Abstract
Fecal pollution of water resources is an environmental problem of increasing importance. Identification of individual host sources of fecal Escherichia coli, such as humans, pets, production animals, and wild animals, is prerequisite to formulation of remediation plans. Ribotyping has been used to distinguish fecal E. coli of human origin from pooled fecal E. coli isolates of nonhuman origin. We have extended application of this technique to distinguishing fecal E. coli ribotype patterns from human and seven individual nonhuman hosts. Classification accuracy was best when the analysis was limited to three host sources. Application of this technique to identification of host sources of fecal coliforms in water could assist in formulation of pollution reduction plans.
Fecal pollution of water resources is a problem of increasing worldwide concern (4, 15). Human population growth, inadequate sewage systems, and management of animal waste (especially related to concentrated animal feeding operations) are some of the issues associated with maintenance of supplies of clean water (17). Counts of commensal coliform bacteria have traditionally been used to indicate the potential presence of pathogenic microbes of intestinal origin (1). Total coliform and fecal coliform numbers (1) are useful for estimating fecal pollution levels but give no indication of the specific sources of microbial pollution, such as humans, production animals, pets, or migratory birds. Examples of methods which have been used as indicators of host sources include phage susceptibility (20) and the ratio of fecal coliforms to streptococci (5). Such indirect measurements are based on unstable parameters and may thereby lead to erroneous conclusions (11). More recently, DNA fingerprinting techniques such as ribotyping (11), pulsed-field gel electrophoresis (9), PCR of repetitive intergenic sequences (3), and 16S ribosomal DNA length heterogeneity PCR with terminal restriction fragment length polymorphism (2) have been described as promising for discriminating between fecal-origin bacteria from humans and animals. Multiple antibiotic resistance phenotype has been used to distinguish between human and nonhuman sources of Escherichia coli (7, 10, 11, 19) and streptococci (6, 18), but genetic instability or changes in antibiotic use can alter the resistance profiles obtained.
Ribotyping has been compared to multiple antibiotic resistance profiles, and both approaches were reportedly complementary in discriminating between human and nonhuman (collective) sources of fecal pollution (11). Ribotyping has since become a popular approach (personal communications) to the problem of differentiating between fecal E. coli pollution from humans and, in particular, that from animals and birds. We describe here the use of ribotyping for the identification of E. coli cultured from feces of humans, cattle, swine, horses, chickens, turkeys, dogs, and migratory geese.
MATERIALS AND METHODS
Fecal E. coli.
Table 1 presents the host sources of feces, the numbers of individuals sampled, and the geographic regions from which samples were collected. E. coli isolates of human origin were isolated directly from anal swabs. Feces of beef cattle, dairy cattle, swine, and horses served as source material for isolates from these species. Composite collections were also made from the excreta of chickens, turkeys, and migratory geese. Samples were incubated overnight in lactose broth at 37°C (Difco Laboratories, Sparks, Md.) and streaked on mEndo (Les) agar (Difco Laboratories). Colonies presenting a gold metallic sheen were transferred to mFC (Difco Laboratories) and cultured overnight at 44.5°C to select for fecal E. coli. Colonies were further characterized as E. coli by subculture on MacConkey-MUG (Difco Laboratories). Pink colonies which fluoresced under UV light were transferred to brain heart infusion (BHI; Difco Laboratories) plates. Individual E. coli isolates were finally confirmed biochemically by growth on Kligler Iron Agar, Simmons Citrate Agar, Methyl Red/VP, and Indol with 1% tryptose (all from Difco Laboratories). A total of 287 isolates were examined, including 40 human, 39 cattle, 44 pig, 37 horse, 29 dog, 23 chicken, 26 turkey, and 49 goose isolates.
TABLE 1.
E. coli isolates used in this study
| Source | No. of isolates | No. of individuals sampleda | Location in Missouri |
|---|---|---|---|
| Human (direct) | 40 | 15 | Central |
| Cattle | 39 | 24 | Central |
| Pig | 44 | 30 | Central |
| Horse | 37 | 10 | Central and southern |
| Dog | 29 | 15 | Central and western |
| Chicken | 23 | C | Central and southern |
| Turkey | 26 | C | Central |
| Goose | 49 | 24 | Central |
C, composite.
