Abstract
Background:
Oncogenic microRNAs, a kind of stable epigenetic inhibitors, often deregulated in Mycosis fungoides (MF) which affect the skin and tend to transform and spread.
Results:
Previous studies investigating the de-expression of microRNA in MF patients skin biopsies identified that they were not only regulated by signaling pathway, but also regulated other signaling pathway. Furthermore, studies have elucidated the molecular mechanisms of the STAT signaling pathway that can promote a great diversity of miRNA expression via cytokine binding receptors, activating Janus kinase-3 and STAT proteins. But some non-STAT signaling pathway with mircoRNA de-expression in MF was incomplete.
Conclusion:
Taken together, these studies demonstrate that microRNA may be used as the prognosis, progression and diagnose of MF, as they can not only control MF cell proliferation, but also induce MF cell apoptosis.
Keywords: de-expression, mircoRNA, mycosis fungoides, signaling pathway, STAT
1. Introduction
Advanced mycosis fungoides (MF) or Sézary syndrome, major variants of cutaneous T-cell lymphoma (CTCL), are associated with–40% to 47% 5-year survival.[1] The annual incidence of primary cutaneous lymphomas is estimated to be 1:100,000, of which CTCL accounts for approximately 75% of cases; therefore, CTCL is a common type of primary cutaneous lymphomas.[2,3] Through the clonal proliferation of skin-invasive mature T-lymphocytes, CTCLs are characterized as non-Hodgkin's lymphomas.[2] Mycosis fungoides (MF) is a common and indolent form of CTCL and is characterized by patches, plaques, or tumors containing epidermotrophic malignant CD4+CD45 RO+ helper/memory T-cells.[4] In the primary stage, MF appears as a flat erythromatous skin lesion and resembles non-malignant psoriasis or eczema and can last for several years.[4] In later stages, tumor cells spread to other parts of the body as a fatal outcome. MF can develop into a leukemic variant, Sézary syndrome, in which cancer T-cells appear in the skin and blood, or shift to large cell lymphoma.[5] Skin cytokines orchestrate inflammation through their impact on the expression and function of other cytokines, and their downstream effectors (such as STAT and SOCS proteins, microRNA) are frequently observed.[6,7] MicroRNAs (miRNAs) are important molecular markers of MF progression and diagnose.[8]
miRNAs are a class of small non-coding RNAs with length of 18 to 22nt that are ubiquitous in eukaryotes and can regulate protein expression at the mRNA level.[9–11] Many studies have reported that MF alters miRNA expression, including reduced miR-191, miR-223, and miR-342, and increased miR-155.[12] These microRNA expression changes influence or influence signaling pathways such as STAT3, STAT5, or p53/Akt et al.[13,14]
2. MicroRNA expression and MF
It is important to understand how microRNA expression changes influence MF cell proliferation in order to develop them as a new target gene for the prevention and treatment of cancer. From Table 1, the expression of about 12 miRNAs whose function and characteristics were further analyzed by reliable experiments up-or down-regulated in MF skin biopsies or cells. Six miRNA expression upregulates and belongs to an oncogenic molecule. Another 6 miRNAs are downregulated and belong to the suppressor gene. Eight miRNAs can be used to evaluate MF progression; miRNA-122 and miRNA-214 are molecular markers of MF prognosis.[14,15] Moreover, miR-155, miR-203, and miR-205 in patient peripheral blood can be used as diagnostic markers,[16,17] that discriminated between malignant and benign skin inflammation with an accuracy of more than 90%.[18]
Table 1.
