Abstract
The tear film protects the terrestrial animal’s ocular surface and the lacrimal gland provides important aqueous secretions necessary for its maintenance. Despite the importance of the lacrimal gland in ocular health, molecular aspects of its development remain poorly understood. We have identified a noncoding RNA (miR-205) as an important gene for lacrimal gland development. Mice lacking miR-205 fail to properly develop lacrimal glands, establishing this noncoding RNA as a key regulator of lacrimal gland development. Specifically, more than half of knockout lacrimal glands never initiated, suggesting a critical role of miR-205 at the earliest stages of lacrimal gland development. RNA-seq analysis uncovered several up-regulated miR-205 targets that may interfere with signaling to impair lacrimal gland initiation. Supporting this data, combinatorial epistatic deletion of Fgf10, the driver of lacrimal gland initiation, and miR-205 in mice exacerbates the lacrimal gland phenotype. We develop a molecular rheostat model where miR-205 modulates signaling pathways related to Fgf10 in order to regulate glandular development. These data show that a single microRNA is a key regulator for early lacrimal gland development in mice and highlights the important role of microRNAs during organogenesis.
Keywords: lacrimal gland, microRNAs, miR-205, Fgf10
Summary statement:
This study establishes a role of miR-205 during embryonic organogenesis, and specifically lacrimal gland development.
Introduction
The lacrimal gland provides the aqueous layer for the tear film to protect the eye from dryness. When lacrimal glands are dysfunctional, dry eye syndrome can develop, a condition that plagues millions of humans (Gayton, 2009). Despite the prevalence of the condition, many of the underlying mechanisms and pathways controlling lacrimal gland development remain unknown.
The lacrimal gland develops by branching morphogenesis to form an interconnected network of secretory acinar units and ducts. In mice, lacrimal gland development begins at embryonic day (E) 13.5 (6 weeks in humans) when the primary bud invaginates from the surface ectoderm and grows toward the periorbital mesenchyme (Govindarajan et al., 2000; Makarenkova et al., 2000). After the bud invades the mesenchyme, it branches extensively to produce the intra- and exorbital lacrimal gland (Govindarajan et al., 2000; Makarenkova et al., 2000). As a mature organ, the lacrimal gland produces tears to lubricate and protect the ocular surface.
Several protein-coding genes are implicated in the early developmental program of the lacrimal gland including members of the fibroblast growth factor (FGF) signaling pathway. FGF10 is secreted from the periocular mesenchyme and binds to its receptor FGFR2IIIb on the surface ectoderm to induce lacrimal gland budding (Govindarajan et al., 2000; Makarenkova et al., 2000). The Pax6 transcription factor behaves as a competence factor that helps establish the transcriptional landscape for these Fgf10 responsive epithelial cells (Makarenkova et al., 2000). Mice that are heterozygous for Fgf10 or that are deficient for the heparan sulfate modifying enzyme Ndst have absent or defective lacrimal glands, indicating the importance of this pathway to lacrimal gland growth (Pan et al., 2008; Qu et al., 2011; Qu et al., 2012). While Fgf10 signaling is clearly required for early lacrimal gland development, the genes that converge to mediate this developmental program remain unclear.
Small noncoding RNAs, known as microRNAs, regulate gene expression post-transcriptionally. MicroRNAs play critical roles in the development and differentiation of many organs, including the heart (Zhao et al., 2007), lung (Ventura et al., 2008), brain (Dugas et al., 2010), and skin (Yi et al., 2008). Recently, several studies have identified microRNAs that regulate branching morphogenesis. Results from these studies provide promising evidence for an important role of microRNAs in the development of branching organs (Carraro et al., 2009; Hayashi et al., 2011; Rebustini et al., 2012; Ucar et al., 2010). However, branching morphogenesis encompasses several developmental decisions, including the induction and elongation of the primary bud, and the budding and clefting of the primary bud to form a branched organ. The majority of microRNA studies in branching organs have been performed ex vivo, providing only a snapshot of how microRNAs control branching morphogenesis. In vivo experiments are necessary to gain a comprehensive understanding of how microRNAs contribute to branching morphogenesis. Furthermore, if and how microRNAs converge with morphogen gradients to govern this process remains an open question.
We recently reported the expression of a microRNA, miR-205, in many epithelial organs, including the murine E18.5 lacrimal gland (Farmer et al., 2013). Conserved between mice and humans, miR-205 is highly enriched in the stem cells of the mammary gland and regulates the proliferation of neonatal skin cells (Ibarra et al. 2007; Greene et al. 2010; Wang et al. 2013). Here, we uncover a role of miR-205 during embryonic lacrimal gland development. Evidence suggests miR-205 represses several targets that may interfere with important pathways, like Fgf10 signaling, to ensure proper lacrimal gland development. This study supports a key role of microRNAs in ensuring proper development by promoting robustness in response to morphogen signals.