DNA extraction.
Fecal E. coli isolates were grown in BHI broth (Difco Laboratories) and DNA extracted by using The Easy DNA Kit (Invitrogen, Carlsbad, Calif.) according to manufacturer's instructions. DNA concentration was measured spectrophotometrically, and 2.5 μg was digested with HindIII (New England Biolabs, Beverly, Mass.) according to the manufacturer's instructions. Fragments were separated in 1% agarose gels in TBE buffer (0.09 M Tris-borate, 0.002 M EDTA) using 30 mV for 16 h.
Southern blot analysis.
Gels were depurinated in 0.25 N HCl for 15 min, rinsed twice with deionized water, denatured in 0.4 M NaOH–0.6 M NaCl, and neutralized in 0.5 M Tris-HCl (pH 7.5)–1.5 M NaCl for 30 min (13). DNA was transferred (16) onto nylon membranes (Boehringer Mannheim Corp., Indianapolis, Ind.) using a vacuum blotter. Membranes were baked at 80°C for 2 h.
Probe preparation.
The probe was a BamHI (New England Biolabs) fragment from plasmid pKK 3535 containing E. coli 16S and 23S rRNA genes (12). Digested DNA was separated in 0.8% agarose gel in TAE buffer for 2 h. at 80 mV. Insert DNA was purified using the Geneclean system (Bio 101, Carlsbad, Calif.) as specified by the manufacturer. The probe was labeled with digoxigenin-dUTP (Boehringer Mannheim Corp.) according to the manufacturer's instructions.
Hybridization.
Membranes were prehybridized at 65°C for 90 min and hybridized with the probe used as 25 ng of DNA per filter (10 by 15 cm) at 65°C for 16 h in a hybridization oven (Hybaid Instruments, Holbrook, N.Y.). Membranes were washed twice for 5 min using 2× SSC (0.3 M NaCl–30 mM sodium citrate)–0.1% sodium dodecyl sulfate (SDS). Two final 15-min washes were performed with 0.5× SSC–0.1% SDS at 65°C (13). Membranes were developed colorimetrically using nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl-phosphate (BCIP; Boehringer Mannheim Corp.) according to the manufacturer's instructions.
Statistical analysis.
The method used for the discrimination of riboprints of E. coli isolated from known-host sources was based on a previously reported procedure (11). Riboprints, developed on membranes after hybridization with the riboprobe, were captured for computer analysis by placement on a flatbed scanner. Each pattern of bands was converted to a line diagram, and fragment sizes (in base pairs) were assigned to each band using DNA Proscan software (Nashville, Tenn.). Riboprint patterns were converted to a binary code for discriminant analysis (8) performed in SAS (SAS/STAT [1989] version 6). All or selected portions of the riboprint patterns were sequentially divided into 8 to 34 equal segments (windows) extending between 0 and 35 kb. The algorithm compared pattern profiles by the presence or absence and number of bands in each window. The number and width of these windows were adjusted until the accuracy of classification of host patterns reached its highest attainable level. Discriminant analysis using SAS software (PROC DISCRIM) was performed as a comparison of riboprints of all eight known-host sources, human versus pooled nonhuman sources, and groups of three to five selected host classes. Cross-validation iterations were performed with each riboprint in the database, and the percent correct classification was determined. Spatial plot display, based on the use of 24 windows, was projected into two principal variables using SAS software (PROC CAN DISC). Plots represented a visual interpretation of pattern analysis. Separation of patterns with respect to the host source is an indication of accuracy of identification.
