Abstract
Background:
NOTCH1 mutation is an essential molecular biologic aberration in chronic lymphocytic leukemia (CLL). CLL patients with NOTCH1 mutation have shown an unfavorable survival and a poor response to chemoimmunotherapy. This study aims to present the mechanisms of adverse prognosis caused by NOTCH1 mutation from the perspective of the splicing factor heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1).
Methods:
The microarray data in Gene Expression Omnibus datasets were analyzed by bioinformatics and the function of hnRNPA1 was checked by testing the proliferation and apoptosis of CLL-like cell lines. Afterward, quantitative reverse transcription-polymerase chain reaction and Western blotting were applied to explore the relationship among NOTCH1, c-Myc, and hnRNPA1.
Results:
RNA splicing was found to play a vital part in NOTCH1-mutated CLL cells; hence, hnRNPA1 was selected as the focus of this study. Higher expression of hnRNPA1 validated in primary NOTCH1-mutated CLL samples could promote proliferation and inhibit apoptosis in CLL. The expression of hnRNPA1 increased when NOTCH1 signaling was activated by transfection with NOTCH1 intracellular domain (NICD)-overexpressed adenovirus vector and declined after NOTCH1 signaling was inhibited by NOTCH1-shRNA. Higher expression of c-Myc was observed in NICD-overexpressed cells and hnRNPA1 expression was downregulated after applying c-Myc inhibitor 10058-F4. Moreover, in NICD-overexpressed cells, hnRNPA1 expression decreased through c-Myc inhibition.
Conclusion:
Overexpression of c-Myc-dependent hnRNPA1 could promote proliferation and inhibit apoptosis in NOTCH1-mutated CLL cells, which might partly account for the poor prognosis of patients with NOTCH1 mutation.
Keywords: Chronic lymphocytic leukemia, NOTCH1 mutation, c-Myc, Heterogeneous nuclear ribonucleoprotein A1
Introduction
Chronic lymphocytic leukemia (CLL) is a common type of mature B-cell chronic lymphoproliferative disease in adults with heterogeneous clinical courses.[1] Many patients have more than one molecular biologic or cytogenetic aberra-tion.[2]NOTCH1 mutation is an essential molecular biologic change in CLL. Previous studies reported that patients with NOTCH1 mutation have shorter treatment-free survival (TFS), progression-free survival, and overall survival (OS), implying NOTCH1 mutation as an adverse prognostic factor.[3,4]
NOTCH1 gene located in chromosome 9q34.3 encodes NOTCH1 protein, whose extracellular receptor binding to its corresponding ligand would trigger two sequential cleavages and eventually release NOTCH1 intracellular domain (NICD), which could form transcriptional activation complexes to activate target gene transcription along with other transcription factors. A majority of NOTCH1 mutations (eg, c.7541_7542delCT) could increase NICD stability by truncating the domain recognized by ubiquitin ligase F-box and WD repeat-containing protein 7 (FBXW7), thus causing sustained NOTCH1 signaling activation and target gene transcription.[5]
Up to now, there has been little research on the mechanisms of poor prognosis caused by NOTCH1 mutation. Arruga et al[6] found that sustained NOTCH1 signaling activation caused by NOTCH1 mutation resulted in increased expression of genes associated with B cell receptor signaling, which, in turn, enhanced NOTCH1 translation. Consequently, NOTCH1 and B cell receptor signaling synergistically facilitated the development of CLL. In addition, NOTCH1 mutation could upregulate surface C-C chemokine receptor 7 of CLL cells by inhibiting DUSP22, thereby facilitating the chemotaxis response to chemokine C-C motif ligand 19 (CCL19). CCL19 would guide CLL cells to lymph nodes and spleen, where the CLL cells would find ideal micro-environments for their proliferation.[7] Pozzo et al[8] discovered that a higher proliferation rate in NOTCH1-mutated CLL cells was related to overexpression of c-Myc related nucleophosmin 1. Moreover, NOTCH1 mutation could downregulate CD20 on the surface of CLL cells by epigenetic dysregulation and cause a poor response to anti-CD20 antibodies.[9]
Different gene expression patterns between NOTCH1-mutated and NOTCH1-wild-type (wt) CLL samples were analyzed by using Gene Expression Omnibus (GEO) dataset GSE75122. It was found that the expression of genes associated with RNA splicing increased in NOTCH1-mutated cells, including the heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1). hnRNPA1 had been proved to affect proliferation, apoptosis, and metabolism in several solid tumors, verifying its importance as a regulator in the development of cancer.[10,11] However, its function in NOTCH1-mutated CLL remains unknown. Therefore, investigating its effect on proliferation and apoptosis and exploring the key mediator between NOTCH1 and hnRNPA1 in CLL were set to be the focus of the present research.
