Abstract
In this study, we used PCR typing methods to assess the presence of tetracycline resistance determinants conferring ribosomal protection in waste lagoons and in groundwater underlying two swine farms. All eight classes of genes encoding this mechanism of resistance [tet(O), tet(Q), tet(W), tet(M), tetB(P), tet(S), tet(T), and otrA] were found in total DNA extracted from water of two lagoons. These determinants were found to be seeping into the underlying groundwater and could be detected as far as 250 m downstream from the lagoons. The identities and origin of these genes in groundwater were confirmed by PCR-denaturing gradient gel electrophoresis and sequence analyses. Tetracycline-resistant bacterial isolates from groundwater harbored the tet(M) gene, which was not predominant in the environmental samples and was identical to tet(M) from the lagoons. The presence of this gene in some typical soil inhabitants suggests that the vector of antibiotic resistance gene dissemination is not limited to strains of gastrointestinal origin carrying the gene but can be mobilized into the indigenous soil microbiota. This study demonstrated that tet genes occur in the environment as a direct result of agriculture and suggested that groundwater may be a potential source of antibiotic resistance in the food chain.
The widespread use of antibiotics in humans and animals has raised several concerns related to human and animal health. The principal area of concern has been the increasing emergence of antibiotic resistance phenotypes in both clinically relevant strains and normal commensal microbiota. In two recent studies, a link between the agricultural use of antibiotics and antibiotic-resistant human infections has been suggested (24, 33). Because consumption of tainted food is considered the main route of transmission of drug resistance, other possible means of antibiotic resistance dissemination have received relatively little attention. One of these possible means could be natural water and soil environments impacted by antibiotics and antibiotic-resistant bacteria from agriculture, where antibiotics are used for disease treatment, prophylaxis, and growth promotion. The concern over the use of antibiotics in agriculture, especially for prophylactic and growth-promoting purposes, has not been limited to the presumed role of antibiotics in selection of antibiotic-resistant bacteria (pathogenic or nonpathogenic) in the animal gut. The more debatable issue arising from chronic low-level exposure to antibiotics is whether this practice contributes significantly to increased gene frequencies and dissemination of resistance genes into other ecosystems.
Furthermore, many antibiotics used in animal agriculture are poorly absorbed in the animal gut. It is estimated that 25% to as much as 75% of the antibiotics administered to feedlot animals could be excreted unaltered in feces (6, 7) and can persist in soil after land application (4, 11). There is little information available concerning the fate of antibiotics in the environment and their link to the emergence of resistant genotypes found there. The annual production of livestock and poultry waste in the United States is nearly 180 million tons (dry weight basis) (13, 34), and coupled with antibiotic usage, this waste is a potentially large source of both antibiotics and antibiotic-resistant bacteria released into the environment.
Lagoons and pit systems are typically used for waste disposal in animal agriculture. Seepage and runoff into watershed systems are of particular concern due to potential mobilization of constituents and exposure of contaminants to humans and other animals. Groundwater, in particular, constitutes about 40% of the water used for public water supplies and provides drinking water for more than 97% of the rural population in the United States (http://water.usgs.gov/wid/html/GW.html). Recent monitoring studies have demonstrated the vulnerability of groundwater to seepage from waste lagoons (19). Over a period of several years, Krapac and coworkers found indicators such as ammonia and fecal enterococci at elevated levels in groundwater samples obtained up to 100 m downstream of from swine waste lagoons, indicating that both long-term impact and environmental migration of contaminants occur (19, 20).
Recent studies have attempted to evaluate the impact of antibiotic use on populations of bacteria in natural waters (12, 23, 39). Antibiotic resistance analysis has been used to identify sources of fecal pollution (14, 18, 27, 38, 39). The traditional method involving cultivation and phenotypic testing is still relied upon but has a clear bias when it is used to determine the representative phenotypes and genotypes in this environment. The use of techniques such as PCR and molecular gene probe analysis has allowed sensitive detection of specific genes in the environment in the absence of cultivation. PCR amplification of small-subunit ribosomal DNA (rDNA) genes coupled with denaturing gradient gel electrophoresis (DGGE) has been used primarily to determine the genetic fingerprints of microbial communities. The repertoire of genes used for such analyses has been extended to specific metabolic genes (2, 8, 17, 31, 37) and, recently, to antibiotic resistance genes (1). In the latter work, we used a phylogenetic approach to design a set of primers to target tetracycline resistance genes encoding ribosomal protection proteins (RPPs). These primers were used to detect the corresponding tetracycline resistance genes in ruminal fluid, swine feed and feces, and pig intestinal fecal streptococci. In the present study, we used these techniques to determine the occurrence and migration of RPP tetracycline resistance genes in lagoons and in groundwater underlying two large swine production facilities known to be impacted by waste seepage (19). The use of tetracycline resistance as the key determinant to monitor resistance genes is relevant due to the common use of this antibiotic in the swine industry. To our knowledge, this is the first study in which the genes for one major class of antibiotic resistance were characterized in natural groundwater that is directly impacted by animal agriculture.
MATERIALS AND METHODS
Description of the sampling sites.
Site A, which began operation in February 1995, is a finishing operation that houses 4,000 pigs (Fig. 1). Antibiotics are used in this facility, but the relevant information concerning such use could not be obtained from the producer. The facility includes a two-stage waste-handling system in which a concrete settling basin collects most of the solids before the supernatant liquid passively enters an earthen lagoon. The area of the lagoon is approximately 1.2 ha, and the lagoon is unlined. No special construction techniques were used to compress the soil during lagoon construction. The average depth of liquid in the lagoon during a study conducted by Krapac et al. (19) was about 1.5 m. The concrete settling basin is periodically pumped, and the manure is applied to crop fields both on-site and off-site.