RESULTS
A total of 287 known-host riboprint patterns were generated for E. coli strains isolated from humans, cattle, pigs, horses, chickens, turkeys, migratory geese, and dogs. These riboprints were composed of 6 to 12 bands ranging in size from approximately 0.5 to 25.0 kb. Lanes containing the patterns were divided into segments (windows) of equal size for computer analysis. Best results were obtained by dividing the 0.5- to 22.0-kb portion of each pattern into 24 equal windows. Rates of correct classification (RCC) for various combinations of host classes are shown in Tables 2 to 6. A comparison of human and nonhuman riboprints resulted in 95.0 and 99.19% correct classifications, respectively (Table 2). The average rate of correct classification (ARCC) for riboprints compared to all eight host classes simultaneously (Table 3) was 73.56%. When comparison was made between a more limited number of classes the ARCC improved. Examples of results obtained from discriminant analyses with three classes included in each exercise are represented by Tables 4 to 6.
TABLE 2.
RCC of riboprints of fecal E. coli from human and nonhuman sources
| Sample source | No. of isolates | No. correctly identified | RCC (%)a |
|---|---|---|---|
| Human | 40 | 38 | 95.00 |
| Nonhuman (pooled) | 247 | 245 | 99.19 |
| Total | 287 | 283 |
The ARRC was 97.10%.
TABLE 6.
RCC of riboprints of fecal E. coli from goose, turkey, and chicken sources
| Sample source | No. of isolates | No. correctly identified | RCC (%)a |
|---|---|---|---|
| Goose | 49 | 47 | 95.92 |
| Turkey | 26 | 25 | 96.15 |
| Chicken | 23 | 22 | 95.65 |
The ARCC was 95.91%.
TABLE 3.
RCC of riboprints of fecal E. coli from eight known-host sources
| Host class | No. of Isolates | No. correctly identified | RCC (%)a | Primary source(s) of misclassification |
|---|---|---|---|---|
| Human (direct) | 40 | 37 | 92.50 | Turkey |
| Cattle | 39 | 29 | 74.36 | Pig |
| Pig | 44 | 29 | 65.91 | Turkey, dog |
| Horse | 37 | 18 | 48.65 | Turkey, pig |
| Dog | 29 | 16 | 55.17 | Cattle, pig |
| Chicken | 23 | 22 | 95.65 | Pig |
| Turkey | 26 | 21 | 80.77 | Pig |
| Goose | 49 | 37 | 75.51 | Dog |
| Total | 287 | 177 |
The ARCC was 73.56%.
TABLE 4.
RCC of riboprints of fecal E. coli from human, dog, and horse sources
| Sample source | No. of isolates | No. correctly identified | RCC (%)a |
|---|---|---|---|
| Human | 40 | 39 | 97.50 |
| Dog | 29 | 27 | 93.10 |
| Horse | 37 | 34 | 91.89 |
| Total | 84 | 79 |
The ARCC was 94.16%.
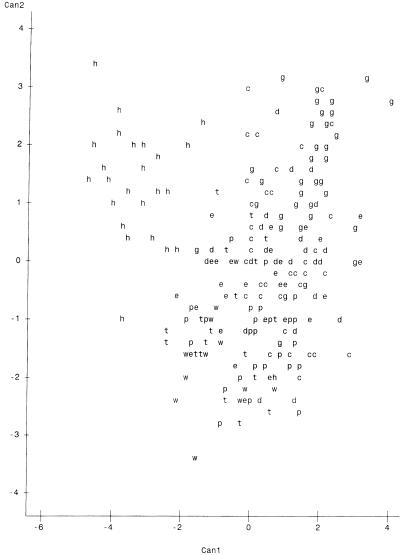
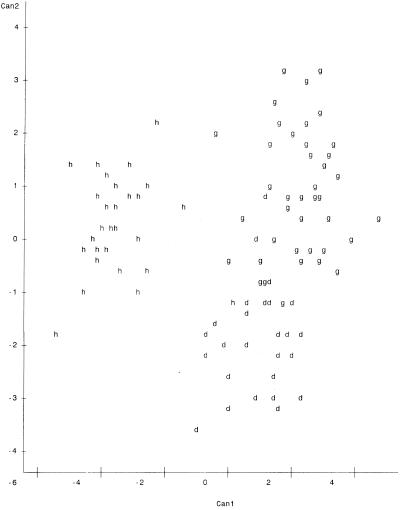
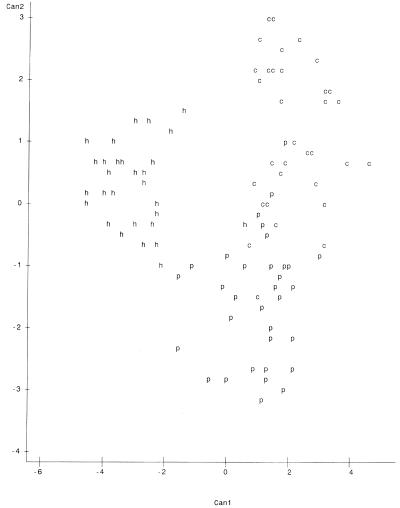
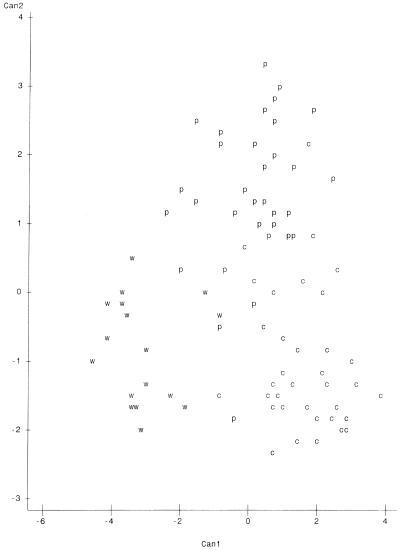