Results
Ocular defects occur in miR-205−/− mice
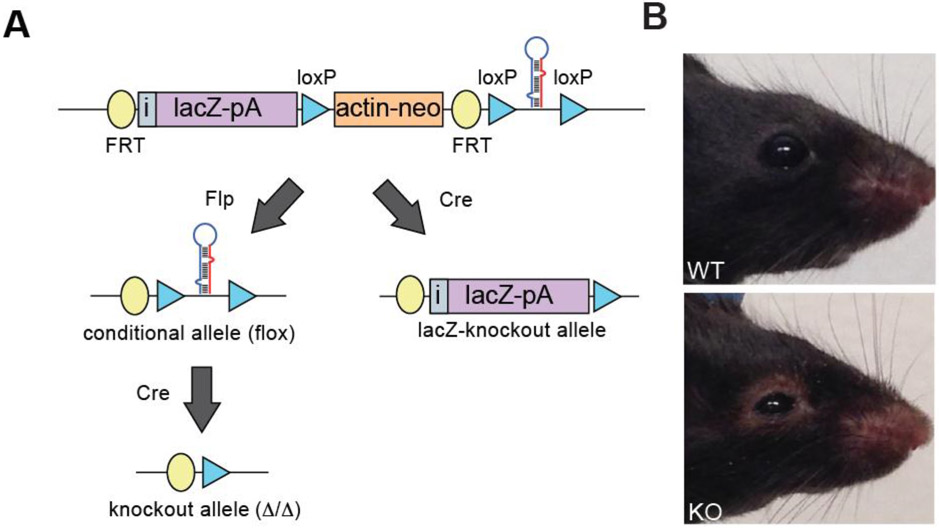
Previously, our lab generated a large cohort of microRNA knockout mice carrying transcriptional reporters for microRNA expression (Park et al., 2012), helping to circumvent the well characterized challenges of microRNA in situ hybridization (Fig. 1A) (Søe et al., 2011; Thomsen et al., 2005). In addition, we recently reported a partially penetrant lethal phenotype when the epithelial-restricted microRNA, miR-205, was deleted in mice (Farmer et al., 2013). While detected in many epithelial organs, the prominent expression of miR-205 in tissues that support the tear film suggested a possible role for miR-205 in ocular surface health. The tear film protects the eye from dryness and is composed of three layers: the mucous layer, the aqueous layer, and the lipid layer. The aqueous and lipid layers are derived from the lacrimal glands and meibomian glands, respectively. We identified miR-205 expression in both lacrimal and meibomian glands as well as within the corneal epithelium. To determine the role of miR-205 in ocular biology, we deleted miR-205 globally in mice using the ACTB-Cre mouse line (Lewandoski et al., 1997) and evaluated surviving miR-205 knockout (205−/−) mice for ocular defects (Fig. 1A). Both wildtype and heterozygous mice served as controls as neither exhibited notable phenotypes. After eyelid opening, miR-205−/− mice displayed a clear ocular phenotype, including a thickening of the eyelid that progressed in severity as the mice aged (Fig. 1B). Histological analysis of the corneal epithelium revealed no noticeable morphological differences between control and miR-205−/− mice (Fig. S1A). To determine if lacrimal gland or meibomian gland dysfunction could account for the observed phenotype, the meibomian glands and the lacrimal glands were evaluated. Meibomian glands were present in miR-205−/− mice and occasionally were enlarged compared to age matched controls (Fig. S1B). Knockout meibomian glands also retained lipid content (Fig. S1C). Thus, miR-205 does not appear to play major roles in the corneal epithelium or the meibomian gland, although further analyses may be warranted to investigate modest defects in these tissues.
Fig. 1. Deletion of miR-205 results in ocular defects.
(A) Construct strategy for mouse generation. (B) Images of representative control and miR-205−/− mice. Mice were imaged at four months of age.
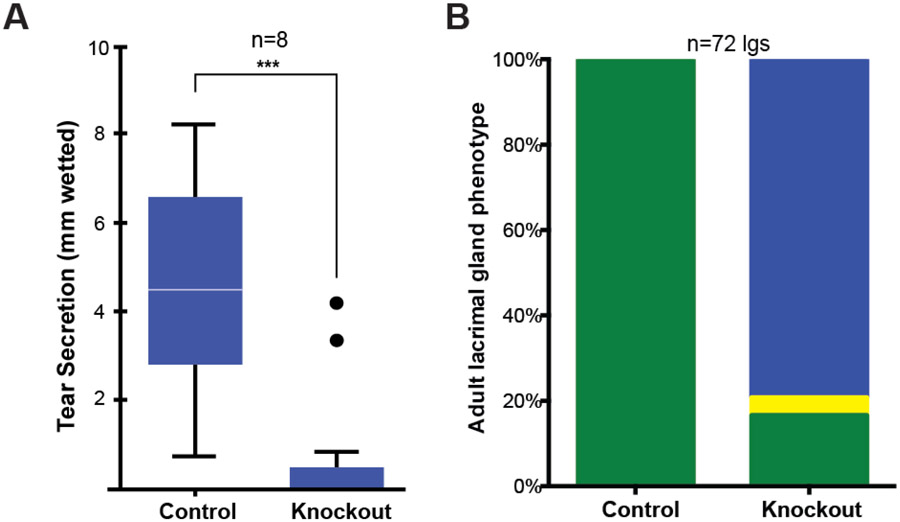
To assess lacrimal gland functionality, tear secretion was assessed in adult control and miR-205−/− mice. On average, after stimulation with pilocarpine, the phenol red thread was wetted 1 millimeter in miR-205−/− mice, a value significantly less than the 6 millimeters wetting observed in control mice (Fig. 2A). Some miR-205−/− mice exhibited results comparable to controls in the tear secretion assay (Fig. 2A, black points in graph), suggesting a partial penetrance in the ocular phenotype. Generally, the failed tear secretion in knockout animals clearly indicated lacrimal gland dysfunction. To further characterize any morphological changes, lacrimal glands were collected from adult control and miR-205−/− mice. Surprisingly, lacrimal glands were largely undetected in miR-205−/− mice (p-value<0.001), and a minor number of glands were considerably smaller than control glands (Fig. 2B). These data suggested a likely mechanism for the observed ocular defects.
Fig. 2. Loss of miR-205 impairs tear secretion.
(A) Whisker plot of tear collection quantification after stimulation (*** p-value <0.0001, t-test, SEM error bars). Black points indicate outliers. (B) Quantification of gland phenotypes in adult mice (p-value, <0.001, Fisher’s exact test). Green: normal, yellow: runted, and blue: absent glands.