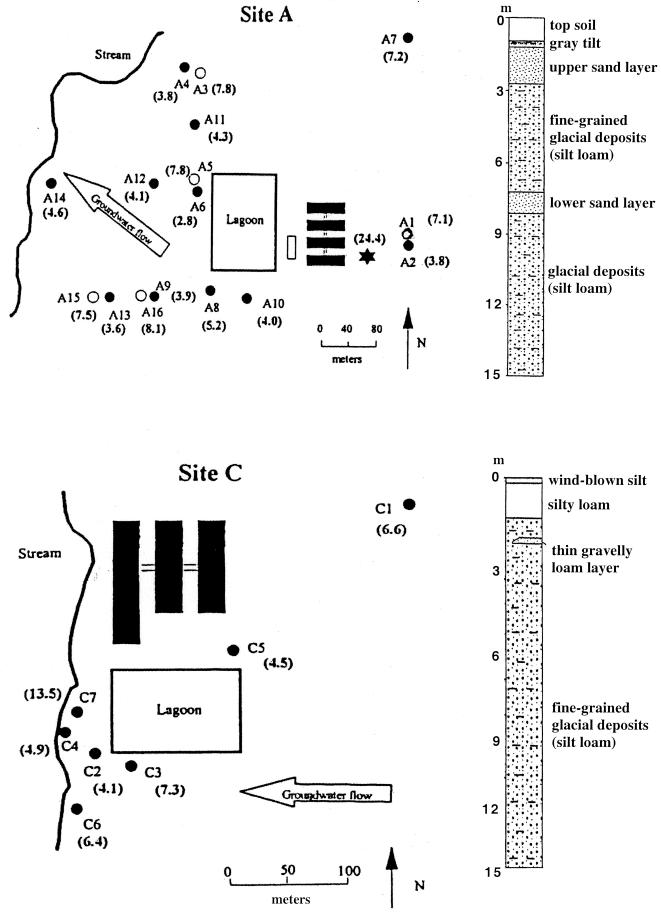
FIG. 1.
Maps of sites A and C and corresponding stratigraphic columns indicating the locations and characteristics of sand layers. The direction of groundwater flow is indicated by large open arrows, and the locations of monitoring wells are indicated by circles; open circles represent nested wells screened in deeper sand layers. Well depths (in meters) are indicated in parentheses. The solid rectangles represent confinement buildings.
Site A is located on glacial outwash and terrace deposits along a stream valley that is incised into a till plain formed during the Illinois Episode of glaciation. The topsoils are silt or silty clay loams developed on alluvial deposits that are 1.3 to 2 m thick (Fig. 1). These deposits overlie a 0.6- to 1.3-m-thick upper layer of fluvial silty sand and gravel outwash, which is continuous across the site. Twelve of the 16 monitoring wells were installed in this upper sand layer (Fig. 1). Slug test results suggested that this upper sand has a saturated hydraulic conductivity of approximately 6.8 × 10−4 m/s. Below the silty sand and gravel is 1.6 to 3 m of silt loam diamicton, which may be colluvial. Below the silt loam diamicton is a 1- to 2-m-thick lower sand layer composed of sand and gravel outwash that is being used locally as an aquifer. Four monitoring wells were installed in this lower sand layer (Fig. 1). The saturated hydraulic conductivity of this deeper sand was estimated to be 8.2 × 10−6 m/s based on slug tests. Below this sand and gravel is more silt loam diamicton. Logs from water wells drilled in the vicinity show the presence of discontinuous sand and gravel outwash units below the diamicton that are used locally as aquifers. The multiple sand layers make this site particularly susceptible to leachate migration from the lagoon.
Site C is a farrowing and nursery operation that houses 1,200 sows (Fig. 1). The facility uses chlortetracycline, which is added once per quarter for about 2 weeks to the feed (400 g per ton). The feed consumption rate is about 3 kg/animal per day, and thus, each animal consumes about 17 g of chlortetracycline over the 2-week period. The facility began operation in the fall of 1992 and every year produces approximately 24,000 pigs. The facility uses a single-stage lagoon. Lagoon water is recycled to partially fill and flush the shallow pits below the confinement buildings. The area of the lagoon is approximately 0.8 ha, and the lagoon is unlined. The average depth of waste in the lagoon was about 6 m. Waste has never been applied to the crop fields surrounding the lagoon.
Site C is located on a glacial till plain formed during the Illinois Episode of glaciation. It is underlain by a silt loam glacial diamicton that is 3 to 15 m thick and overlies shale bedrock (Fig. 1). Thin (less than 30-cm-thick) glacial gravelly loam layers were found in two of the seven borings at the site. An intermittent stream borders the site on the west, and a small pond is located about 100 m south of the lagoon. Large-diameter bored and dug wells and ponds are the predominant sources of drinking water in the area.
Groundwater and lagoon sampling.
Groundwater was collected from 14 monitoring wells at site A and from six monitoring wells at site C (Fig. 1). Water samples from the waste lagoons were also taken from each site. A polyethylene bailer dedicated to each sample was sterilized with alcohol and rinsed with sterile water prior to use for sample collection. Between 1.5 and 3 well volumes of groundwater was removed from each well before collection as described previously (19). Samples were stored in clean, sterile plastic bottles and kept on ice in the field. Samples were refrigerated at 4°C in the laboratory until they were analyzed.
Bacterial identification and isolation.
Tetracycline-resistant bacteria from lagoon and groundwater samples were grown aerobically on Enterococcosel agar (BBL, Cockeysville, Md.) and MR2A agar (9); 20 μg of tetracycline (Sigma, St. Louis, Mo.) per ml was added to each medium. Undiluted groundwater samples (100 μl) were directly plated onto agar media, while lagoon samples were plated by using 10-fold serial dilutions up to 1,000-fold. Media lacking tetracycline were inoculated similarly. Cultures grown on Enterococcosel agar were incubated at 37°C for 48 h. Cultures grown on MR2A agar were incubated at room temperature for up to 14 days. Tetracycline-resistant colonies were further purified by restreaking on the same media and were used for PCR screening as described previously (1). Gram staining was performed with a Protocol kit (Biochemical Sciences, Swedesboro, N.J.).
DNA extraction.
Groundwater samples (250 ml) were centrifuged at 17,700 × g for 20 min at 4°C. The supernatants were discarded, and the pellets were washed three times with 0.1 volume of phosphate-buffered saline (120 mM NaH2PO4 [pH 8.0], 0.85% NaCl) before extraction of total DNA by the method of Tsai and Olsen (36). Lagoon samples (50 ml) were centrifuged at 10,000 × g for 10 min at 4°C before DNA extraction as described above. DNA (final concentration, 125 ng/μl) was stored in TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]) at −20°C.
PCR amplification.
