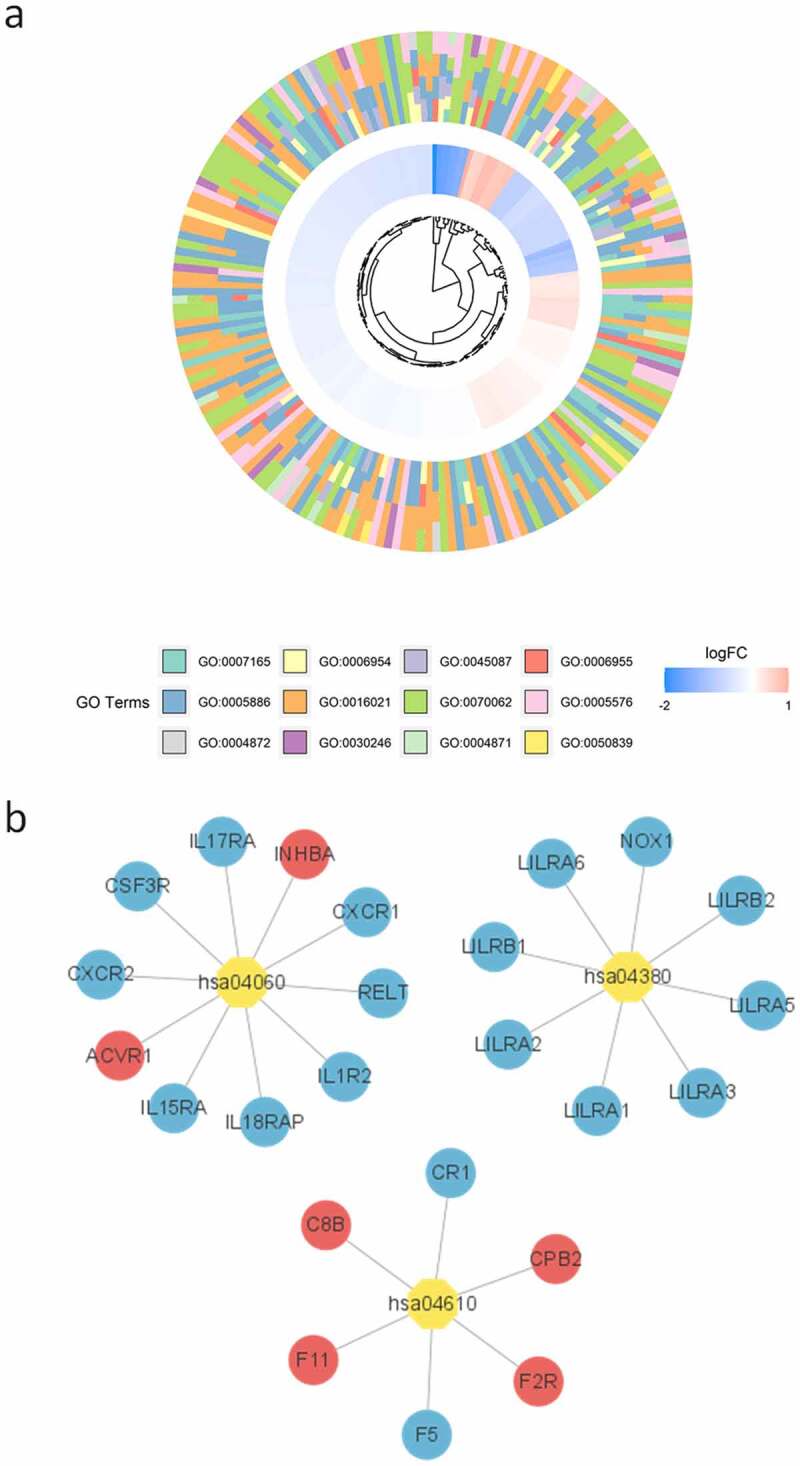
Functional analysis of candidate genes. (a) Circle chart of GO items (first four terms in each category); (b) KEGG enrichment of candidate genes. The yellow octagon represents the KEGG term, the circle of genes. Red in circles and ellipses indicates the up-regulation of genes and blue indicates the down-regulation of genes.
Figure 4.