miR-205 controls lacrimal gland development
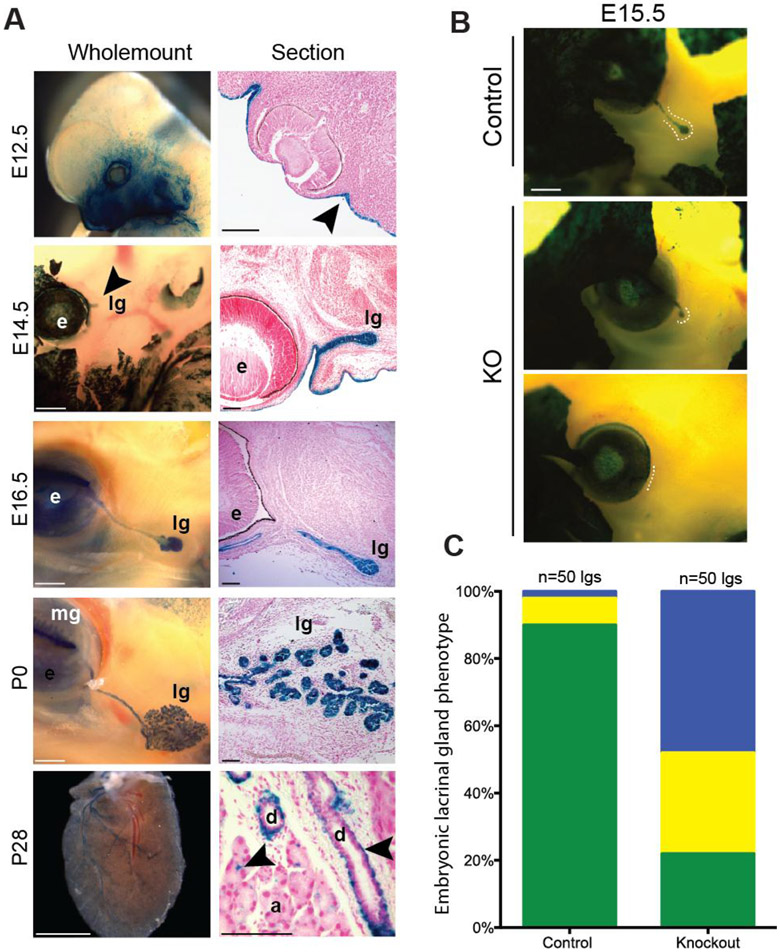
The absence of lacrimal glands in adult miR-205−/− mice led us to focus on earlier stages of lacrimal gland development. Embryonic lacrimal gland development can be divided into three phases: lacrimal gland initiation (E12.5-E13.5), early bud elongation (E14.5-E15.5), and successive rounds of epithelial branching (E16.5-P0). Previously, we reported the miR-205 LacZ reporter (Fig. 1A) as an exquisite readout for mature microRNA levels using both in situ hybridization and qPCR as validations (Farmer et al., 2013). Using this reporter, we identified miR-205 expression in the embryonic (E) day 18.5 lacrimal gland. To expand our understanding of the timing and localization of mir-205 throughout lacrimal gland development, temporal analyses of miR-205 transcription was performed using these miR-205 LacZ reporter mice. X-gal staining was readily detected before lacrimal gland initiation at E12.5, specifically in the surface ectoderm neighboring the developing eye (Fig. 3A). After initiation, miR-205 expression was present within the epithelium of the lacrimal gland throughout all stages of embryonic development (Fig. 3A). Unlike in the embryonic lacrimal gland, widespread expression of miR-205 in the postnatal lacrimal gland diminished and became restricted to cell populations around the ducts and scattered between acinar cells (Fig. 3A). These data opened up the possibility that miR-205 could play an early developmental role in the lacrimal gland.
Fig. 3. miR-205 controls lacrimal gland initiation.
(A) Expression analysis of miR-205 in the lacrimal gland. Lacrimal gland (lg), meibomian gland (mg), eye (e). Surface ectoderm is indicated by arrowhead. Scattered lacZ staining around ducts (d) and acini cells (a) is indicated by arrowhead. Scale bars are 100 μM. (B) Representative images of E15.5 embryos with lacZ reporter. Dashed lines indicate the location of the lacrimal gland. Scale bars are 500 μM. (C) Quantification of phenotypes in control and miR-205−/− mice (p-value<0.001, Fisher’s exact test). Green: normal glands, yellow: runted glands, and blue: absent glands.
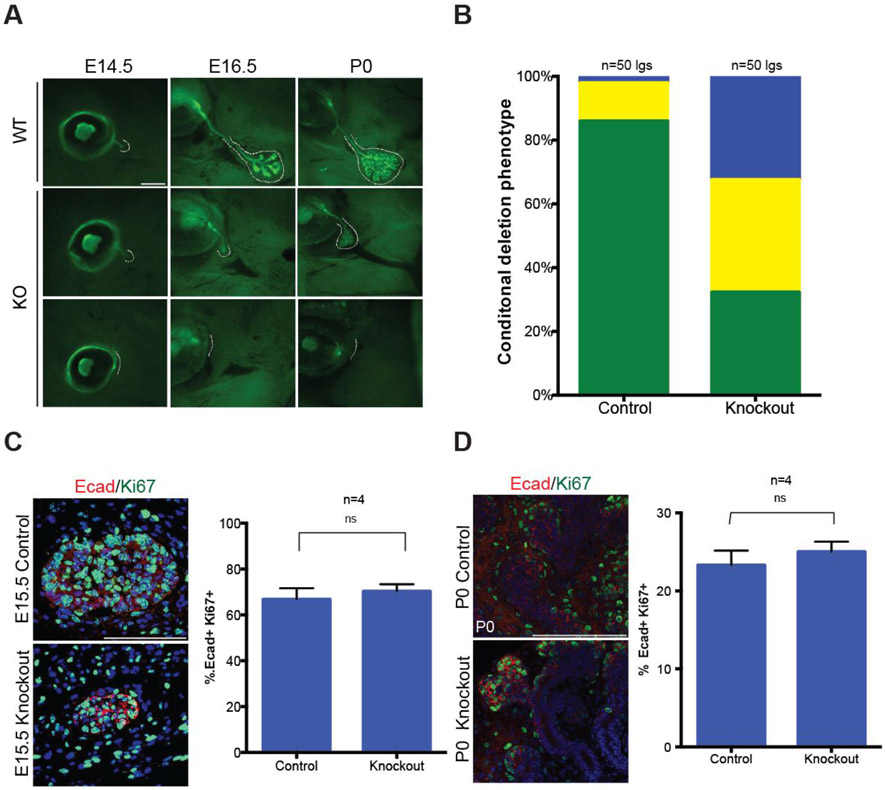
To decipher how miR-205 controls the lacrimal gland, lacrimal glands from E15.5 mice carrying the miR-205 lacZ reporter allele or a single Krt5GFP reporter allele were assessed (Bruen et al., 2004). For these experiments, only heterozygous mice were used as controls. Strikingly, in greater than 50% of knockout animals (p-value<0.001), no lacrimal glands were identified, indicative of an initiation failure (Fig. 3B and C). In addition, approximately 25% of miR-205−/− lacrimal glands initiated but were smaller and poorly extended compared to littermate controls. We attribute the increased number of small yet runted lacrimal glands at E15.5 compared to adult stages to the use of reporter lines that enable easy visualization of these tiny glands. To conclusively evaluate whether the lacrimal gland defects observed in miR-205−/− embryos (Fig. 3B) originated from the loss of miR-205 in the lacrimal gland epithelia, miR-205 floxed animals were crossed to the epithelial restricted Le-Cre line (miR-205flox/flox:cre) (Ashery-Padan et al., 2000). All evaluated animals carried a single copy of the Le-Cre allele to avoid deleterious effects caused by homozygosity and to ensure phenotypes were specific to miR-205 deletion (Dorà et al., 2014). Efficient miR-205 deletion was confirmed by qPCR of P0 lacrimal glands (Fig. S2). While control animals (205flox/+:cre mice) developed lacrimal glands as expected, miR-205flox/flox:cre mice recapitulated the previously observed lacrimal gland phenotype with 60% of lacrimal glands absent or runted at E15.5 (p-value<0.001) (Fig. 4A and 4B). Combined, these data demonstrate a specific requirement for miR-205 within the lacrimal gland epithelia and support a model for a role of miR-205 in lacrimal gland initiation and early development.