Recent microRNA and mycosis fungoides Studies.
| MiRNA type | Year | Biomarker use | Sample | Target/Pathway | Expression | Function | Ref. |
| miR-93 | 2021 | Progression | Malignant T cells lines | p21 | down | Oncogenic molecule | [40] |
| miR-195–5p | 2020 | Progression | Skin biopsies and cell lines | ARL2 | down | Suppressor gene | [41] |
| miR-106b | 2020 | Prognosis | Skin biopsies | P21/TXNIP | up | Oncogenic function | [42] |
| miR-337 | 2019 | / | Malignant and non-malignant T cell lines | JAK/STAT | up | / | [36] |
| miR-214 | 2019 | Diagnose | CD4+ T cells | TWIST1/BRD4/miR-214 | up | Oncogenic molecule | [31] |
| miR-155 | 2018 | Diagnose | Malignant T-cell lines (MyLa2059, MyLa3675 and MyLa2000) and HH cells | SATB1/GATA3 | up | Oncogenic molecule | [25] |
| miR-155 | 2018 | Diagnose | HUT102, HUT78 and HH cell lines | JAK/STAT, MAPK/ERK and PI3K/AKT | up | Oncogenic molecule | [16] |
| miR-155 | 2017 | Diagnose | Peripheral blood | / | up | oncogenic Molecule | [16] |
| miR-203, R-205 | 2017 | Diagnose | Peripheral blood | / | down | Suppressor gene | [43] |
| miR-150 | 2017 | Progression | Skin biopsy, HH, HUT78, and MJ cell lines | CCR6 | down | Suppressor gene | [43] |
| miR-214 | 2017 | Prognosis | Skin biopsies | FCRL-3, Tox | up | / | [15] |
| miR-21 | 2016 | Progression | Skin biopsies | STAT5/JAK3 | up | Oncogenic Molecule | [27] |
| miR-34a | 2016 | Progression | Skin biopsies | p53 | up | oncogenic Molecule | [32] |
| miR-29a | 2016 | Progression | Skin biopsies | p53 | down | Suppressor gene | [32] |
| miR-17–92 cluster | 2016 | Progression | Skin biopsies | / | up | Oncogenic function | [44] |
| miR-16 | 2016 | Progression | Skin, My-La, MJ, HUT102, HH, and HUT78 | miR-16/p21/Bmi1 | down | Suppressor gene | [35] |
| miR-22 | 2015 | / | MyLa2059, MyLa2000, PB2B, MyLa1850 and MySi cell lines | Jak3/STAT3/STAT5 | down | Suppressor gene | [13] |
| miR-223 | 2014 | Progression | Skin biopsies, HH and Hut-78 cell lines | TOX | down | Suppressor gene | [30] |
| miR-155 | 2014 | Progression | Skin biopsies | STAT4 | up | Oncogenic molecule | [21] |
| miR-155 | 2013 | / | Skin biopsies | STAT5/BIC | up | Oncogenic molecule | [20] |
| miR-122 | 2012 | Prognosis | Skin, MyLa2000, SeAx and Hut-78 cell lines | p53/Akt | up | Oncogenic molecule | [14] |
| miR-21 | 2011 | Progression | SeAx cell line | STAT3 | up | Oncogenic molecule | [28] |
3. MicroRNA and STAT signaling pathway
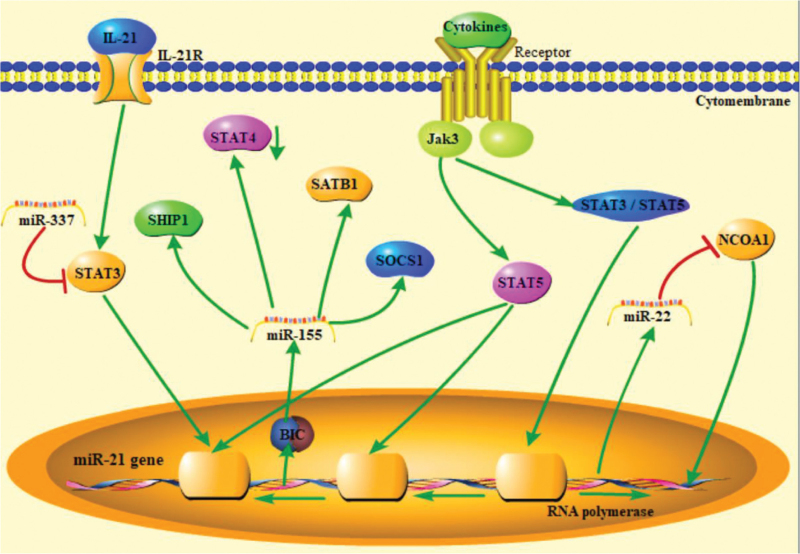
The expression and function of STAT3, STAT4, and STAT5 have been extensively studied in MF, and these genes appear to play an important role in disease pathogenesis and can be used as important prognostic markers. The effectors (STAT3 and STAT5 et al) and the up-stream Interleukin-2 receptor common gamma chain, the associated Janus kinase-3 (Jak3) have attracted substantial interest. Interleukin-2 receptor common gamma chain-signaling cytokines, including IL-2, IL-4, IL-7, IL-15, and IL-21 are implicated in early pathogenesis and constitutive.[13,19] As shown in Figure 1, the deregulation of Jak3/STAT3/STAT5 signaling in MF cells can repress the expression of the gene encoding miR-22, which targets the transcriptional co-activator NCoA1.[13]
Figure 1.