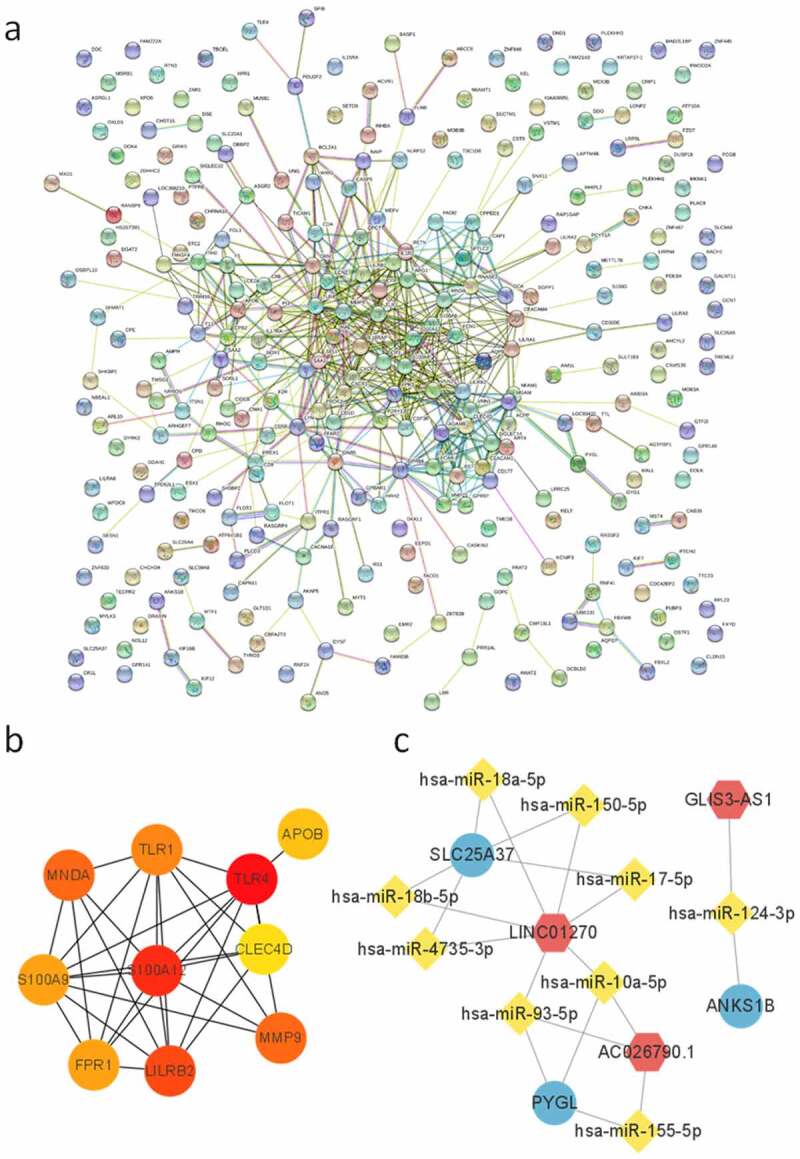
PPI and ceRNA network construction. (a) PPI of candidate genes, nodes represent genes; (b) 10 hub genes predicted by Cytoscape (scores were ranked by color from red to yellow); (c) the ceRNA network of candidate genes (red hexagon denotes lncRNA, yellow diamond denotes miRNA, and blue ellipse denotes protein coding gene).
3.3. miR-155-5p and PYGL expression in hypoxia-treated PASMCs
It was demonstrated that the cell viability of PASMCs was remarkably enhanced under hypoxia through CCK-8 assay (Figure 5(a)), and hypoxia stimulation caused a significant increase in and VEGF (Figure 5(b,c)). The level of miR-155-5p was markedly up-regulated in hypoxia-treated PASMCs (Figure 5(d)), and PYGL expression was down-regulated by qRT-PCR and western blot (Figure 5(b,c)). The results stated that hypoxia stimulated the proliferation of PASMCs and increased the levels of and VEGF. Meanwhile, miR-155-5p was upregulated, and PYGL was downregulated in hypoxia-treated PASMCs.
Figure 5.