The primers and PCR protocol used to amplify tetracycline resistance genes in this study were described previously (1). Briefly, PCR was performed with 25 pmol of each primer and an ExTaq PCR kit (purchased from PanVera Corporation, Madison, Wis.) by using a final reaction mixture volume of 25 μl. Purified DNA (125 ng) or one-half of a 1- to 2-mm-diameter individual colony that was resuspended in sterile water was used as the PCR template. PCR amplification was performed with a GeneAmp PCR system 2400 thermocycler (Perkin-Elmer, Norwalk, Conn.). The temperature program consisted of denaturation at 94°C for 5 min, followed by 30 cycles consisting of 94°C for 30 s, annealing for 30 s, and extension at 72°C for 30 s and a final extension at 72°C for 7 min. The annealing temperatures used for amplification of different RPP genes were as follows: tet(O), 60°C; tet(Q), 63°C; tet(W), 64°C; tet(M), 55°C; tetB(P), 47°C; tet(S), 50°C; otrA, 66°C; and tet(T), 46°C. The PCR protocol was modified for DNA obtained from groundwater samples due to the presence of unidentified PCR-inhibiting substances. For these samples, a second, nested PCR was performed by using 1 μl of the first PCR mixture as the template and amplifying the template for 25 or 30 cycles as described above. Nested PCR was also performed with DNA from lagoon samples when necessary. The control reactions included PCR amplification with bacterial 16S rDNA primers 27F and 1525R (21) and sterile water or DNA of tetracycline-sensitive Escherichia coli EPEC as the negative control template for all primer sets and the positive control strains for each primer set as described previously (1). PCR product aliquots (5 μl) were analyzed by electrophoresis on a 2.5% (wt/vol) agarose gel (NuSieve; FMC Bioproducts, Rockland, Maine) and were stained with ethidium bromide. The sizes of the PCR products obtained from amplification were 167 bp for tet(O), tet(Q), tet(W), tet(M), tetB(P), tet(S), and tet(T) and 212 bp for otrA.
DGGE analysis.
DNA from groundwater, lagoons, and bacterial isolates was PCR amplified as described above, except that in the nested PCR the forward primer included a GC clamp at the 5′ end. Primers that targeted tetracycline resistance genes and the V3 variable region of 16S rDNA (25) were used. DGGE analyses of the amplified tetracycline resistance genes were performed as described previously (1). Briefly, electrophoretic separation of the PCR products was accomplished by using a polyacrylamide gel with a gradient containing urea and formamide (8% acrylamide, 30 to 60% urea–formamide, 0.5× TAE buffer [pH 7.4]). Electrophoresis was performed at 60°C for 2 h at 150 V and then for 1 h at 200 V, using a D-Code system (Bio-Rad, Hercules, Calif.). The gels were silver stained, and images were captured by using the Bio-Rad Diversity Database fingerprinting software on a G3 Macintosh computer equipped with a Bio-Rad GS-710 calibrated imaging densitometer. The gel standard markers consisted of known mixtures of PCR-amplified RPP tetracycline resistance genes or the V3 variable region of 16S rDNA.
Cloning and sequencing.
DNA bands were excised from DGGE gels, crushed, equilibrated in 50 μl of sterile water, and then subjected to three cycles of freezing and thawing (at −20°C and room temperature). Eluted DNA (1 μl) was reamplified with the corresponding primers and reelectrophoresed on a DGGE gel as described above. PCR products were cloned by using a TOPO-TA Cloning kit (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. White colonies harboring the corresponding tet fragments were identified by PCR by using the small amounts of colony biomass and the corresponding primer sets. Representative bacterial isolates from the groundwater and lagoons were identified by sequencing of the cloned 16S rRNA genes, which were amplified with the bacterial primer set (21). Sequence analysis of the cloned tet and small-subunit rDNA fragments was performed by the University of Illinois Keck Center for Functional and Comparative Genomics. Sequences were analyzed on-line by using the BLAST (Basic Local Alignment Search Tool) family of programs of GenBank (22).
Nucleotide sequence accession number.
The 16S rDNA sequence data reported in this paper have been deposited in the GenBank database under accession no. AY017049 to AY017063.
RESULTS
PCR amplification of tet genes from lagoon and groundwater samples.
All of the known ribosomal protection tetracycline resistance determinants were detected in the lagoon samples (Table 1). Most of them were detected in total DNA from the lagoons after the initial 30 cycles of PCR; the only exception was otrA, which was detected only after a second, nested PCR. The most frequently detected determinants in groundwater samples from both sites were Tet Q, Tet W, Tet M, Tet T, and Tet O. Groundwater from well A8 contained Tet S, and well C6 water contained detectable otrA. None of the groundwater samples contained detectable tetB(P). Background well A7 at site A contained tet(T), which was also detected in more than one-half of the wells located downstream of the site lagoon in the direction of groundwater flow. Site C background well C1 did not contain any of the ribosomal protection tetracycline resistance determinants. Two of the shallow wells at site A, wells A10 and A4, also did not contain any detectable levels of these determinants. Well C3, located close to the lagoon but perpendicular to the general direction of groundwater flow away from the lagoon, did not contain any determinant. Sample A14 from the well located farthest away from the site A lagoon contained Tet Q, which was the most frequently detected determinant.
TABLE 1.
Tetracycline resistance genes detected in total DNA from lagoons and groundwater
| Sample | Tetracycline resistance genes
|
|||||||
|---|---|---|---|---|---|---|---|---|
| tet(W) | tet(O) | tet(Q) | tet(M) | tet(S) | tet(T) | tetB(P) | otrA | |
| Site A lagoon | + | + | + | + | + | + | + | + |
| A7 bkga | − | − | − | − | − | + | − | − |
| A10 | − | − | − | − | − | − | − | − |
| A8 | + | + | + | + | + | + | − | − |
| A9 | + | − | + | + | − | + | − | − |
| A16 | + | + | + | + | − | − | − | − |
| A13 | + | + | + | + | − | − | − | − |
| A15 | + | − | + | + | − | + | − | − |
| A6 | + | − | + | − | − | − | − | − |
| A5 | + | + | + | + | − | + | − | − |
| A12 | + | − | + | − | − | + | − | − |
| A14 | − | − | + | − | − | − | − | − |
| A11 | + | − | + | + | − | + | − | − |
| A3 | + | + | + | + | − | + | − | − |
| A4 | − | − | − | − | − | − | − | − |
| Site C lagoon | + | + | + | + | + | + | + | + |
| C1 bkga | − | − | − | − | − | − | − | − |
| C3 | − | − | − | − | − | − | − | − |
| C2 | + | − | + | + | − | + | − | − |
| C4 | − | − | − | − | − | − | − | − |
| C6 | + | + | + | + | − | + | − | + |
| C7 | + | + | + | + | − | − | − | − |
Background control well located upstream of the lagoon.