Fig. 4. miR-205 is required within the epithelium of the lacrimal gland.
(A) Representative images of phenotypes observed in miR-205flox:flox;cre embryos and timed controls (p-value<0.001, Fisher’s exact test). Dashed lines highlight the lacrimal gland position. Scale bars are 500 μM (B) Quantification of phenotypes in conditional miR-205 knockout mice. Green: normal glands, yellow: runted glands, and blue: absent glands. (C) Confocal imaging of E15.5 lacrimal glands and quantification of Ki67+ Ecad+ positive cells. Scale bars are 100 μM (ns=not significant, t-test, SEM error bars). (D) Confocal imaging of P0 lacrimal glands and quantification of Ki67+ Ecad+ positive cells. Scale bars are 100 μM (ns=not significant, t-test, SEM error bars).
Initiated knockout glands retain proliferative capacity
While a large subset of lacrimal glands never initiated, another subset initiated but remained smaller than controls (Fig. 3B and 4A). At later stages of embryonic lacrimal gland development, these smaller glands continued to branch and formed highly elaborate organs that remained proximal to the eye by postnatal day (P) 0 (Fig. 4A). To determine the proliferative capacity of these small knockout glands, E15.5 and P0 lacrimal glands were co-labeled with Ki67, to mark cells in the cell cycle, and E-cadherin, to specifically visualize the epithelia where miR-205 is normally expressed. Immunofluorescence indicated that miR-205−/− lacrimal glands, as assessed by the combinatorial staining of Ki67 and E-cadherin, retained highly proliferative end buds at both time points and quantification of the fraction of double positive cells throughout the epithelia confirmed no significant impairment in the proliferative rate of miR-205−/− runted glands (Fig. 4C and 4D). Similar results were obtained using EdU or pH3 coupled with E-cadherin to label proliferating epithelial cells (data not shown). In addition, no differences in proliferation were detected in the neighboring non-epithelial cells (data not shown). Furthermore, there were no notable differences in cell death, pERK, or pAKT staining between wildtype and knockout glands (data not shown). Taken together, these data implicate miR-205 as an important regulator of early lacrimal gland development and suggests that successfully initiated glands may continue to proliferate normally despite their smaller size.
Multiple targets are up-regulated in miR-205−/− mice
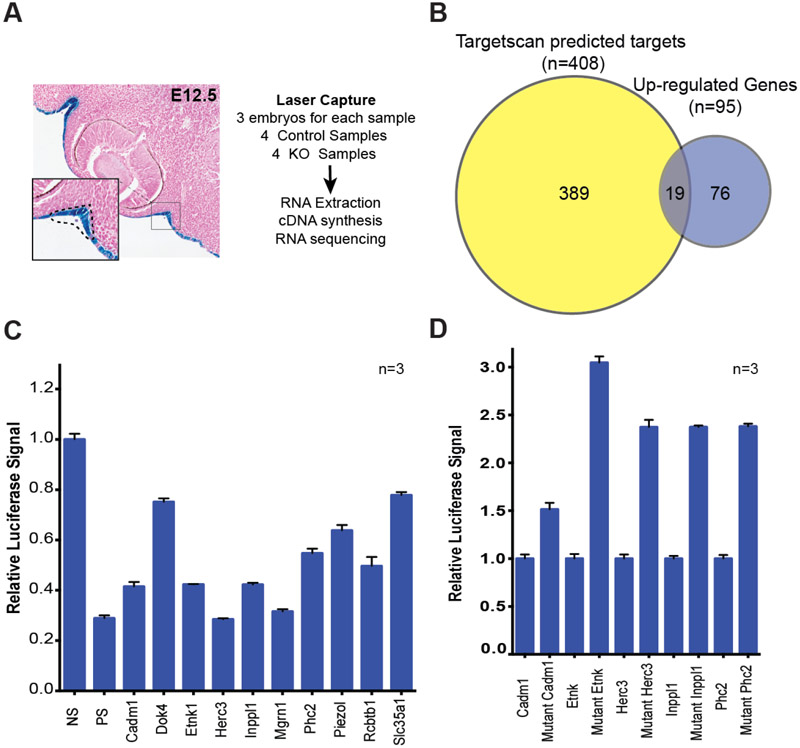
Lacrimal gland initiation is known to be controlled by Fgf10 signaling (Ashery-Padan et al., 2000; Makarenkova et al., 2000; Pan et al., 2008; Pan et al., 2010; Qu et al., 2012). To uncover whether miR-205 might contribute to these early signals, we sought to identify differentially regulated miR-205 targets prior to initiation. To accomplish this, surface ectoderm, from which the lacrimal gland is derived, was isolated from miR-205−/− E12.5 embryos and their wildtype littermates using laser capture, and RNA-seq analysis was independently performed in quadruplicate on collected samples (Fig. 5A). Importantly, this time point represents a time when putative miR-205 targets can interfere with FGF signaling, and as a result, lacrimal bud initiation. No significant differences were observed in any genes previously identified to control lacrimal gland development, including Pax6 and Fgfr2 (Table S1). Of the 95 genes significantly up-regulated in miR-205−/− samples, 19 were putative miR-205 targets based on Targetscan (Agarwal et al., 2015) (chi-square, p-value<0.001, Fig. 5B, and Table S2). A subset of these up-regulated miR-205 targets were validated by conventional 3’-UTR experiments (Fig. 5C). Indeed, a majority of the tested UTRs demonstrated sensitivity to miR-205 levels, further implicating these genes as direct miR-205 targets. Furthermore, mutating the miR-205 seed sequence in the 3’ UTR of target genes augmented their response to miR-205 expression (Fig. 5D). Thus, miR-205 appears to modestly regulate several targets simultaneously to control lacrimal gland initiation and development (Table S2). These data are consistent with many reports suggesting that microRNAs can act as ‘micromanagers’ and adjust gene dosage for large numbers of genes in concert (Bartel and Chen, 2004). In this model, miR-205 micromanages gene expression within the surface ectoderm, repressing many targets simultaneously to ensure proper lacrimal gland initiation. We did note that among the 19 up-regulated targets, we identified genes that antagonize AKT signaling (Inppl1 and Cadm1), which has been shown to be activated downstream of Fgf signaling and regulated by miR-205 (Chen et al., 2000; Kawano et al., 2009; Pesesse et al., 1997; Wang et al., 2013; Yu et al., 2008; Yu et al., 2010) . Together, these data suggested a possible intersection between miR-205 and lacrimal gland developmental signaling pathways. Thus, given the established role of Fgf10 as an inducer of lacrimal gland initiation (Makarenkova et al., 2000), we hypothesized that miR-205 controls the fidelity of lacrimal gland initiation by repressing targets that interfere with or antagonize Fgf10 signaling.