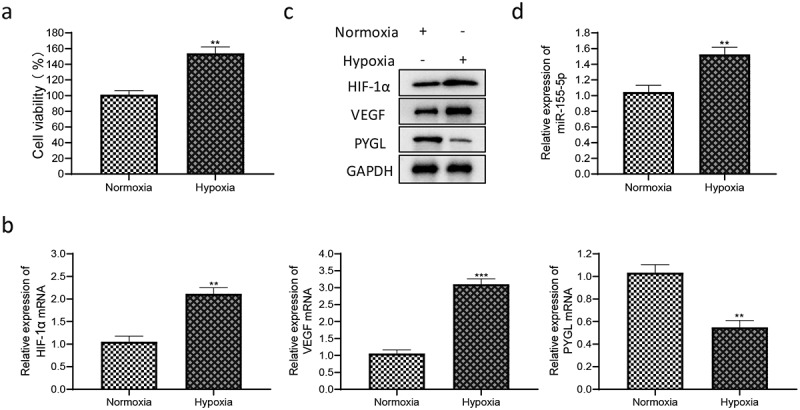
Cell proliferation and related gene expression in PASMCs under hypoxia. (a) Detection of cell viability of normoxic and hypoxic PASMCs by CCK-8 assay; (b) the mRNA levels of , VEGF and PYGL; (c) the protein expression of , VEGF and PYGL in normoxic and hypoxic PASMCs; (d) the expression of miR-155-5p in normoxic and hypoxic PASMCs. All experiments were conducted 3 times. The normoxia group was used as the control group. **P < 0.01, ***P < 0.01.
3.4. miR-155-5p regulated the proliferation, migration, and cell cycle of PASMCs
To explore the effect of miR-155-5p on hypoxia-treated PASMCs, PASMCs were transfected with miRNA-NC/miR-155-5p inhibitor, and then cultured under hypoxia for 24 hours. It showed that hypoxia enhanced the proliferation of PASMCs, while miR-155-5p inhibitor restrained the cell proliferation under hypoxia (Figure 6(a)). It was revealed by the transwell migration assay that hypoxia promoted the migration of PASMCs, while miR-155-5p inhibitor inhibited the cell migration under hypoxia (Figure 6(b)). To illustrate the regulation of miR-155-5p on the cell cycle progression of PASMCs under hypoxia, flow cytometry was utilized to detect the distribution of G0/G1, S, and G2/M phase cells. Compared with the normoxia group, the proportion of G0/G1 phase cells in hypoxia-treated PASMCs was remarkably reduced, while miR-155-5p inhibitor eliminated this change (Figure 6(c,d)). The ratios of S and G2/M phase cells in the hypoxia group were prominently higher than in the normoxia group, and miR-155-5p inhibitor eliminated this change, but miRNA-NC did not (Figure 6(c,d)). No difference was discovered in p27KIP (Figure 6(e)), and the expression of other cycle proteins (Cyclin E, Cyclin D1, and CDK2) in the hypoxia group was notably higher compared with the normoxia group, while miR-155-5p inhibitor reversed this kind of change (Figure 6(e)). Based on all the above findings, it can be concluded that miR-155-5p stimulated the cell proliferation, cell migration, and cell cycle progression in hypoxia-induced PASMCs, and this may substantially affect the vascular remodeling of PAHs.
Figure 6.
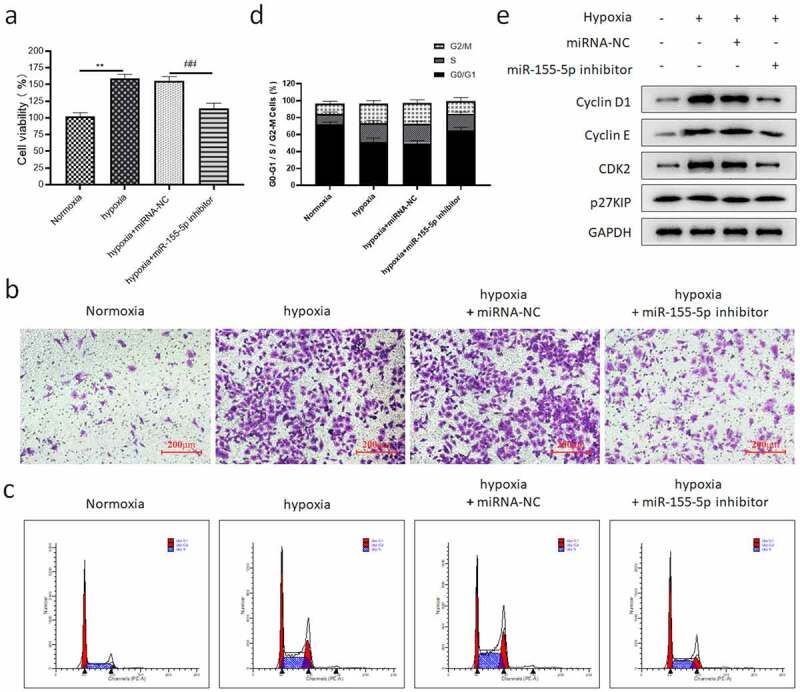
The regulation of miR-155-5p on the cell function of hypoxia-induced PASMCs. PASMCs were transfected with miRNA-NC/miR-155-5p inhibitor and cultured in hypoxia for 24 hours. (a) The cell activity; (b) transwell migration assay results (200 ); (c) the cell cycle distribution of PASMCs; (d) the quantitative results of the ratio of PASMCs in three phases; (e) the expression of cyclin-related protein. All tests were conducted 3 times. When compared with the normoxia group, **P < 0.01. When compared with the hypoxia + miRNA-NC group, ##P < 0.01.