MicroRNA and STAT signaling pathway.
The expression of miR-155 is enhanced in the affected skin of MF patients compared to that in healthy individuals.[8,18] The host gene of miR-155 is BIC, which is transcribed by STAT5.[20] Upregulated miR-155 binds and cleaves its target gene, STAT4, in advanced MF patients skin biopsies.[21] The PI3K/AKT, JAK/STAT, and MAPK signaling pathways negatively regulate signaling through miR-155, directing inositol phosphatase SHIP1 and SOCS1.[22–24] When JAK3 phosphorylates STAT5, it translocates to the nucleus and initiates the transcription of miR-155. Upregulation of miR-155 leads to inhibition of SATB1 and inhibition of IL-5 and IL-9 expression.[25] Oncogenic miR-155 appears to contribute to the cancerous phenotype of MyLa and MJ cells by interrupting the activation of the G2/M checkpoint.[26] STAT5 is not only a driver of miR-155 but also regulates miR-21 expression, which has been linked to disease progression interconnecting the JAK/STAT pathways as a key regulator of miRNAs in CTCL.[27] STAT3 has been regarded as a promising therapeutic target for many years in Se’zary syndrome and can increase miR-21 expression by IL-21 activating IL-21R and STAT3, and then miR-21 inhibits Se’zary syndrome cell apoptosis.[28]
4. MicroRNA and other signaling pathway
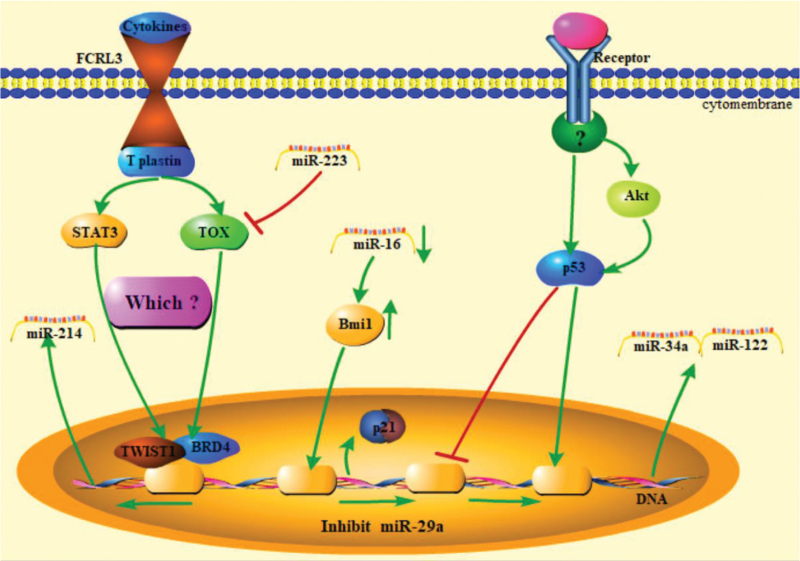
The deregulation of signaling pathways, including STAT, Src kinases, c-Myc, COX-2, NFκB, GATA3, TOX, and embryonic stem cell regulators, appears to play an important role in pathogenesis.[13,29] As shown in Figure 2, Fc receptor-like protein 3, along with T plastin, GATA-3, Tox, and miR-214, was significantly higher in Se’zary syndrome patients.[15] Based on the above results, we hypothesized that GATA-3 or/and Tox were activated by activating Fc receptor-like protein 3 and T plastin, and then they or/and enhanced miR-214 transcription. The protein level of Tox is regulated by miR-223.[30] Kohnken et al showed that miR-214 levels are significantly higher in purified CD4þ neoplastic T cells from patients with MF than from healthy donors. Further studies have proposed that the TWIST1 and BRD4 complex regulate miR-214 expression.[31]
Figure 2.