Plots of canonical variables 1 and 2 (Can1 and Can2) on the x and y axes, respectively, are presented in Fig. 1 to 4. The variables represent major characteristics used as criteria for the comparison of riboprints. The resulting spatial display of riboprints from all eight host classes displayed simultaneously appears in Fig. 1. In this instance there is a complex and complicated array without distinct clustering of host-associated patterns. However, comparison of three host classes at a time resulted in distinct separation of patterns from each class (Fig. 2 to 4). These visual displays of cluster association reflect the level of accuracy of classification of the riboprints of respective host species.
FIG. 1.
Two-dimensional spatial plot of riboprints of fecal E. coli from all eight host sources studied. Hosts are identified by the following letters: h, human; p, pig; c, cattle; e, horse; d, dog; w, chicken; t, turkey; and g, migratory geese. Can1 is on the x axis, and Can2 is on y axis. There were 84 hidden observation points which were invisible due to overlapping.
FIG. 4.
Spatial plot of riboprints of fecal E. coli from humans, dogs, and geese. Positions of patterns are represented as follows: h, humans; d, dogs; and g, geese. Can1 is on the x axis, and Can2 is on the y axis. There were 24 hidden observations.
FIG. 2.
Spatial plot of riboprints of fecal E. coli from cattle, pigs, and humans. Positions of patterns are represented as follows: c, cattle; p, pigs; and h, humans. Can1 is on the x axis, and Can2 is on the y axis. There were 28 hidden observations.
DISCUSSION
We have tested the use of riboprinting for identification of fecal E. coli from specific sources. This method proved to be quite accurate for discriminating between riboprint patterns of human and nonhuman origin, with an ARCC of 97.10%. The RCC of riboprints, from each of the eight known-host sources studied, ranged between 48.65 and 95.65% when compared simultaneously (Table 3). However, it was shown that the accuracy of classification can be greatly increased by limiting the number of classes compared (Tables 2 to 6). Similarly, the spatial analysis of all patterns simultaneously did not distinctly cluster the host sources. When each clustering iteration was limited to three host sources, such as cattle, pigs, and humans (Fig. 2), chickens, pigs, and cattle (Fig. 3), or geese, humans, and dogs (Fig. 4), it became possible to distinguish riboprints representing E. coli from particular hosts.
FIG. 3.
Spatial plot of riboprints of fecal E. coli from cattle, pigs, and chickens. Positions of patterns are represented as follows: c, cattle; p, pigs; and w, chickens. Can1 is on the x axis, and Can2 is on the y axis. There were 21 hidden observations.