3.5. miR-155-5p targeted PYGL directly
According to the analysis of an online database, the PYGL 3ʹUTR region has a binding site with miR-155-5p sequence (Figure 7(a)). To test whether miR-155-5p acts on PYGL to affect the cell function of hypoxia-induced PASMCs, we regulated the expression via miRNA-NC/miR-155-5p mimics/miR-155-5p inhibitor transfection. It was found that miR-155-5p mimics significantly declined the protein and mRNA levels of PYGL (Figure 7(b,c)), while the downregulation of miR-155-5p evidently elevated PYGL levels (Figure 7(b,c)). In addition, pGL3-PYGL-3’-UTR vector was applied to detect whether PYGL reacts with miR-155-5p. In 293 T cells co-transfected with miR-155-5p mimics and the pGL3-PYGL-3’-UTR (WT) plasmids, the luciferase activity decreased significantly (Figure 7(d)). However, in 293 T co-transfected with pGL3-PYGL mut-3’-UTR (MT) plasmid and miRNA-NC/miR-155-5p mimics, there was no difference in the luciferase activity (Figure 7(d)). These outcomes suggested that miR-155-5p directly targeted PYGL.
Figure 7.
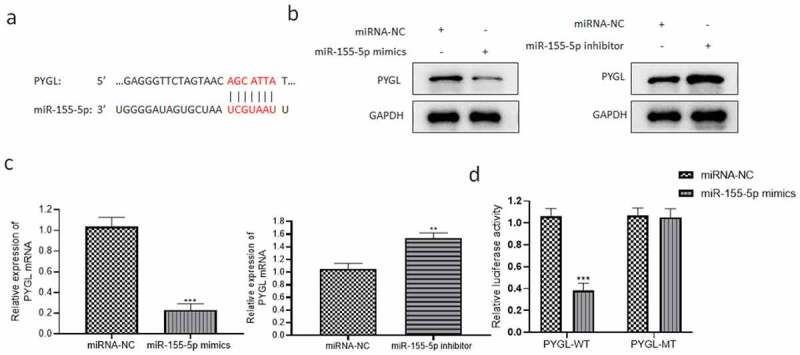
miR-155-5p directly targeted PYGL. (a) Binding sites between 3ʹUTR region of PYGL and miR-155-5p sequence revealed through bioinformatics analysis; (b) PYGL protein level detected by western blot; (c) PYGL mRNA level detected by qRT-PCR; (d) the dual-luciferase reporter assay results. All the tests were done 3 times. The hypoxia + miRNA-NC group as the control group, **P < 0.01, ***P < 0.001.
3.6. PYGL knockdown reversed the beneficial effect of miR-155-5p inhibitor on hypoxia-treated PASMCs
To prove that the positive effects of miR-155-5p inhibitor on hypoxia-induced PASMCs were achieved by targeting PYGL, we co-transfected miR-155-5p inhibitor and siPYGL into PASMCs, and cultured them under hypoxia for 24 hours. We found that the knockdown of PYGL reversed the effects of miR-155-5p inhibitor on cell proliferation (Figure 8(a)), migration (Figure 8(b)) and cell cycle (Figure 8(c,d)). Figure 8(e) showed the expression of cyclin-related protein and PYGL.
Figure 8.