One of the objectives of this study was to determine whether the genetic diversity of tetracycline resistance genes uncovered by in vitro analysis of total DNA overlapped the genetic diversity of the cultivable isolates. Also, amplification of tet genes from total-DNA preparations is not discriminatory in terms of the host strains in which tet genes occur, and our aim was to determine whether tet gene dissemination to groundwater and indigenous soil microbiota could occur. The experiments which we performed are described below.
Phenotypic characterization and tetracycline resistance genotypes of bacterial isolates.
Approximately 4 × 103 and 3 × 104 CFU per ml were obtained on Enterococcosel agar containing 20 μg of tetracycline per ml after direct plating of 100 μl of slurry from the lagoons at sites A and C, respectively. Several presumptive enterococci (gram-positive, catalase-negative cocci) were randomly selected and isolated (Table 2). All of the enterococcal isolates contained tet(M) as determined by PCR, and one isolate (CLE3) also carried tet(S). Another randomly selected isolate obtained from the Enterococcosel medium was a gram-positive rod (CLE4) which also harbored the tet(M) gene. Few tetracycline-resistant colonies were obtained after direct plating of groundwater on Enterococcosel agar.
TABLE 2.
Characteristics of bacterial isolates from lagoons and groundwater
| Isolatea | Source | Phenotypic description | tet(M)b | tet(S) |
|---|---|---|---|---|
| ALE1 | Site A lagoon | Gram-positive coccus | + | − |
| ALE2 | Site A lagoon | Gram-positive coccus | + | − |
| ALE3 | Site A lagoon | Gram-positive coccus | + | − |
| ALE4 | Site A lagoon | Gram-positive coccus | + | − |
| CLE1 | Site C lagoon | Gram-positive coccus | + | − |
| CLE2 | Site C lagoon | Gram-positive coccus | + | − |
| CLE3 | Site C lagoon | Gram-positive coccus | + | + |
| CLE4 | Site C lagoon | Gram-positive, large, bent rod | + | − |
| A7-2 | Bkg well A7c | Gram-negative, short, motile rod | − | − |
| A8-2 | Well A8 | Gram-positive, short, slender, nonmotile rod | + | − |
| A8-3 | Well A8 | Gram-negative, short, motile rod | + | − |
| A8-4 | Well A8 | Gram-negative, short, plump, motile rod | + | − |
| AL-1 | Site A lagoon | Gram-negative, short, plump, motile rod | − | − |
| AL-2 | Site A lagoon | Gram-negative, short, nonmotile rod | + | − |
| AL-3 | Site A lagoon | Gram-negative, short, plump, motile rod | + | − |
| AL-4 | Site A lagoon | Gram-negative, short, motile, rod | + | − |
| AL-5 | Site A lagoon | Gram-negative, short, nonmotile rod | − | − |
| CL-1 | Site C lagoon | Gram-negative, oval, motile | + | − |
| CL-2 | Site C lagoon | Gram-negative, short, motile rod | − | − |
| CL-3 | Site C lagoon | Gram-negative, short, plump, motile rod | + | − |
| C2-1 | Well C2 | Gram-negative, short, nonmotile rod | + | − |
| A3-1 | Well A3 | Gram-negative, small, nonmotile rod | − | − |
| C7-1 | Well C7 | Gram-negative, plump, nonmotile rod | − | − |
| A16-2 | Well A16 | Gram-negative, short, plump, nonmotile rod | + | − |
Isolates ALE1, ALE2, ALE3, ALE4, CLE1, CLE2, CLE3, and CLE4 were isolated on Enterococcosel agar containing 20 μg of tetracycline per ml. All other isolates were obtained from MR2A agar containing 20 μg of tetracycline per ml.
No tet(W), tet(O), tet(Q), tet(T), tetB(P), or otrA was detected in any isolate.
Background control well located upstream of the lagoon.
Cultivation of aerobic heterotrophs from undiluted groundwater on MR2A medium yielded counts between approximately 1.3 × 103 and >105 CFU per ml. In the presence of 20 μg of tetracycline per ml, only groundwater from well A8 (1.7 × 102 CFU per ml) and the lagoons (>105 CFU per ml) yielded significant numbers of tetracycline-resistant colonies. Tetracycline-resistant colonies were obtained sporadically from direct plating of groundwater from several wells (wells A7, A3, A16, C2, and C7). These colonies, along with several randomly selected tetracycline-resistant colonies from well A8 and lagoon samples, were isolated. All of the MR2A medium isolates from groundwater and lagoon samples were distinct gram-negative rods. Of the 16 tetracycline-resistant isolates, 10 contained the tet(M) gene. The remaining six resistant isolates did not harbor any of the ribosomal protection tetracycline resistance determinants and presumably harbored the efflux mechanism of resistance.
DGGE analysis.
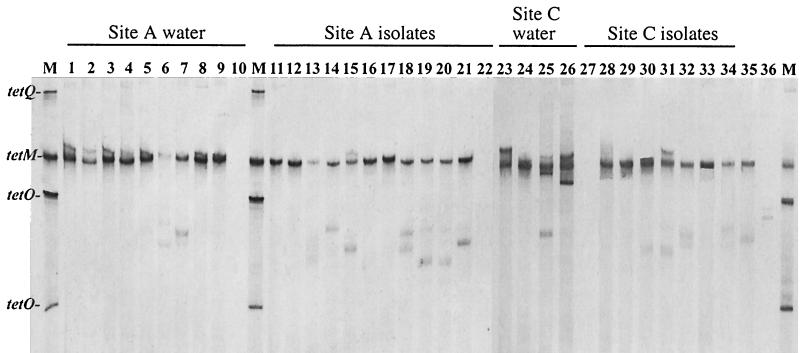
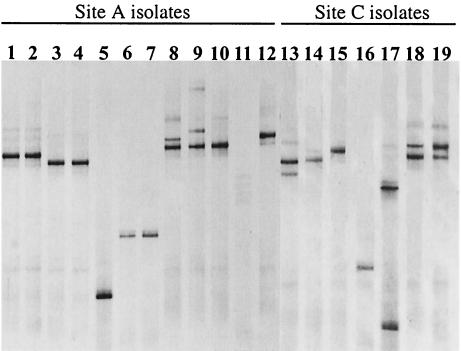
DGGE analysis of the tet(M) determinant was used to assess conservation of the gene in both environmental samples and individual isolates. One major band corresponding to tet(M) was present in all of the groundwater and lagoon samples (Fig. 2). Multiple minor bands were present in several samples (e.g., well C7); however, when these bands were extracted from the gel, reamplified, and reelectrophoresed on a DGGE gel, the migration was identical to the migration of the major band and the positive control for tet(M) (data not shown). DGGE analysis for the tet(M) gene in isolates also demonstrated that there was one principal band corresponding to the tet(M) control band. The isolates were also analyzed by DGGE by using primers specific for the V3 region of the 16S rDNA (Fig. 3). The results indicated that isolates harboring the same tet(M) gene were representatives of at least 10 different phylotypes at sites A and C.