MicroRNA and signaling pathway participated by p53, STAT3, and TOX.
Deregulation of the p53 signaling pathway was also found in MF patient skin biopsies or cells, including My-La, MJ, HUT102, HH, and HUT78.[32] The up-regulated miR-122 inhibits tumor cell apoptosis by Akt/p53 signaling pathway,[14] similarly miR-34a expression level was also increased by p53 signaling pathway in Se’zary syndrome patients skin biosies.[32] However, miR-29a expression was lower in MF patients than in healthy patients because of p53 signaling pathway inhibition.[32]
Earlier studies did not detect changes in p16 expression in all MF cells until 2016.[33,34] Kitadate et al speculated that miR-16 directly or indirectly suppresses Bmi1, thereby enhancing p21 expression in MF cells based on their experiments results.[35] It has been confirmed that microRNAs can directly regulate receptor proteins. The miR-150 in MF cells was upregulated and combined with the C-C chemokine receptor 6 “seed sequence” mRNA of the 3-untranslated region (3-UTR) in advanced MF.
STAT3 expression can be significantly downregulated following transfection with the miR-337 mimic, which potentially targets the 3-UTR of STAT3.[36] MF progression can be estimated by analyzing the upregulated expression of miR-155, miR-146a, 146b-5p, miR-342–3p, and let-7i∗ and downregulated expression of miR-203 and miR-205.[37] Subsequent studies confirmed that miRNAs are potentially valuable tools for the evaluation of disease progression in MF.[38,39]
5. Conclusions and perspectives
Collectively, the findings discussed in the present review provide novel insights into the effect of microRNAs on MF cells, which supports new concepts for the prognosis, progression, and diagnosis of MF. As shown in Table 1, some miRNAs were upregulated in the skin biosies of MF patients; however, some miRNAs were downregulated. These miRNAs can be used as biomarkers for disease diagnosis and as therapeutic targets.
MF cell proliferation and apoptosis involve the functional cooperation of many signaling molecules. As summarized in Figure 1, STAT signaling pathways, including STAT3, STAT4, and STAT5, can promote a great diversity of miRNA expression via cytokine binding receptors, activating Jak3 and STAT proteins. Transcripted microRNAs regulate MF cell proliferation and apoptosis via other signaling pathways or target molecules.
MicroRNA transcription can also be regulated by another signaling pathway, as shown in Figure 2. However, these signaling pathways can regulate microRNA expression and can be regulated by microRNAs. It is not clear at the moment that some important molecules are involved in this pathway. STAT3, p53, TOX, and Bmi1 participate in these pathways and play an important role.
These results indicate that cocktail therapy with microRNA and other drugs may greatly reduce the risk of MF cell metastasis in all types. The significance of miRNA as a molecular marker may be used for prognosis, progression, and diagnosis of MF in the future. Further studies are required to determine which of the signaling pathways or miRNAs is most important for the treatment or diagnosis of MF. In addition, many previous studies have not established an integrated non-STAT signaling pathway. Future studies investigating whole signal regulation in MF may provide further insight into the mechanisms underlying its activity.