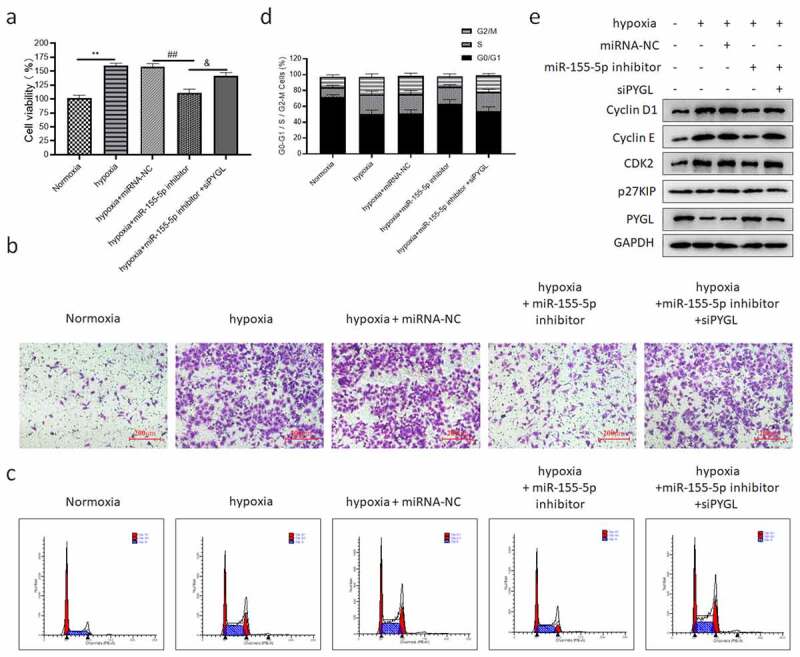
PYGL knockdown reversed the beneficial effect of miR-155-5p inhibitor on hypoxia-induced PASMCs. PASMCs were transfected with miRNA-NC/miR-155-5p inhibitor/siPYGL and cultured in hypoxia for 24 h. (a) The CCK-8 assay results; (b) transwell migration assay (200); (c) the cell cycle distribution of PASMCs in five groups; (d) the quantitative results of the ratio of PASMCs in three phases; (e) the expression of cyclin-related protein and PYGL in five groups. All the tests were done 3 times. When compared with the normoxia group, **P < 0.01. When compared with the hypoxia + miRNA-NC group, ##P < 0.01. When compared with the hypoxia + miR-155-5p inhibitor group, &P < 0.05.
4. Discussion
PAH is an irreversible disease with the clinical manifestations of tachypnea, fatigue, chest pain, and syncope [3,39]. The abnormal proliferation of PASMCs can be induced through different signaling pathways, such as hypoxia stress and activation of inflammatory signal transduction [40], which ultimately brings about the thickening of the pulmonary vascular middle layer, narrowing or occlusion of lumen, and participating in PAH formation [41]. Inflammation and abnormal proliferation of SMCs lead to media hypertrophy and adventitial fibroblast proliferation, which affects vascular remodeling of PAH, and in turn results in coaxial neointimal damage and plexiform lesions [42]. We simulated the pathological process of PAH in vitro by hypoxia treatment of PASMCs and found that the viability of PASMCs was enhanced, and and VEGF were significantly elevated. Studies have shown that the excessively increased proliferation and migration of hypoxia-induced PASMCs is in a close connection with the incidence and progression of PAH [43], which is in accord with our findings.
Based on the data from the GEO database (GSE117261), the PAH-related genes were screened out by WGCNA and DEGs analysis. And functional enrichment analysis was carried out. Combined with the clinical data (gender and group (PAH patients or healthy controls)), gene modules significantly associated with clinical features were selected. We found that miR-155-5p/PYGL contributed greatly in the occurrence and progress of PAH. Meanwhile, it showed that the miR-155-5p level increased and PYGL decreased in hypoxia-treated PASMCs.
miR-155 has already been confirmed by previous studies to be of vital importance for the carcinogenesis and progression of diverse cancers, for instance, liver cancer, colorectal cancer, and gastric cancer [35–37,44]. Furthermore, miR-155-5p stimulates oxalate- and calcium-induced renal oxidative stress suffering by inhibiting MGP expression [45]. The miR-155-5p levels in arterial and coronary sinus plasma were significantly elevated in sufferers with advanced heart failure because of cardiovascular disease [46]. It was reported that cold exposure aggravated monocrotaline-induced PAH with an increase in miR-155-5p level [47]. However, miR-155-5p has not yet been studied deeply in PAH.
This study found that in the hypoxia-induced PASMC cells, miR-155-5p was significantly up-regulated, and miR-155-5p inhibitor attenuated the proliferation and migration of hypoxia-stimulated PASMCs. Therefore, miR-155-5p may be relevant to the vascular remodeling of PAH. Flow cytometry showed that the proportion of G0/G1 phase cells was reduced, and the S and G2/M phase cells were remarkably increased in the hypoxia-induced PASMCs. There was no remarkable difference in cell cycle protein p27KIP. The expression of Cyclin D1, Cyclin E, and CDK2 in the hypoxia group was remarkably more than that in the normoxia group. It has been reported that cyclin E and CDK2 complexes are of great significance in the cell cycle from G1 to S phase and are the crux kinase complex regulatory factors of cell cycle. Cyclin D1 may reduce the time to get into the S phase, quicken the G1/S conversion procedure, and promote the cell proliferation [48]. As a negative regulator, p27KIP expression was unchanging in PAH [49], which is in line with the findings of this study.