Application of this technology to testing water quality is our ultimate goal. This approach would require riboprinting of unknown-source E. coli in water samples and comparison of the resultant patterns with known-host patterns in the database. For example, Fig. 3 provides a distinct display of fecal coliform riboprints from three of the most common animal species involved in concentrated feeding operations—cattle, chickens, and pigs—indicating that unknowns compared to particular known-host patterns may be accurately characterized. In analyses of municipal storm water the suspected pollution sources may include humans, dogs, or migratory geese. The associated patterns in Fig. 4 form fairly distinct clusters, indicating good probability for correct classification. In field situations it is also recommended that additional samples of host sources in the local landscape be included in the database for discriminant analysis of unknown E. coli isolated from waters under study. This measure could prove to be important if the geographic location affects host intestinal flora.
Undoubtedly, there are instances where fecal E. coli pollutants in water may come from a large number of contributing host sources, consequently increasing the rates of misclassification by ribotyping. Commonly, there will also be instances of application of ribotyping for microbial source tracking where there will be only a few obvious potential sources of pollution. In these latter situations we would propose that the fingerprinting scheme presented here will be more accurate in the rate of correct classification of riboprints.
As with other DNA fingerprinting methods, ribotyping produces various patterns that represent the genomic similarity or dissimilarity between isolates. Certain fecal E. coli riboprints appear to be associated with (if not unique) to certain host classes. We can only speculate as to why this phenomenon may occur and suppose that factors affecting the microenvironment of particular host intestines, including temperature, pH, and diet may be associated with strain selection or enrichment. Seasonal, geographic, or genetic variation in natural fecal E. coli populations may also occur, but these issues were not examined in the present study. Based on a study of host-associated E. coli strains by multiple antibiotic resistance profiles, there is reportedly little or no cross-colonization (9). It has also been reported that different host classes harbor E. coli which have very similar or identical riboprints (11), and consequently misclassification may occur in these instances.
It appears that typical enteric populations of E. coli within each host species studied are significantly different for ribotyping to serve as a valuable means for identification of sources of fecal pollution. Reported results are based on a modest database of 287 patterns. The accuracy of results is expected to increase as the database of known-host samples is expanded. For practical application of this technology to water quality, it will also be valuable to include additional host sources of E. coli in our database.
TABLE 5.
RCC of riboprints of fecal E. coli from cow, pig, and chicken sources
| Sample source | No. of isolates | No. correctly identified | RCC (%)a |
|---|---|---|---|
| Cow | 39 | 33 | 84.62 |
| Pig | 44 | 41 | 93.18 |
| Chicken | 23 | 21 | 91.30 |
The ARCC was 89.70%.
ACKNOWLEDGMENTS
This work was supported by the University of Missouri Outreach and Extension, Columbia, Mo., and the U.S. Geological Survey, Rolla, Mo.
We thank Salina Parveen (University of Florida) for advice and methods information and Shaun Tyler (Health Canada) for plasmid pKK3535 used in preparing the riboprobe. We are also grateful to John Schumacher and Don Wilkison (U.S. Geological Survey), Mike Monda (U.S. Army Corps of Engineers), and John Knudsen and Michael Heaton (Missouri Department of Natural Resources) for assistance in sample collection.