FIG. 2.
DGGE analysis of tet(M) in water samples and in bacterial isolates. Lane 1, site A lagoon; lane 2, A8; lane 3, A9; lane 4, A16; lane 5, A3; lane 6, A13; lane 7, A15; lane 8, A5; lane 9, A11; lane 10, A7 (site A background well); lane 11, ALE1; lane 12, ALE2; lane 13, ALE3; lane 14, ALE4; lane 15, A8-2; lane 16, A8-3; lane 17, A8-4; lane 18, AL-2; lane 19, AL-3; lane 20, AL-4; lane 21, A16-2; lane 22, AL-1; lane 23, site C lagoon; lane 24, C2; lane 25, C6; lane 26, C7; lane 27, C1 (site C background well); lane 28, CLE1; lane 29, CLE2; lane 30, CLE3; lane 31, CLE4, lane 32, CL-1; lane 33, CL-3; lane 34, C2-1; lane 35, tet(M) positive control strain; lane 36, CL-2. Lanes M contained markers consisting of tet(Q), tet(M), and tet(O).
FIG. 3.
DGGE analysis of V3 variable region of 16S rDNA from tetracycline-resistant bacterial isolates. Lane 1, ALE1; lane 2, ALE2; lane 3, ALE3; lane 4, ALE4; lane 5, A8-2; lane 6, A8-3; lane 7, A8-4; lane 8, AL-2; lane 9, AL-3; lane 10, AL-4; lane 11, A16-1 (fungal isolate, negative control); lane 12, AL-1; lane 13, CLE1; lane 14, CLE2; lane 15, CLE3; lane 16, CLE4; lane 17, CL-1; lane 18, CL-3; lane 19, CL-2.
Identification of tetracycline-resistant bacteria.
Several tetracycline-resistant isolates were selected for further identification by 16S rDNA sequencing. The strains that were selected on Enterococcosel medium belonged to the low-G+C-content gram-positive bacterial phylum, and the closest relatives were Enterococcus sp., Staphylococcus sp., and Lactobacillus reuteri (Table 3). Growth on MR2A medium resulted in a wider range of taxonomic affiliations, including members of the alpha and gamma subdivisions of the class Proteobacteria and the Actinobacteridae subclass of high-G+C-content gram-positive bacteria (Table 3). The only tetracycline-resistant isolate obtained from the background well at site A was identified as a member of the genus Bosea, which includes bacteria mostly of soil origin. With the exception of L. reuteri, all of the isolates had levels of similarity with database entries between 91 and 96%, which allowed only approximate taxonomic identification at the genus level and higher taxa levels.
TABLE 3.
Identification of tetracycline-resistant isolates by using 16S rDNA sequences
| Isolate | Size of 16S rDNA fragment analyzed (bp) | Closest taxon (% similarity) | Taxonomic affiliation |
|---|---|---|---|
| ALE1 | 1,438 | Enterococcus hirae (94) | Low-G+C-content gram-positive bacteria |
| ALE3 | 1,103 | Staphylococcus cohnii (96) | Low-G+C-content gram-positive bacteria |
| CLE2 | 520 | Enterococcus sp. (91) | Low-G+C-content gram-positive bacteria |
| CLE4 | 1,312 | Lactobacillus reuteri (97) | Low-G+C-content gram-positive bacteria |
| A7-2 | 1,445 | Bosea thiooxidans (95) | Proteobacteria, alpha subdivision |
| A8-2 | 1,424 | Microbacterium oxydans (95) | High-G+C-content gram-positive bacteria, subclass Actinobacteridae |
| A8-3 | 1,102 | Afipia genospecies 9 (96) | Proteobacteria, alpha subdivision |
| CL-2 | 1,363 | Pseudomonas pseudoalcaligenes (93) | Proteobacteria, gamma subdivision |
DISCUSSION
Tetracycline is commonly used in animal agriculture, particularly in the swine industry (16). Because of this use, selection for tetracycline resistance occurs in the normal swine gut microflora, and in our previous study we demonstrated that the reservoir of tetracycline resistance determinants in the swine intestinal microbiota is substantial and diverse (1). However, the details of what happens with this pool of antibiotic resistance genes further downstream remain largely unknown. Since animal production systems are not closed ecosystems, this pool may be released into the environment. In this study, we demonstrated that a broad range of tetracycline resistance genes occur in two swine waste lagoons and that upon release into the environment these genes can potentially mobilize and persist.
Detection of all of the RPP genes in the waste lagoons at both sites clearly indicated that selection for this drug resistance trait occurs and that a number of resistance genes can be maintained in the microbial populations present. A wide range of RPP genes were detected in groundwater downstream of the waste lagoons. Greater occurrence of RPP genes was detected in wells proximal to both lagoons in the direction of groundwater flow, and the detection of tet(Q) in well A14, located more than 250 m downstream of the lagoon at site A (Fig. 1), suggested that the mobility of resistance genes in the environment can be substantial. Two of the locations at site A had nested wells (Fig. 1, wells A4 and A6), and second wells were screened in deeper sand layers (wells A3 and A5). The occurrence of RPP tetracycline resistance genes was greater in the deep wells than in the corresponding shallow wells, demonstrating that contaminants may be vertically mobile depending on the hydrogeology of the location. Although this study was based on a single sampling event, the data suggest that the presence of the tetracycline resistance genes is due to seepage and movement of groundwater underlying the lagoons. Wells A10 and C3 did not contain any of the tetracycline resistance determinants, as expected due to the locations of the wells relative to the lagoons and the direction of groundwater flow. Thus, there were clear gradients of relative frequency and diversity of tetracycline resistance genes, with the maximal values occurring at waste lagoons and a gradual decline in the direction of groundwater flow; however, the genes were still detectable 250 m downstream. These observations may have important implications for understanding the circulation and acquisition of antibiotic resistance genes. Groundwater constitutes a substantial part of the public water supply in the United States (http://water.usgs.gov/wid/html/GW.html), and at both study sites it is used as the predominant source of untreated drinking water. Thus, along with other ways of acquiring antibiotic resistance, such as consumption of tainted food, the occurrence of antibiotic resistance genes in drinking water provides a possible way for antibiotic resistance to enter the animal and human food chain.