Analysis of the online database indicated that the PYGL 3ʹUTR region had a binding site with the miR-155-5p. miR-155-5p mimics decreased the PYGL levels, while miR-155-5p silencing notably increased PYGL levels. Dual-luciferase assay also indicated that miR-155-5p directly targeted PYGL. Currently, there are few studies on PYGL. It is known that PYGL mutation can cause liver phosphorylase deficiency and lead to glycogen decomposition disorders, namely glycogen storage disease (GSD) VI [50]. In this study, PYGL knockdown changed the function of miR-155-5p inhibitor on cell viability, migration, and cell cycle of hypoxia-induced PASMCs.
To sum up, miR-155-5p/PYGL was revealed to play a key role in PAH, and this finding may supply novel treatment strategies for PAH.
5. Conclusion
This study identified miR-155-5p/PYGL as important pathway related to PAH through bioinformatics analysis, and this was validated by in vitro experiments, providing novel ideas for the treatment of PAH. However, the lack of in vivo data (animal or clinical samples) is the main limitation of the current study, so we will continue to investigate in depth in subsequent researches.
Funding Statement
This work was supported by grants from the Science and Technology Innovation Project of Shaoxing Health and Family Planning (2017CX010).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Author contribution
Guowen Wang: Conceptualization, Formal analysis, Investigation, Methodology, Writing—original draft.
Xuefang Tao: Funding acquisition, Resources, Methodology, Data curation.
Linlin Peng: Methodology, Supervision, Project administration, Writing—review and editing.
Data availability statement
The datasets used and/or analyzed during the current study are available from the GEO database (https://www.ncbi.nlm.nih.gov/geo/).
References
- [1].Bienertova-Vasku J, Novak J, Vasku A.. MicroRNAs in pulmonary arterial hypertension: pathogenesis, diagnosis and treatment. J Am Soc Hypertens. 2015;9:221–234. [DOI] [PubMed] [Google Scholar]
- [2].Prewitt AR, Ghose S, Frump AL, et al. Heterozygous null bone morphogenetic protein receptor type 2 mutations promote SRC kinase-dependent caveolar trafficking defects and endothelial dysfunction in pulmonary arterial hypertension. J Biol Chem. 2015;290:960–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Galie N, Humbert M, Vachiery JL, et al. 2015 ESC/ERS guidelines for the diagnosis and treatment of pulmonary hypertension: the joint task force for the diagnosis and treatment of pulmonary hypertension of the European Society of Cardiology (ESC) and the European Respiratory Society (ERS): endorsed by: association for European Paediatric and Congenital Cardiology (AEPC), International Society for Heart and Lung Transplantation (ISHLT). Eur Heart J. 2016;37:67–119. [DOI] [PubMed] [Google Scholar]
- [4].Morrell NW, Adnot S, Archer SL, et al. Cellular and molecular basis of pulmonary arterial hypertension. J Am Coll Cardiol. 2009;54:S20–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chang YT, Tseng CN, Tannenberg P, et al. Perlecan heparan sulfate deficiency impairs pulmonary vascular development and attenuates hypoxic pulmonary hypertension. Cardiovasc Res. 2015;107:20–31. [DOI] [PubMed] [Google Scholar]
- [6].Iswariya GT, Paital B, Padma PR, et al. microRNAs: epigenetic players in cancer and aging. Front Biosci (Schol Ed). 2019;11:29–55. [DOI] [PubMed] [Google Scholar]
- [7].Chen JQ, Papp G, Szodoray P, et al. The role of microRNAs in the pathogenesis of autoimmune diseases. Autoimmun Rev. 2016;15:1171–1180. [DOI] [PubMed] [Google Scholar]
- [8].Falk E, Nakano M, Bentzon JF, et al. Update on acute coronary syndromes: the pathologists’ view. Eur Heart J. 2013;34:719–728. [DOI] [PubMed] [Google Scholar]
- [9].Papathanasiou G, Zerva E, Zacharis I, et al. Association of high blood pressure with body mass index, smoking and physical activity in healthy young adults. Open Cardiovasc Med J. 2015;9:5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Nie X, Chen Y, Tan J, et al. MicroRNA-221-3p promotes pulmonary artery smooth muscle cells proliferation by targeting AXIN2 during pulmonary arterial hypertension. Vascul Pharmacol. 2019;116:24–35. [DOI] [PubMed] [Google Scholar]