Author contributions
Conceptualization: Xiaona Yao, zhiyuan Sun.
Data curation: Xiaona Yao, zhiyuan Sun.
Formal analysis: Xiaona Yao, zhiyuan Sun.
Funding acquisition: Xiaona Yao, Xuewen Tian, Xun Li.
Resources: Xing Ding.
Writing – review & editing: Xiaona Yao, zhiyuan Sun.
Footnotes
Abbreviations: CTCL = cutaneous T-cell lymphoma, JAK3 = Janus kinase-3, MF = mycosis fungoides.
How to cite this article: Sun Z, Yao X, Ding X, Li X, Tian X. MicroRNAs and their signaling pathway in mycosis fungoides. Medicine. 2022;101:25(e29248).
ZS and XY contributed equally to this work.
This work was supported by grants from the Taishan Scholars Program of Shandong Province (tsqn201909148); Shandong Province Key R&D Program (2020CXGC010902) and Planning Research Project of Philosophy and Social Science of Shandong Province (21CTYJ19).
The authors have no conflicts of interests to disclose.
The datasets generated during and/or analyzed during the current study are not publicly available, but are available from the corresponding author on reasonable request.
Contributor Information
Zhiyuan Sun, Email: 1213225245@qq.com.
Xiaona Yao, Email: 2359301658@qq.com.
Xing Ding, Email: dingxing@163.com.
Xun Li, Email: lixun@163.com.
References
- [1].Agar NS, Wedgeworth E, Crichton S, et al. Survival outcomes and prognostic factors in mycosis fungoides/Sezary syndrome: validation of the revised International Society for Cutaneous Lymphomas/European Organisation for Research and Treatment of Cancer staging proposal. J Clin Oncol 2010;28:4730–9. [DOI] [PubMed] [Google Scholar]
- [2].Willemze R, Cerroni L, Kempf W, et al. The 2018 update of the WHO-EORTC classification for primary cutaneous lymphomas. Blood 2019;133:1703–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Neelis KJ, Schimmel EC, Vermeer MH, et al. Low-dose palliative radiotherapy for cutaneous B- and T-cell lymphomas. Int J Radiat Oncol Biol Phys 2009;74:154–8. [DOI] [PubMed] [Google Scholar]
- [4].Kim EJ, Hess S, Richardson SK, et al. Immunopathogenesis and therapy of cutaneous T cell lymphoma. J Clin Invest 2005;115:798–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kim YH, Willemze R, Pimpinelli N, et al. TNM classification system for primary cutaneous lymphomas other than mycosis fungoides and Sezary syndrome: a proposal of the International Society for Cutaneous Lymphomas (ISCL) and the Cutaneous Lymphoma Task Force of the European Organization of Research and Treatment of Cancer (EORTC). Blood 2007;110:479–84. [DOI] [PubMed] [Google Scholar]
- [6].Persson JL. miRNA in mycosis fungoides and skin inflammation. Apmis 2013;121:1017–9. [DOI] [PubMed] [Google Scholar]
- [7].Krejsgaard T, Lindahl LM, Mongan NP, et al. Malignant inflammation in cutaneous T-cell lymphoma-a hostile takeover. Semin Immunopathol 2017;39:269–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].van Kester MS, Ballabio E, Benner MF, et al. miRNA expression profiling of mycosis fungoides. Mol Oncol 2011;5:273–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jiang XI, Luo Y, Zhao S, et al. Clinical significance and expression of microRNA in diabetic patients with erectile dysfunction. Exp Ther Med 2015;10:213–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jia W, Wu Y, Zhang Q, et al. Expression profile of circulating microRNAs as a promising fingerprint for cervical cancer diagnosis and monitoring. Mol Clin Oncol 2015;3:851–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Graziano A, Lo MG, Piva I, et al. Diagnostic findings in adenomyosis: a pictorial review on the major concerns. Eur Rev Med Pharmacol Sci 2015;19:1146–54. [PubMed] [Google Scholar]