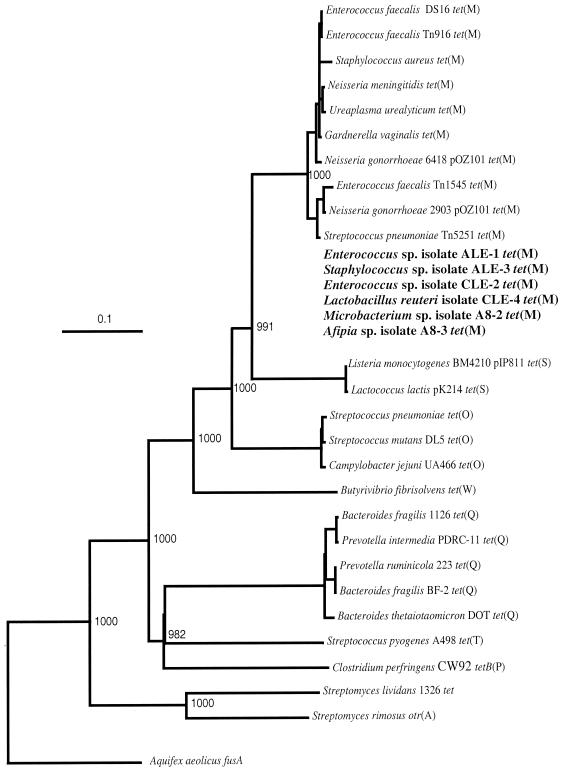
Several tet genes, particularly tet(M), tet(O), tet(Q), and tet(W), were dominant in the total communities in the lagoons and groundwater. These genes were found to be predominant in the gastrointestinal tracts of pigs and steers (1), and in this study the elevated frequencies of these genes in the environment surrounding the farms were consistent with the hypothesis that this occurrence was the result of gene flow from the animals. Tetracycline-resistant bacteria obtained from the lagoon and groundwater samples primarily harbored the tet(M) gene, probably because the cultivation technique was biased towards aerobic growth and the group of bacteria containing the known tet(M) genes includes members of a broad range of genera, including genera of gastrointestinal origin (Fig. 4). Furthermore, it is known that alleles of tet(M) are present in a broad range of bacteria, and their transfer is presumably mediated by conjugative transposons Tn916, Tn1545, and Tn5251 and large conjugative plasmids. It is notable, however, that the use of two different media and random selection of tetracycline-resistant colonies resulted primarily (75%) in bacteria harboring tet(M). No groundwater isolates were correlated with a high occurrence of Tet T and Tet O, the two determinants which occur in aerotolerant Streptococcus spp. (Fig. 4). None of the other RPP tetracycline resistance genes were detected in any of the tet(M)-harboring isolates except CLE3, which contained both tet(M) and tet(S). In some tetracycline-resistant isolates (A7-2, AL-1, AL-5, CL-2, A3-1, and C7-1) no genes conferring ribosomal protection were detected, and these isolates most likely have an alternative mechanism of tetracycline resistance, such as energy-dependent efflux of tetracycline. Anaerobic genera such as Prevotella, Bacteroides, and Butyrivibrio were not targeted for cultivation in our study, and consequently, isolates containing the two most prominent lagoon and groundwater markers [tet(Q) and tet(W)] were not represented. Few groundwater samples contained tet(S), otrA, and tetB(P); however, these determinants were detected in the lagoons, which suggested that they were present at significantly lower levels than the other RPP tetracycline resistance genes.
FIG. 4.
Phylogenetic placement of tetracycline resistance genes encoding RPPs. The sequence of the Aquifex aeolicus fusA gene encoding translation elongation factor EF-G is used as the outgroup for rooting the tree. The numbers above nodes indicate the number of times that a tree configuration occurred among 1,000 bootstrap trials. Scale bar = 0.1 fixed nucleotide substitution per sequence position. The tet(M)-harboring strains isolated in this work are indicated by boldface type. These strains were not incorporated into the phylogenetic analysis and were placed in the Tet M cluster arbitrarily based on sequence similarity.
Enterococci were the dominant fecal bacteria detected in lagoons and in several groundwater samples. Tetracycline-resistant Enterococcus spp. were isolated along with an organism known to be of gastrointestinal origin, L. reuteri. The presence of identical tet(M) alleles in these bacteria suggests that lateral transfer of the gene occurred; however, it cannot be determined if this event occurred in the gut of an animal, in the lagoon environment, or in the groundwater. This gene and tet(O) were found to be circulating among the fecal streptococci in the pig gut (1). In this work, more significantly, the potential for transfer of the tet(M) allele involving groundwater and commensal soil bacteria was evident. Taxonomic analysis of isolates carrying the same tet(M) gene and belonging to the genera Afipia and Microbacterium confirmed that they probably originated from soil. Very little is known about the occurrence of tet(M) and other tetracycline resistance genes in phenotypically tetracycline-resistant bacteria in soil. Most of the tet(M) genotyping work has been done with clinical isolates, and interestingly, the genes are genetically quite diverse (92 to 96.8% identity) and display a mosaic structure consistent with homologous recombination between different lineages (3, 10, 15, 26). In our experiments, we observed extreme homogeneity of these genes in taxonomically (and ecologically) diverse bacteria, including intestinal lactobacilli, enterococci, and staphylococci, as well as soil species of Microbacterium and Afipia (Fig. 4). Consistent with this observation, we hypothesize that in our case we found a point source of genetic contamination originating on farms with the selective pressure of tetracycline. It is likely that transfer events occur in part because of the high density of bacteria in the lagoons, but it is not known to what extent such events occur in less populated groundwater environments. The observed horizontal spread of tet(M) genes to soil bacteria may have several notable implications. First, such spread suggests that antibiotic resistance gene dissemination is not restricted to the release of antibiotic-resistant gastrointestinal microbiota, which may have limited viability outside the gastrointestinal tract. The dissemination vector is not interrupted even if the original antibiotic resistance-harboring bacteria are not viable anymore. Second, when the resistance gene pool is mobilized into the indigenous bacteria in the environment, it has a much better chance of survival, persistence, and mobility, effectively increasing the gene frequency in local populations and having increased potential for reaching other ecosystems.