- [11].Bonnet S, Boucherat O, Paulin R, et al. Clinical value of non-coding RNAs in cardiovascular, pulmonary, and muscle diseases. Am J Physiol Cell Physiol. 2020;318:C1–C28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Miano JM, Long X.. The short and long of noncoding sequences in the control of vascular cell phenotypes. Cell Mol Life Sci. 2015;72:3457–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yuan C, Xu M, Rong R, et al. miR-200c regulates endothelin-1 induced PASMCs abnormal proliferation and apoptosis. IUBMB Life. 2017;69:877–886. [DOI] [PubMed] [Google Scholar]
- [15].Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Iancu OD, Colville A, Oberbeck D, et al. Cosplicing network analysis of mammalian brain RNA-Seq data utilizing WGCNA and Mantel correlations. Front Genet. 2015;6:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Pei G, Chen L, Zhang W. WGCNA application to proteomic and metabolomic data analysis. Methods Enzymol. 2017;585:135–158. [DOI] [PubMed] [Google Scholar]
- [18].Presson AP, Sobel EM, Papp JC, et al. Integrated weighted gene co-expression network analysis with an application to chronic fatigue syndrome. BMC Syst Biol. 2008;2:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Miller JA, Horvath S, Geschwind DH. Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc Natl Acad Sci U S A. 2010;107:12698–12703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].D’Haeseleer P, Liang S, Somogyi R. Genetic network inference: from co-expression clustering to reverse engineering. Bioinformatics. 2000;16:707–726. [DOI] [PubMed] [Google Scholar]
- [21].Hirose O, Yoshida R, Imoto S, et al. Statistical inference of transcriptional module-based gene networks from time course gene expression profiles by using state space models. Bioinformatics. 2008;24:932–942. [DOI] [PubMed] [Google Scholar]
- [22].Romanoski CE, Qi X, Sangam S, et al. Transcriptomic profiles in pulmonary arterial hypertension associate with disease severity and identify novel candidate genes. Pulm Circ. 2020;10:2045894020968531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wathes DC, Cheng Z, Salavati M, et al. Relationships between metabolic profiles and gene expression in liver and leukocytes of dairy cows in early lactation. J Dairy Sci. 2021;104:3596–3616. [DOI] [PubMed] [Google Scholar]
- [25].Liu J, Zhou S, Li S, et al. Eleven genes associated with progression and prognosis of endometrial cancer (EC) identified by comprehensive bioinformatics analysis. Cancer Cell Int. 2019;19:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wu X, Sui Z, Zhang H, et al. Integrated analysis of lncRNA-Mediated ceRNA network in lung adenocarcinoma. Front Oncol. 2020;10:554759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhang F, Jiang H, Wang N, et al. Comprehensive network analysis of different subtypes of molecular disorders in lung cancer. Am J Transl Res. 2021;13:9248–9259. [PMC free article] [PubMed] [Google Scholar]
- [28].Xing XQ, Li B, Xu SL, et al. MicroRNA-214-3p regulates hypoxia-mediated pulmonary artery smooth muscle cell proliferation and migration by targeting ARHGEF12. Med Sci Monit. 2019;25:5738–5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Li Q, Zhou X, Zhou X. Downregulation of miR98 contributes to hypoxic pulmonary hypertension by targeting ALK1. Mol Med Rep. 2019;20:2167–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang D, Xu H, Wu B, et al. Long noncoding RNA MALAT1 sponges miR1243p.1/KLF5 to promote pulmonary vascular remodeling and cell cycle progression of pulmonary artery hypertension. Int J Mol Med. 2019;44:871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Luo L, Xiao L, Lian G, et al. miR-125a-5p inhibits glycolysis by targeting hexokinase-II to improve pulmonary arterial hypertension. Aging (Albany NY). 