- [12].McGirt LY, Baerenwald DA, Vonderheid EC, Eischen CM. Early changes in miRNA expression are predictive of response to extracorporeal photopheresis in cutaneous T-cell lymphoma. J Eur Acad Dermatol Venereol 2015;29:2269–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sibbesen NA, Kopp KL, Litvinov IV, et al. Jak3, STAT3, and STAT5 inhibit expression of miR-22, a novel tumor suppressor microRNA, in cutaneous T-Cell lymphoma. Oncotarget 2015;6:20555–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Manfe V, Biskup E, Rosbjerg A, et al. miR-122 regulates p53/Akt signalling and the chemotherapy-induced apoptosis in cutaneous T-cell lymphoma. Plos One 2012;7:e29541.1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Benoit BM, Jariwala N, O’Connor G, et al. CD164 identifies CD4(+) T cells highly expressing genes associated with malignancy in Sezary syndrome: the Sezary signature genes, FCRL3, Tox, and miR-214. Arch Dermatol Res 2017;309:11–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Dusilkova N, Basova P, Polivka J, et al. Plasma miR-155, miR-203, and miR-205 are biomarkers for monitoring of primary cutaneous T-cell lymphomas. Int J Mol Sci 2017;18:2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ralfkiaer U, Hagedorn PH, Bangsgaard N, et al. Diagnostic microRNA profiling in cutaneous T-cell lymphoma (CTCL). Blood 2011;118:5891–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Marstrand T, Ahler CB, Ralfkiaer U, et al. Validation of a diagnostic microRNA classifier in cutaneous T-cell lymphomas. Leuk Lymphoma 2014;55:957–8. [DOI] [PubMed] [Google Scholar]
- [19].Abraham RM, Zhang Q, Odum N, Wasik MA. The role of cytokine signaling in the pathogenesis of cutaneous T-cell lymphoma. Cancer Biol Ther 2011;12:1019–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kopp KL, Ralfkiaer U, Gjerdrum LM, et al. STAT5-mediated expression of oncogenic miR-155 in cutaneous T-cell lymphoma. Cell Cycle 2013;12:1939–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Litvinov IV, Cordeiro B, Fredholm S, et al. Analysis of STAT4 expression in cutaneous T-cell lymphoma (CTCL) patients and patient-derived cell lines. Cell Cycle 2014;13:2975–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yang Y, Yang L, Liang X, Zhu G. MicroRNA-155 Promotes Atherosclerosis Inflammation via Targeting SOCS1. Cell Physiol Biochem 2015;36:1371–81. [DOI] [PubMed] [Google Scholar]
- [23].O’Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci U S A 2009;106:7113–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Seto AG, Beatty X, Lynch JM, et al. Cobomarsen, an oligonucleotide inhibitor of miR-155, co-ordinately regulates multiple survival pathways to reduce cellular proliferation and survival in cutaneous T-cell lymphoma. Br J Haematol 2018;183:428–44. [DOI] [PubMed] [Google Scholar]
- [25].Fredholm S, Willerslev-Olsen A, Met O, et al. SATB1 in Malignant T Cells. J Invest Dermatol 2018;138:1805–15. [DOI] [PubMed] [Google Scholar]
- [26].Moyal L, Yehezkel S, Gorovitz B, et al. Oncogenic role of microRNA-155 in mycosis fungoides: an in vitro and xenograft mouse model study. Br J Dermatol 2017;177:791–800. [DOI] [PubMed] [Google Scholar]
- [27].Lindahl LM, Fredholm S, Joseph C, et al. STAT5 induces miR-21 expression in cutaneous T cell lymphoma. Oncotarget 2016;7:45730–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].van der Fits L, van Kester MS, Qin Y, et al. MicroRNA-21 expression in CD4+ T cells is regulated by STAT3 and is pathologically involved in Sezary syndrome. J Invest Dermatol 2011;131:762–8. [DOI] [PubMed] [Google Scholar]