Fig. 5. RNA-seq of miR-205−/− tissues uncovers relevant miR-205 targets and pathways.
(A) Schematic overview of experimental method for tissue isolation and RNA sequencing. (B) Overlap between up-regulated genes and predicted miR-205 targets expressed in the surface ectoderm. (C) Luciferase UTR assays for subset of genes up-regulated in miR-205 knockout samples (SEM error bars). No binding site (NS), a perfect binding site (PS) control. (D) Luciferase UTR assays for miR-205 targets with wildtype UTRs (normalized to one) and mutated UTRs.
Ablation of miR-205 exacerbates the Fgf10 phenotype
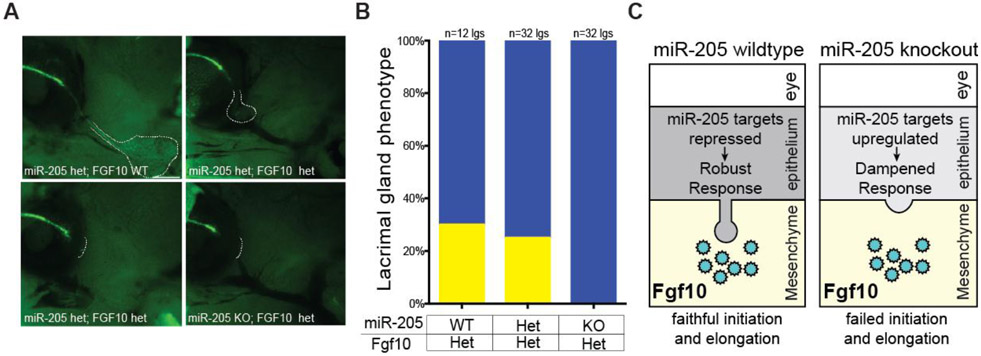
Glandular initiation occurs in a complex environment that may not necessarily easily be recapitulated in an in vitro setting, making in vivo models optimal to study this developmental process. To test whether deletion of miR-205 impairs the fidelity of FGF signaling, miR-205−/− mice were crossed to Fgf10 heterozygous mice (Min et al., 1998) and imaged at P0 to capture possible defects in both lacrimal gland initiation and branching. Deletion of a single copy of Fgf10 in either wildtype mice (data not shown) or miR-205+/− mice resulted in ~75% of lacrimal glands absent at P0 and the remaining 25% of lacrimal glands were stunted, consistent with a dosage sensitive model for Fgf10 (Fig. 6A and B) (Qu et al., 2011). Strikingly, no lacrimal glands were present in miR-205−/−; Fgf10+/− animals. The absence of lacrimal glands at P0 indicated a complete impairment in lacrimal gland initiation and this was supported by identical observations made at earlier embryonic stages (E15, data not shown). These data suggest a role of miR-205 in modulating the fidelity of Fgf10 signaling. Taken together, these data support a model where miR-205 reduces the levels of its targets to enhance proper signaling and ensure the fidelity of lacrimal gland initiation (Fig. 6C).
Fig. 6. miR-205 controls the fidelity of Fgf signaling during lacrimal gland development.
(A) Representative images of combinatorial deletions in P0 mice imaged with Krt5GFP reporter. Dashed lines indicate the location of the lacrimal gland. (B) Quantification of lacrimal gland defects in the Fgf10 heterozygous background (p=0.0048, Fisher’s exact test). Green: normal, yellow: runted, and blue: absent glands. (C) Working model for the role of miR-205 in lacrimal gland development. In wildtype mice, FGF10 is secreted by the mesenchyme to stimulate epithelial budding and miR-205 targets several genes to ensure efficient lacrimal gland initiation. In the absence of the microRNA, miR-205 targets are up-regulated, interfere with Fgf10 signaling and impair lacrimal gland development.
Discussion
Few single gene microRNA knockouts have readily identifiable phenotypes in mouse models (Park et al., 2012). Our lab and others established miR-205 as a critical regulator of postnatal development in mice (Farmer et al., 2013; Wang et al., 2013). In this study, we expand the role of miR-205 and establish the microRNA as a critical regulator of lacrimal gland development. Remarkably, the observed ocular phenotype is likely secondary to absent lacrimal glands. Unlike previous studies, this work highlights a role of miR-205 during embryonic development rather than postnatal development (Farmer et al., 2013; Wang et al., 2013). Furthermore, it uncovers a previously unappreciated role of miR-205 in organogenesis and identifies the first noncoding RNA necessary for lacrimal gland development.
The prevalence of lacrimal gland initiation defects in miR-205−/− mice highlights the role of miR-205 at the earliest phases of lacrimal gland development. Indeed, failure to initiate indicates an inability of the surface ectoderm to robustly respond to external stimuli. Furthermore, although some miR-205−/− glands successfully form, they remain significantly smaller throughout all stages of development, never recovering after lacrimal gland initiation. These small glands, however, retained normal proliferative capacity and eventually formed highly branched structures. It should be noted that Pax6, a competence factor for lacrimal gland development, is unaffected by miR-205 as evidenced by both RNAseq and immunostaining (data not shown) (Makarenkova et al., 2000). Thus, it is likely that the initiation defects are a consequence of signaling rather than cellular identity. Smaller glands may reflect fewer successfully initiated epithelial cells or very early and short-term proliferative defects before E14.5, when smaller glands are already detected.