2020;12:9014–9030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Shang L, Wang K, Liu D, et al. TMEM16A regulates the cell cycle of pulmonary artery smooth muscle cells in high-flow-induced pulmonary arterial hypertension rat model. Exp Ther Med. 2020;19:3275–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Xu J, Liu D, Niu H, et al. Resveratrol reverses Doxorubicin resistance by inhibiting epithelial-mesenchymal transition (EMT) through modulating PTEN/Akt signaling pathway in gastric cancer. J Exp Clin Cancer Res. 2017;36:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhao CC, Jiao Y, Zhang YY, et al. Lnc SMAD5-AS1 as ceRNA inhibit proliferation of diffuse large B cell lymphoma via Wnt/beta-catenin pathway by sponging miR-135b-5p to elevate expression of APC. Cell Death Dis. 2019;10:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Mohamed MA, Mohamed EI, El-Kaream SAA, et al. Underexpression of miR-486-5p but not overexpression of miR-155 is associated with lung cancer stages. Microrna. 2018;7:120–127. [DOI] [PubMed] [Google Scholar]
- [36].Ren XY, Han YD, Lin Q. Long non-coding RNA MIR155HG knockdown suppresses cell proliferation, migration and invasion in NSCLC by upregulating TP53INP1 directly targeted by miR-155-3p and miR-155-5p. Eur Rev Med Pharmacol Sci. 2020;24:4822–4835. [DOI] [PubMed] [Google Scholar]
- [37].He XH, Zhu W, Yuan P, et al. miR-155 downregulates ErbB2 and suppresses ErbB2-induced malignant transformation of breast epithelial cells. Oncogene. 2016;35:6015–6025. [DOI] [PubMed] [Google Scholar]
- [38].Fang H, Shuang D, Yi Z, et al. Up-regulated microRNA-155 expression is associated with poor prognosis in cervical cancer patients. Biomed Pharmacother. 2016;83:64–69. [DOI] [PubMed] [Google Scholar]
- [39].Hoeper MM, Bogaard HJ, Condliffe R, et al. Definitions and diagnosis of pulmonary hypertension. J Am Coll Cardiol. 2013;62:D42–50. [DOI] [PubMed] [Google Scholar]
- [40].Chen T, Zhou G, Zhou Q, et al. Loss of microRNA-17 approximately 92 in smooth muscle cells attenuates experimental pulmonary hypertension via induction of PDZ and LIM domain 5. Am J Respir Crit Care Med. 2015;191:678–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Maron BA, Loscalzo J. Pulmonary hypertension: pathophysiology and signaling pathways. Handb Exp Pharmacol. 2013;218:31–58. [DOI] [PubMed] [Google Scholar]
- [42].Kim J, Kang Y, Kojima Y, et al. An endothelial apelin-FGF link mediated by miR-424 and miR-503 is disrupted in pulmonary arterial hypertension. Nat Med. 2013;19:74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Marsboom G, Archer SL. Pathways of proliferation: new targets to inhibit the growth of vascular smooth muscle cells. Circ Res. 2008;103:1047–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zhu HZ, Hou J, Guo Y, et al. Identification and imaging of miR-155 in the early screening of lung cancer by targeted delivery of octreotide-conjugated chitosan-molecular beacon nanoparticles. Drug Deliv. 2018;25:1974–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Jiang K, Hu J, Luo G, et al. miR-155-5p promotes oxalate- and calcium-induced kidney oxidative stress injury by suppressing MGP expression. Oxid Med Cell Longev. 2020;2020:5863617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Marques FZ, Vizi D, Khammy O, et al. The transcardiac gradient of cardio-microRNAs in the failing heart. Eur J Heart Fail. 2016;18:1000–1008. [DOI] [PubMed] [Google Scholar]
- [47].Sanchez-Gloria JL, Carbo R, Buelna-Chontal M, et al. Cold exposure aggravates pulmonary arterial hypertension through increased miR-146a-5p, miR-155-5p and cytokines TNF-alpha, IL-1beta, and IL-6. Life Sci. 2021;287:120091. [DOI] [PubMed] [Google Scholar]
- [48].Liu J, Liu Y, Ren Y, et al. Transmembrane protein with unknown function 16A overexpression promotes glioma formation through the nuclear factor-kappaB signaling pathway. Mol Med Rep. 2014;9:1068–1074. [DOI] [PubMed] [Google Scholar]
- [49].Wang M, Yang H, Zheng LY, et al. Downregulation of TMEM16A calcium-activated chloride channel contributes to cerebrovascular remodeling during hypertension by promoting basilar smooth muscle cell proliferation. Circulation. 2012;125:697–707. [DOI] [PubMed] [Google Scholar]
- [50].Luo X, Hu J, Gao X, et al. Novel PYGL mutations in Chinese children leading to glycogen storage disease type VI: two case reports. BMC Med Genet. 2020;21:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the GEO database (https://www.ncbi.nlm.nih.gov/geo/).