Another explanation for the elevated occurrence of tet(M) in soil bacteria may be the selective pressure of tetracycline, which may move through the soil layers to the groundwater and select for tetracycline resistance genes residing in soil microbiota. Few previous studies have addressed the fate of antibiotics in the environment and, if they are present, whether they exert any selective pressure for resistance. Tetracycline is not known to be readily degradable and is reportedly strongly absorbed in various soil types (28). This, along with the constant input of tetracycline via animal waste, can potentially provide a concentrated environment in which selection can occur. However, because of the strong sorption of tetracycline in soil (28), this antibiotic may have limited ability to reach distant groundwater layers and is concentrated primarily in soil layers close to lagoons. Despite the possibility of accumulation of tetracycline in soil environments, even low levels can substantially increase the transfer frequency of large conjugative transposons, such as Tn916 (32) and Tn1545 (5), Tn916-like elements (29, 35), or Bacteroides transposons (30) on which tetracycline resistance genes reside. Other factors conducive to resistance persistence and acquisition in the environment are not known, and groundwater is a unique environment for study. In particular, these factors could be naturally occurring compounds with structural similarity to tetracyclines, such as certain plant flavonoids.
In this study, we estimated the pool, diversity, and possible transmission of one group of tetracycline resistance genes in two animal production systems and their surrounding environments. Although previous studies have addressed the occurrence of specific antibiotic resistance characteristics in the environment, none of these studies included a more thorough genotypic characterization of a specific antibiotic resistance trait. While the set of genes which we targeted in our study includes only a limited portion of the bacterial resistance genes that may be present, it provides the basis for using a similar approach in order to combine molecularly and phenotypically based methods to characterize the occurrence, diversity, and routes of transmission of antibiotic genes in nature. The data obtained can be used to address the question of whether environments such as groundwater and soil are significant reservoirs for dissemination of resistance genes. In addition, long-term surveillance can provide an indication of the mobility, persistence, and transfer of resistance genes and begin to address the issue of agriculture as a source of resistance genes for entry into the environment.
REFERENCES
- 1.Aminov R I, Garrigues-Jeanjean N, Mackie R I. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl Environ Microbiol. 2001;67:22–32. doi: 10.1128/AEM.67.1.22-32.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deplancke B, Hristova K R, Oakley H A, McCracken V J, Aminov R I, Mackie R I, Gaskins H R. Molecular ecological analysis of the succession and diversity of sulfate-reducing bacteria in the mouse gastrointestinal tract. Appl Environ Microbiol. 2000;66:2166–2174. doi: 10.1128/aem.66.5.2166-2174.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doherty N, Trzcinski K, Pickerill P, Zawadzki P, Dowson C G. Genetic diversity of the tet(M) gene in tetracycline-resistant clonal lineages of Streptococcus pneumoniae. Antimicrob Agents Chemother. 2000;44:2979–2984. doi: 10.1128/aac.44.11.2979-2984.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donoho A L. Biochemical studies on the fate of monensin in animals and in the environment. J Anim Sci. 1984;58:1528–1539. doi: 10.2527/jas1984.5861528x. [DOI] [PubMed] [Google Scholar]
- 5.Doucet-Populaire F, Trieu-Cuot P, Dosbaa I, Andremont A, Courvalin P. Inducible transfer of conjugative transposon Tn1545 from Enterococcus faecalis to Listeria monocytogenes in the digestive tracts of gnotobiotic mice. Antimicrob Agents Chemother. 1991;35:185–187. doi: 10.1128/aac.35.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elmund G K, Morrison S M, Grant D W, Nevins M P. Role of excreted chlortetracycline in modifying the decomposition process in feedlot waste. Bull Environ Contam Toxicol. 1971;6:129–135. doi: 10.1007/BF01540093. [DOI] [PubMed] [Google Scholar]
- 7.Feinman S E, Matheson J C. Draft environmental impact statement: subtherapeutic antibacterial agents in animal feeds. Food and Drug Administration Department of Health, Education and Welfare Report. Washington, D.C.: Food and Drug Administration; 1978. p. 372. [Google Scholar]
- 8.Flanagan D A, Gregory L G, Carter J P, Karakas-Sen A, Richardson D J, Spiro S. Detection of genes for periplasmic nitrate reductase in nitrate respiring bacteria and in community DNA. FEMS Microbiol Lett. 1999;177:263–270. doi: 10.1111/j.1574-6968.1999.tb13742.x. [DOI] [PubMed] [Google Scholar]
- 9.Fries M R, Zhou J, Chee-Sanford J, Tiedje J M. Isolation, characterization, and distribution of denitrifying toluene degraders from a variety of habitats. Appl Environ Microbiol. 1994;60:2802–2810. doi: 10.1128/aem.60.8.2802-2810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gascoyne-Binzi D M, Heritage J, Hawkey P M. Nucleotide sequences of the tet(M) genes from the American and Dutch type tetracycline resistance plasmids of Neisseria gonorrhoeae. J Antimicrob Chemother. 1993;32:667–676. doi: 10.1093/jac/32.5.667. [DOI] [PubMed] [Google Scholar]
- 11.Gavalchin J, Katz S E. The persistence of fecal-borne antibiotics in soil. J Assoc Off Anal Chem Int. 1994;77:481–485. [Google Scholar]
- 12.Goni-Urriza M, Capdepuy M, Arpin C, Raymond N, Caumette P, Quentin C. Impact of an urban effluent on antibiotic resistance of riverine Enterobacteriaceae and Aeromonas spp. Appl Environ Microbiol. 2000;66:125–132. doi: 10.1128/aem.66.1.125-132.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haapapuro E R, Barnard N D, Simon M. Review—animal waste used as livestock feed: dangers to human health. Prev Med. 1997;26:599–602. doi: 10.1006/pmed.1997.0220. [DOI] [PubMed] [Google Scholar]