Despite the widespread expression of miR-205 in various glands that are governed by similar developmental pathways, deleting miR-205 has a remarkably specific effect on lacrimal gland development. One plausible rationale for this lacrimal gland-specific phenotype is the distinct dosage sensitivity of lacrimal gland development to a morphogen cue, Fgf10 (Qu et al., 2011). We propose that miR-205 in the surface ectoderm functions to ensure robust activation of pathways associated with Fgf10 signaling. This model is supported by the exacerbated lacrimal gland phenotype in miR-205−/−: Fgf10+/− mice, where lacrimal gland initiation is completely blocked. While we appreciate the limitation of the narrow phenotypic window in Fgf10+/− mice, the absence of lacrimal glands in double mutants, rather than defects in bud length or number, highlights the specific requirement for miR-205 during early lacrimal gland development. In particular, we suspect that initiation represents an especially sensitive stage of development, as the epithelium at this stage is maximally distal from the FGF10 diffusion gradient and is not yet intimately surrounded by the FGF10 secreting mesenchyme (Qu et al., 2012). Supporting this idea, several studies have illustrated potent initiation defects when Fgf signaling is impaired (Makarenkova et al., 2000; Pan et al., 2010; Qu et al., 2011; Qu et al., 2012). Likewise, following lacrimal gland initiation, other signals (e.g. BMPs) has been observed to facilitate normal lacrimal gland growth and development (Dean et al., 2004; Liu and Lin, 2014). Thus, in contrast to later stages when the epithelium is in direct contact with the mesenchyme, the earliest stages of lacrimal gland development may be particularly susceptible to molecular noise caused by the deletion of miR-205. Independent of a potential role in Fgf10 signaling, it is possible that miR-205 controls the cellular activity of surface ectoderm and early lacrimal gland epithelial cells. While no other pathway has been identified to control lacrimal gland initiation, it remains a formal possibility that miR-205 can antagonize other as yet uncharacterized pathway(s) to perturb lacrimal gland development.
While it is clear that miR-205 impairs lacrimal bud initiation, a process driven by Fgf10, future research will be required to resolve the molecular details of this complex process. Using RNA-seq, we identified and validated several up-regulated targets in the surface ectoderm of miR-205−/− mice, suggesting that miR-205 regulates gene expression to ensure efficient signaling within the surface ectoderm. Minimally, these data are consistent with a role for miR-205 as a micromanager during lacrimal gland initiation. Thus, we postulate that miR-205 acts as a molecular rheostat to fine-tune the landscape of the surface ectoderm in order to ensure proper lacrimal gland initiation. In this idea, disrupting miR-205 expression likely exposes the gland to greater molecular noise, reducing the competence and rigor of pathways that regulate early developmental decisions including Fgf10 signaling. Overall, this work highlights the necessity of microRNAs for organogenesis and identifies the first non-coding RNA important during lacrimal gland development.
Methods
Ethics Statement.
All mouse experiments were performed according to the protocols approved by the Animal Care and Use Committee of the University of California, San Francisco.
Mouse generation.
miR-205 mice (including the miR-205 flox, miR-205 null, and miR-205 LacZ reporter alleles) were generated as previously reported (Farmer et al., 2013; Park et al., 2012) and are available at The Jackson Laboratory (34650-JAX). ACTB-Cre mice (003376) were backcrossed 7 generations to C57/B6 mice (Lewandoski et al., 1997). Le-Cre mice were obtained from Jackson Laboratories and backcrossed 7 generations to C57/B6 mice (Ashery-Padan et al., 2000). For Le-Cre experiments, only one parent carried the Cre allele to avoid defects caused by homozygosity. Krt5GFP reporter mice were kindly provided by Dr. Jason Rock (Bruen et al., 2004). Fgf10 heterozygous mice were kindly provided by Dr. Ophir Klein (Min et al., 1998).
X-Gal staining.
Xgal staining was performed as previously described (Farmer et al., 2013). Briefly, embryos were dissected and fixed in 4% formaldehyde and 0.2% glutaraldehyde in PBS and permeabilized in 0.02% NP40, 0.01% sodium deoxycholate, and 2 mM MgCl2 in PBS. X-gal staining was completed overnight at 37°C. Embryos were washed to remove background, post-fixed in 4% paraformaldehyde in PBS to retain Xgal staining and subsequently dehydrated and processed for paraffin embedding using Citrosolv. Sections were dewaxed and counterstained with Nuclear fast red.
Tear Secretion Measurement.
Mice were weighed and pilocarpine diluted in saline was injected by intraperitoneal injection (IP) at a dosage of 4.5 mg/kg. After ten minutes, injected mice were anesthetized with isoflurane and tear secretion was measured using Zone-Quick phenol red thread (Showa Yakuhin Kako Co. Ltd., Tokyo, Japan). Tear secretion was determined by measuring the length (in millimeters) of absorption along the thread.
Immunofluorescence.
Whole lacrimal glands were isolated and fixed for 20 minutes at room temperature in 4% paraformaldehyde. Glands were permealized in 0.2% Triton in PBS and blocked in 10% goat serum and 3% BSA for two hours. Glands were then incubated in primary antibodies for 2 hours at room temperature followed by an overnight incubation at 4°C. Antibodies included 1:300 for E-cadherin (Sigma U3294) and 1:200 for Ki67 (Abcam ab15580). Glands were thoroughly washed in 0.05% Tween in PBS followed by a 2-hour incubation in appropriate secondary antibodies (1:500, Life Technologies) and stained with DAPI. Glands imaged on a Leica SP5 confocal. Ki67 and DAPI were quantified using ImageJ (Abràmoff et al., 2004). Paraffin sections were de-paraffinized and boiled for 20 minutes in Citrate buffer, following by 1 hour blocking solution (10% chicken serum in PBST) and an overnight antibody at 4°C. Antibodies included 1:500 for E-cadherin (Sigma U3294) and 1:500 for Ki67 (Abcam ab15580). After washes in PBS, slides were incubated with secondary for 1 hour (1:500, Life Technologies) and stained with DAPI.
qPCR analysis.
P0 lacrimal glands were isolated and immediately flash frozen. After mechanical disruption with pestle, RNA was isolated from lacrimal glands using the Qiagen miRNeasy Mini Kit Print. cDNA synthesis was performed using Applied Biosystems’ TaqMan® MicroRNA Reverse Transcription Kit (4366596) and Thermo microRNA primers for miR-205 and U6 snRNA. qPCR was performed in technical triplicates of 3 independently isolated sets of lacrimal glands using TaqMan® Universal PCR Master Mix. miR-205 levels were normalized to U6 snRNA levels.