- [29].Huang Y, Su MW, Jiang X, Zhou Y. Evidence of an oncogenic role of aberrant TOX activation in cutaneous T-cell lymphoma. Blood 2015;125:1435–43. [DOI] [PubMed] [Google Scholar]
- [30].McGirt LY, Adams CM, Baerenwald DA, et al. miR-223 regulates cell growth and targets proto-oncogenes in mycosis fungoides/cutaneous T-cell lymphoma. J Invest Dermatol 2014;134:1101–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kohnken R, McNeil B, Wen J, et al. Preclinical targeting of MicroRNA-214 in cutaneous T-cell lymphoma. J Invest Dermatol 2019;139:1966–74.e3. [DOI] [PubMed] [Google Scholar]
- [32].Papadavid E, Braoudaki M, Bourdakou M, et al. Aberrant microRNA expression in tumor mycosis fungoides. Tumour Biol 2016;37:14667–75. [DOI] [PubMed] [Google Scholar]
- [33].Zhang C, Toulev A, Kamarashev J, et al. Consequences of p16 tumor suppressor gene inactivation in mycosis fungoides and Sezary syndrome and role of the bmi-1 and Ras oncogenes in disease progression. Hum Pathol 2007;38:995–1002. [DOI] [PubMed] [Google Scholar]
- [34].Tosca A, Linardopoulos S, Malliri A, et al. Implication of the Ras and myc oncoproteins in the pathogenesis of mycosis fungoides. Anticancer Res 1991;11:1433–8. [PubMed] [Google Scholar]
- [35].Kitadate A, Ikeda S, Teshima K, et al. MicroRNA-16 mediates the regulation of a senescence-apoptosis switch in cutaneous T-cell and other non-Hodgkin lymphomas. Oncogene 2016;35:3692–704. [DOI] [PubMed] [Google Scholar]
- [36].Xia L, Wu L, Xia H, et al. miR-337 suppresses cutaneous T-cell lymphoma via the STAT3 pathway. Cell Cycle 2019;18:1635–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ralfkiaer U, Lindahl LM, Litman T, et al. MicroRNA expression in early mycosis fungoides is distinctly different from atopic dermatitis and advanced cutaneous T-cell lymphoma. Anticancer Res 2014;34:7207–17. [PubMed] [Google Scholar]
- [38].Lindahl LM, Besenbacher S, Rittig AH, et al. Prognostic miRNA classifier in early-stage mycosis fungoides: development and validation in a Danish nationwide study. Blood 2018;131: [DOI] [PubMed] [Google Scholar]
- [39].Shen X, Wang B, Li K, et al. MicroRNA signatures in diagnosis and prognosis of cutaneous T-cell lymphoma. J Invest Dermatol 2018;138:2024–32. [DOI] [PubMed] [Google Scholar]
- [40].Gluud M, Fredholm S, Blumel E, et al. MicroRNA-93 targets p21 and promotes proliferation in mycosis fungoides T cells. Dermatology 2021;237:277–82. [DOI] [PubMed] [Google Scholar]
- [41].Rittig AH, Johansen C, Celis P, et al. Suppressed microRNA-195-5p expression in mycosis fungoides promotes tumor cell proliferation. Exp Dermatol 2021;30:1141–9. [DOI] [PubMed] [Google Scholar]
- [42].Lindahl LM, Gluud M, Emmanuel T, et al. MicroRNA-106b regulates expression of the tumour suppressors p21 and TXNIP and promotes tumour cell proliferation in mycosis fungoides. Acta Derm Venereol 2020;100:adv00270.1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Abe F, Kitadate A, Ikeda S, et al. Histone deacetylase inhibitors inhibit metastasis by restoring a tumor suppressive microRNA-150 in advanced cutaneous T-cell lymphoma. Oncotarget 2017;8:7572–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Garaicoa FH, Roisman A, Arias M, et al. Genomic imbalances and microRNA transcriptional profiles in patients with mycosis fungoides. Tumour Biol 2016;37:13637–47. [DOI] [PubMed] [Google Scholar]