- 14.Hagedorn C, Robinson S L, Filtz J R, Grubbs S M, Angier T A, Reneau R B. Determining sources of fecal pollution in a rural Virginia watershed with antibiotic resistance patterns in fecal streptococci. Appl Environ Microbiol. 1999;65:5522–5531. doi: 10.1128/aem.65.12.5522-5531.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang R, Gascoyne-Binzi D M, Hawkey P M, Yu M, Heritage J, Eley A. Molecular evolution of the tet(M) gene in Gardnerella vaginalis. J Antimicrob Chemother. 1997;40:561–565. doi: 10.1093/jac/40.4.561. [DOI] [PubMed] [Google Scholar]
- 16.Jonsson E, Conway P. Probiotics for pigs. In: Fuller R, editor. Probiotics: the scientific basis. New York, N.Y: Chapman and Hall; 1992. pp. 260–316. [Google Scholar]
- 17.Karkhoff-Schweizer R R, Huber D P, Voordouw G. Conservation of the genes for dissimilatory sulfite reductase from Desulfovibrio vulgaris and Archaeoglobus fulgidus allows their detection by PCR. Appl Environ Microbiol. 1995;61:290–296. doi: 10.1128/aem.61.1.290-296.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaspar C W, Burgess J L, Knight I T, Colwell R R. Antibiotic resistance indexing of Escherichia coli to identify sources of fecal contamination in water. Can J Microbiol. 1990;36:891–894. doi: 10.1139/m90-154. [DOI] [PubMed] [Google Scholar]
- 19.Krapac I G, Dey W S, Smyth C A, Roy W R. Proceedings of the National Ground Water Association Animal Feeding Operations and Groundwater: Issues, Impacts, and Solutions—A Conference for the Future. St. Louis, Mo: National Groundwater Association; 1998. Impacts of bacteria, metals, and nutrients on groundwater at two hog confinement facilities; pp. 29–50. [Google Scholar]
- 20.Krapac I G, Dey W S, Roy W R, Jellerichs B G, Smyth C. 8th International Symposium on Animal, Agricultural and Food Processing Wastes. Des Moines, Iowa: American Society for Agricultural Engineering; 2000. Groundwater quality near livestock manure pits; pp. 710–718. [Google Scholar]
- 21.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley and Sons; 1991. pp. 115–175. [Google Scholar]
- 22.Madden T L, Tatusov R L, Zhang J. Application of network BLAST server. Methods Enzymol. 1996;266:131–141. doi: 10.1016/s0076-6879(96)66011-x. [DOI] [PubMed] [Google Scholar]
- 23.McKeon D M, Calabrese J P, Bissonnette G K. Antibiotic resistant gram-negative bacteria in rural groundwater supplies. Water Res. 1995;29:1902–1908. [Google Scholar]
- 24.Molbak K, Baggesen D L, Aarestrup F M, Ebbesen J M, Engberg J, Frydendahl K, Gerner-Smidt P, Petersen A M, Wegener H C. An outbreak of multidrug-resistant, quinolone-resistant Salmonella enterica serotype Typhimurium DT104. N Engl J Med. 1999;341:1420–1425. doi: 10.1056/NEJM199911043411902. [DOI] [PubMed] [Google Scholar]
- 25.Muyzer G, de Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oggioni M R, Dowson C G, Smith J M, Provvedi R, Pozzi G. The tetracycline resistance gene tet(M) exhibits mosaic structure. Plasmid. 1996;35:156–163. doi: 10.1006/plas.1996.0018. [DOI] [PubMed] [Google Scholar]
- 27.Pillai S D, Widmer K W, Maciorowski K G, Ricke S C. Antibiotic resistance profiles of Escherichia coli isolated from rural and urban environments. J Environ Sci Health Part A Environ Sci Eng. 1997;32:1665–1675. [Google Scholar]
- 28.Rabolle M, Spliid N H. Sorption and mobility of metronidazole, olaquindox, oxytetracycline and tylosin in soil. Chemosphere. 2000;40:715–722. doi: 10.1016/s0045-6535(99)00442-7. [DOI] [PubMed] [Google Scholar]
- 29.Rice L B, Marshall S H, Carias L L. Tn5381, a conjugative transposon identifiable as a circular form in Enterococcus faecalis. J Bacteriol. 1992;174:7308–7315. doi: 10.1128/jb.174.22.7308-7315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salyers A A, Shoemaker N B, Stevens A M, Li L-Y. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol Rev. 1995;59:579–590. doi: 10.1128/mr.59.4.579-590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schramm A, Santegoeds C M, Nielsen H K, Ploug H, Wagner M, Pribyl M, Wanner J, Amann R, de Beer D. On the occurrence of anoxic microniches, denitrification, and sulfate reduction in aerated activated sludge. Appl Environ Microbiol. 1999;65:4189–4196. doi: 10.1128/aem.65.9.4189-4196.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Showsh S A, Andrews R E. Tetracycline enhances Tn916-mediated conjugal transfer. Plasmid. 1992;28:213–224. doi: 10.1016/0147-619x(92)90053-d. [DOI] [PubMed] [Google Scholar]
- 33.Smith K E, Besser J M, Hedberg C W, Leano F T, Bender J B, Wicklund J H, Johnson B P, Moore K A, Osterholm M T. Quinolone-resistant Campylobacter jejuni infections in Minnesota, 1992–1998. Investigation team. N Engl J Med. 1999;340:1525–1532. doi: 10.1056/NEJM199905203402001. [DOI] [PubMed] [Google Scholar]
- 34.Sweeten J M. Livestock and poultry waste management: a national overview. In: Blake J, Donald J, Magette W, editors. National livestock, poultry, and aquaculture waste management. St. Joseph, Mich: American Society for Agricultural Engineering; 1992. pp. 4–15. [Google Scholar]
- 35.Torres O R, Korman R Z, Zahler S A, Dunny G M. The conjugative transposon Tn925: enhancement of conjugal transfer by tetracycline in Enterococcus faecalis and mobilization of chromosomal genes in Bacillus subtilis and E. faecalis. Mol Gen Genet. 1991;225:395–400. doi: 10.1007/BF00261679. [DOI] [PubMed] [Google Scholar]
- 36.Tsai Y-L, Olsen B H. Rapid method for direct extraction of DNA from soil and sediments. Appl Environ Microbiol. 1991;57:1070–1074. doi: 10.1128/aem.57.4.1070-1074.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner M, Roger A J, Flax J L, Brusseau G A, Stahl D A. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J Bacteriol. 1998;180:2975–2982. doi: 10.1128/jb.180.11.2975-2982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiggins B A. Discriminant analysis of antibiotic resistance patterns in fecal streptococci, a method to differentiate human and animal sources of fecal pollution in natural waters. Appl Environ Microbiol. 1996;62:3997–4002. doi: 10.1128/aem.62.11.3997-4002.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiggins B A, Andrews R W, Conway R A, Corr C L, Dobratz E J, Dougherty D P, Eppard J R, Knupp S R, Limjoco M C, Mettenburg J M, Rinehardt J M, Sonsino J, Torrijos R L, Zimmerman M E. Use of antibiotic resistance analysis to identify nonpoint sources of fecal pollution. Appl Environ Microbiol. 1999;65:3483–3486. doi: 10.1128/aem.65.8.3483-3486.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]