Histology.
Eyes, eyelids, meibomian glands, and lacrimal glands were dissected from mice and fixed overnight in 4% PFA in PBS. Organs were then transferred to 70% EtOH and processed for paraffin embedding by either the Mouse Pathology Core at UCSF or the Gladstone Histology and Light Microscopy Core and stained with haematoxylin and eosin. Lipid Oil O staining was completed at the Gladstone Histology and Light Microscopy Core.
Laser capture and RNA isolation.
The laser capture staining protocol was adapted from the Laser Capture Molecular Core at Ohio State University. Briefly, E12.5 embryo heads were dissected and flash frozen in OCT for cyrosectioning. Sections were collected at 10 μM onto RNase-Zap treated 1.0 PEN membrane slides from Zeiss and stored at −80°C. Frozen sections were immediately submerged into ice cold RNase free 70% ethanol, stained in Vector® Hematoxylin QS for 30 sec, and then rinsed in RNase water, following by 95% ethanol and 100% ethanol. Slides were then air dried and returned to −80°C before laser capture. The surface ectoderm between the eye and brain was isolated using the Zeiss PALM into 0.5 mL tubes containing Qiagen RLT buffer. Three embryos were pooled together per sample. RNA was isolated using a Qiagen RNeasy Plus Micro kit and assessed on an Agilent 2100 Bioanalyzer using Agilent RNA Pico Chips. All samples had a RIN above 7.0.
cDNA synthesis and library preparation.
cDNA was generated using Clontech’s SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing according to manufacturer’s recommendations. Sequencing libraries were prepared with 1 ng of cDNA using the Nextera XT DNA Library Prep Kit according to manufacturer’s recommendations. The final product was then run on a Novex 4-12% TBE gel and DNA between 300-500 nt was selected. Samples were run on an Illumina HiSeq 2500 (UCSF Institute for Human Genetics Genomics Core). Raw files have been uploaded to GEO (GSE95108).
RNA-seq data analysis.
Reads were mapped to the mouse GRCm38 genome using Tophat v.2.0.9 (Trapnell et al., 2009). Gene expression was measured from the mapped reads by using HT-seq-count (Anders et al., 2014) in intersection-strict mode, which counts the reads aligning to each annotated gene (gene set, Ensembl.org). Differentially expressed genes were called using the DESeq2 R package, (Anders and Huber, 2010) considering genes differentially expressed with FDR < 0.1. Only genes with normalized count values above 10 in all samples were evaluated. One WT samples was removed due to contamination of non-surface ectoderm tissues, as evident by decreased markers of Pax6, Krt5, and others that constant across all other samples.
UTR analysis.
Putative miR-205 targets were identified using Targetscan (Agarwal et al., 2015). UTRs were amplified using Phusion High Fidelity polyermase (Thermo) and included a minimal of 300 nt on each end of the miR-205 seed sequence. UTRs were cloned into the psiCHECK™-2 vector from Promega. psiCHECK™-2 vectors were transfected into 293T cells with a vector expressing wildtype miR-205 (TCCTTCATTCCACCGGAGTCTG) or a mutant miR-205 (TCgTTCATTaCACCGGAGTCTG). UTR mutants were generated using PCR and seed sequences were changed from the WT seed sequence (ATGAAGG) to a mutant version (AaGtAGG). Each condition was transfected independently for biological triplicates. After 48 hours, firefly and renilla expression was detected using Promega’s Dual-Glo® Luciferase Assay System. Seed deletions in Inppl1 were created using PCR with a primer including two mutations in the miR-205 seed sequence.
Supplementary Material
Acknowledgements:
We thank the members of the Knox, Marson, Ku, and Al-Sady labs for their considerable feedback and recommendations. In addition, the Black lab and the Bruneau lab provided valuable advice for optimizing staining protocols. We also thank Jason Rock for sharing the Krt5GFP mice. The Mouse Pathology Core at UCSF and the Gladstone Histology and Light Microscopy Core provided their services in histological experiments. Farmer was funded by the NIH IMSD fellowship and the NSF GRFP fellowship during this project. Funding for this project was provided by the W. M. Keck Foundation and the National Institutes of Health (1U19CA179513).
Footnotes
Competing interests
The authors declare no competing or financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abràmoff MD, Magalhães PJ and Ram SJ (2004). Image processing with imageJ. Biophotonics Int. 11, 36–41. [Google Scholar]
- Agarwal V, Bell GW, Nam JW and Bartel DP (2015). Predicting effective microRNA target sites in mammalian mRNAs. Elife 4,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S and Huber W (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT and Huber W (2014). HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashery-Padan R, Marquardt T, Zhou X and Gruss P (2000). Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes Dev. 14, 2701–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP and Chen C-Z (2004). Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet 5, 396–400. [DOI] [PubMed] [Google Scholar]
- Bruen KJ, Campbell C. a, Schooler WG, deSerres S, Cairns B. a, Hultman CS, Meyer A. a and Randell SH (2004). Real-time monitoring of keratin 5 expression during burn re-epithelialization. J. Surg. Res 120, 12–20. [DOI] [PubMed] [Google Scholar]
- Carraro G, El-Hashash A, Guidolin D, Tiozzo C, Turcatel G, Young BM, De Langhe SP, Bellusci S, Shi W, Parnigotto PP, et al. (2009). miR-17 family of microRNAs controls FGF10-mediated embryonic lung epithelial branching morphogenesis through MAPK14 and STAT3 regulation of E-Cadherin distribution. Dev. Biol 333, 238–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Li X, Eswarakumar VP, Seger R and Lonai P (2000). Fibroblast growth factor (FGF) signaling through PI 3-kinase and Akt/PKB is required for embryoid body differentiation. Oncogene 19, 3750–3756. [DOI] [PubMed] [Google Scholar]
- Dean C, Ito M, Makarenkova HP, Faber SC and Lang R. a (2004). Bmp7 regulates branching morphogenesis of the lacrimal gland by promoting mesenchymal proliferation and condensation. Development 131, 4155–65. [DOI] [PubMed] [Google Scholar]