REFERENCES
- 1.American Public Health Association. Standard methods for the examination of water and wastewater. 19th ed. Washington, D.C.: American Public Health Association; 1995. [Google Scholar]
- 2.Bernhard A E, Field K G. A PCR assay to discriminate human and animal feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl Environ Microbiol. 2000;66:4571–4574. doi: 10.1128/aem.66.10.4571-4574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dombek P E, Johnson L K, Zimmerley S J, Sadowsky M J. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl Environ Microbiol. 2000;66:2572–2577. doi: 10.1128/aem.66.6.2572-2577.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fleisher J M, Kay D, Salmen R L, Jones F, Wyer M D, Godfree A F. Marine waters, contaminated with domestic sewage: nonenteric illnesses associated with bather exposure in the United Kingdom. Am J Public Health. 1996;86:1228–1234. doi: 10.2105/ajph.86.9.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geldreich E E, Kenner B A. Concepts of fecal streptococci in stream pollution. J Water Pollut Control Fed. 1969;41:R336–R352. [PubMed] [Google Scholar]
- 6.Hagedorn C, Robinson S L, Filtz J R, Grubbs S M, Angier T A, Reneau R B., Jr Determining sources of fecal pollution in rural Virginia watershed with antibiotic resistance patterns in fecal streptococci. Appl Environ Microbiol. 1999;65:5522–5531. doi: 10.1128/aem.65.12.5522-5531.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harwood V J, Whitlock J, Withington V. Classification of antibiotic resistance patterns of indicator bacteria by discriminant analysis: use in predicting the source of fecal contamination in subtropical waters. Appl Environ Microbiol. 2000;66:3698–3704. doi: 10.1128/aem.66.9.3698-3704.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson R A, Wichern D W. Applied multivariate statistical analysis. 3rd ed. Englewood Cliffs, N.J: Prentice-Hall, Inc.; 1992. [Google Scholar]
- 9.Kariuki S, Gilks C, Kimari J, Obanda A, Muyodi J, Waiyaki P, Hart C A. Genotype analysis of Escherichia coli strains isolated from children and chickens living in close contact. Appl Environ Microbiol. 1999;65:472–476. doi: 10.1128/aem.65.2.472-476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parveen S, Murphree R L, Edmiston L, Kaspar C W, Portier K M, Tamplin M L. Association of multiple-antibiotic resistance profiles with point and nonpoint sources of Escherchia coli in Apalachicola Bay. Appl Environ Microbiol. 1997;63:2607–2612. doi: 10.1128/aem.63.7.2607-2612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parveen S, Portier K M, Robinson K, Edmiston L, Tamplin M L. Discriminant analysis of ribotype profiles of Escherichia coli for differentiating human and nonhuman sources of fecal pollution. Appl Environ Microbiol. 1999;65:3142–3147. doi: 10.1128/aem.65.7.3142-3147.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Regnault B, Grimont F, Grimont P A D. Universal ribotyping method using a chemically labelled oligonucleotide probe mixture. Res Microbiol. 1997;148:649–659. doi: 10.1016/S0923-2508(99)80064-3. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 14.SAS Institute, Inc. SAS/STAT user's guide, version 6. 4th ed. Vol. 1. Cary, N.C: SAS Institute, Inc.; 1989. [Google Scholar]
- 15.Sauer T J, Daniel T C, Nichols D J, West C P, Moore P A, Jr, Wheeler G L. Runoff water quality from poultry litter-treated pasture and forest sites. J Environ Qual. 2000;29:515–521. [Google Scholar]
- 16.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 17.U.S. Environmental Protection Agency. TMDL tracking system data, version 1.0. Total maximum daily load program. Washington, D.C.: U.S. Environmental Protection Agency Office of Water; 1998. 1998.http: //www.epa.gov/owow/tmdl/trcksys.html [Online.] http: //www.epa.gov/owow/tmdl/trcksys.html. [Google Scholar]
- 18.Wiggins B A. Discriminant analysis of antibiotic resistance patterns in fecal streptococci, a method to differentiate human and animal sources of fecal pollution in natural waters. Appl Environ Microbiol. 1996;62:3997–4002. doi: 10.1128/aem.62.11.3997-4002.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiggins B A, Andrews R W, Conway R A, Corr C L, Dobratz E J, Dougherty D P, Eppard F R, Knupp S R, Limjoco M C, Mettenburg J M, Rinehardt J M, Sonsino J, Torrijos R L, Zimmerman M E. Use of antibiotic resistance analysis to identify nonpoint sources of fecal pollution. Appl Environ Microbiol. 1999;65:3483–3486. doi: 10.1128/aem.65.8.3483-3486.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zierdt C H, Robertson E A, Williams R L, MacLowry J D. Computer analysis of Staphylococcus aureus phage typing data from 1957 to 1975, citing epidemiological trends and natural evolution within phage typing system. Appl Environ Microbiol. 1980;39:623–629. doi: 10.1128/aem.39.3.623-629.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]