- Dorà NJ, Collinson JM, Hill RE and West JD (2014). Hemizygous Le-Cre transgenic mice have severe eye abnormalities on some genetic backgrounds in the absence of LoxPSites. PLoS One 9,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas JC, Cuellar TL, Scholze A, Ason B, Ibrahim A, Emery B, Zamanian JL, Foo LC, McManus MT and Barres B. a. (2010). Dicer1 and miR-219 Are Required for Normal Oligodendrocyte Differentiation and Myelination. Neuron 65, 597–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer DT, Shariat N, Park CY, Liu HJ, Mavropoulos A and McManus MT (2013). Partially penetrant postnatal lethality of an epithelial specific MicroRNA in a mouse knockout. PLoS One 8, e76634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayton JL (2009). Etiology, prevalence, and treatment of dry eye disease. Clin. Ophthalmol 3, 405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajan V, Ito M, Makarenkova HP, Lang R. a and Overbeek P. a (2000). Endogenous and ectopic gland induction by FGF-10. Dev. Biol 225, 188–200. [DOI] [PubMed] [Google Scholar]
- Greene SB, Gunaratne PH, Hammond SM and Rosen JM (2010). A putative role for microRNA-205 in mammary epithelial cell progenitors. J. Cell Sci 123, 606–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Koyama N, Azuma Y and Kashimata M (2011). Mesenchymal miR-21 regulates branching morphogenesis in murine submandibular gland in vitro. Dev. Biol 352, 299–307. [DOI] [PubMed] [Google Scholar]
- Ibarra I, Erlich Y, Muthuswamy SK, Sachidanandam R and Hannon GJ (2007). A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev. 21, 3238–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano S, Ikeda W, Kishimoto M, Ogita H and Takai Y (2009). Silencing of ErbB3/ErbB2 signaling by immunoglobulin-like Necl-2. J. Biol. Chem 284, 23793–23805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandoski M, Meyers EN and Martin GR (1997). Analysis of Fgf8 gene function in vertebrate development. In Cold Spring Harbor Symposia on Quantitative Biology, pp. 159–168. [PubMed] [Google Scholar]
- Liu Y and Lin D (2014). Necessity of Smad4 for the normal development of the mouse lacrimal gland. Jpn. J. Ophthalmol 58, 298–306. [DOI] [PubMed] [Google Scholar]
- Makarenkova HP, Ito M, Govindarajan V, Faber SC, Sun L, McMahon G, Overbeek P. a and Lang R. a (2000). FGF10 is an inducer and Pax6 a competence factor for lacrimal gland development. Development 127, 2563–72. [DOI] [PubMed] [Google Scholar]
- Min H, Danilenko DM, Scully SA, Bolon B, Ring BD, Tarpley JE, DeRose M and Simonet WS (1998). Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev. 12, 3156–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Carbe C, Powers A, Zhang EE, Esko JD, Grobe K, Feng G-S and Zhang X (2008). Bud specific N-sulfation of heparan sulfate regulates Shp2-dependent FGF signaling during lacrimal gland induction. Development 135, 301–10. [DOI] [PubMed] [Google Scholar]
- Pan Y, Carbe C, Powers A, Feng G-S and Zhang X (2010). Sprouty2-modulated Kras signaling rescues Shp2 deficiency during lens and lacrimal gland development. Development 137, 1085–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CY, Jeker LT, Carver-Moore K, Oh A, Liu HJ, Cameron R, Richards H, Li Z, Adler D, Yoshinaga Y, et al. (2012). A resource for the conditional ablation of microRNAs in the mouse. Cell Rep. 1, 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesesse X, Deleu S, De Smedt F, Drayer L and Erneux C (1997). Identification of a Second SH2-Domain-Containing Protein Closely Related to the Phosphatidylinositol Polyphosphate 5-Phosphatase SHIP. Biochem. Biophys. Res. Commun 239, 697–700. [DOI] [PubMed] [Google Scholar]
- Qu X, Carbe C, Tao C, Powers A, Lawrence R, van Kuppevelt TH, Cardoso WV, Grobe K, Esko JD and Zhang X (2011). Lacrimal gland development and Fgf10-Fgfr2b signaling are controlled by 2-O- and 6-O-sulfated heparan sulfate. J. Biol. Chem 286, 14435–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Pan Y, Carbe C, Powers A, Grobe K and Zhang X (2012). Glycosaminoglycan-dependent restriction of FGF diffusion is necessary for lacrimal gland development. Development 139, 2730–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebustini IT, Hayashi T, Reynolds AD, Dillard ML, Carpenter EM and Hoffman MP (2012). miR-200c regulates FGFR-dependent epithelial proliferation via Vldlr during submandibular gland branching morphogenesis. Development 139, 191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Søe MJ, Møller T, Dufva M and Holmstrøm K (2011). A sensitive alternative for microRNA in situ hybridizations using probes of 2’-O-methyl RNA + LNA. J. Histochem. Cytochem 59, 661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen R, Nielsen PS and Jensen TH (2005). Dramatically improved RNA in situ hybridization signals using LNA-modified probes. RNA 11, 1745–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L and Salzberg SL (2009). TopHat Manual. Bioinformatics 25, 1105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ucar A, Vafaizadeh V, Jarry H, Fiedler J, Klemmt PAB, Thum T, Groner B and Chowdhury K (2010). miR-212 and miR-132 are required for epithelial stromal interactions necessary for mouse mammary gland development. Nat. Genet 42, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. (2008). Targeted Deletion Reveals Essential and Overlapping Functions of the miR-17~92 Family of miRNA Clusters. Cell 132, 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zhang Z, O’Loughlin E, Wang L, Fan X, Lai EC and Yi R (2013). MicroRNA-205 controls neonatal expansion of skin stem cells by modulating the PI(3)K pathway. Nat. Cell Biol 15, 1153–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Poy MN, Stoffel M and Fuchs E (2008). A skin microRNA promotes differentiation by repressing “stemness.” Nature 452, 225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Ryan DG, Getsios S, Oliveira-Fernandes M, Fatima A and Lavker RM (2008). MicroRNA-184 antagonizes microRNA-205 to maintain SHIP2 levels in epithelia. Proc. Natl. Acad. Sci. U. S. A 105, 19300–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Peng H, Ruan Q, Fatima A, Getsios S and Lavker RM (2010). MicroRNA-205 promotes keratinocyte migration via the lipid phosphatase SHIP2. FASEB J. 24, 3950–3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ and Srivastava D (2007). Dysregulation of Cardiogenesis, Cardiac Conduction, and Cell Cycle in Mice Lacking miRNA-1-2. Cell 129, 303–317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.