Methods
Primary CLL cells
Seven NOTCH1-mutated and eight NOTCH1-wt CLL peripheral blood samples were collected from the patients who were admitted to our department. Detailed characteristics of patients were shown in Supplementary Table 1. The diagnosis was based on the International Workshop on CLL-National Cancer Institute criteria. Ficoll-Hypaque solution (Tbdscience, Tianjin, China) and B-CLL cell isolation kit (Miltenyi Biotec, Bergisch Gladbach, Germany) were applied to obtain purified B cells (>95%). NOTCH1 mutation was detected by Sanger sequencing or next-generation sequencing. Moreover, this study was approved by the hospital ethics committee (No. 2019–SR-307) and the informed consent of all patients was provided in line with the Helsinki Declaration of 1975.
Cell lines and cell culture
CLL-like cell lines MEC-1 and JVM-3 were purchased from Cobioer Biological Technology Co., Ltd. (Nanjing, China). Short tandem repeat authentication was conducted by Shanghai Biowing Applied Biotechnology Co., Ltd. (Shanghai, China). MEC-1 was established from the peripheral blood of a 61-year-old Caucasian man with chronic B cell leukemia. JVM-3 was constructed from the peripheral blood of a 73-year-old man with B-prolymphocytic leukemia. Neither of these two cell lines harbored NOTCH1 mutation. The culture medium of MEC-1 consisted of 90% Iscove's modified Dulbecco's Modified Eagle's Medium (Gibco, Invitrogen, Grand Island, NY, USA) and 10% fetal bovine serum (Gibco). JVM-3 was cultured in 90% RPMI 1640 culture medium (Gibco) and 10% fetal bovine serum (Gibco). All the cells were incubated in a 5% CO2 humidified incubator at 37°C. All experiments were conducted with mycoplasma-free cells.
In vitro treatment with a c-Myc inhibitor
To inhibit c-Myc expression, cells were treated with c-Myc inhibitor 10058–F4 (100 mmol/L, Selleck, Shanghai, China) for 24 hours, which could inhibit c-Myc-max hetero-dimerization and transcription of c-Myc target genes.[12] As ethanol was chosen as the solvent for 10058–F4, it was chosen as the control in the experiments of this study.
Vector constructs, virus production, and cell transfection
Commercially available adenovirus vectors were applied to construct the HBAD-NOTCH1-NICD-Null-GFP vector and corresponding negative control vector (Hanbio, Shanghai, China). Commercially available lentiviral vectors were applied to construct LV-NOTCH1-RNAi vector (Genechem, Shanghai, China), HBLV-h-HNRNPA1–3xflag-ZsGreen-PURO vector (Hanbio), HBLV-h-HNRNPA1 shRNA-ZsGreen-PURO vector (Hanbio), and corresponding negative control vectors. These constructions were verified by DNA sequencing before being used. An appropriate multiplicity of infection was chosen to infect MEC-1 and JVM-3. Stable cell lines were screened by adding 2 μg/mL puromycin (InvivoGen, San Diego, CA, USA) for 2 days. Transfection efficiency was confirmed by flow cytometry and expression was validated by real-time quantitative polymerase chain reaction and Western blot.
Total RNA extraction and real-time quantitative polymerase chain reaction
Total RNA was extracted from purified B cells or cell lines applying an RNA-quick purification kit (Yishan Biotec, Shanghai, China) in line with instructions. Complementary DNA was generated by using PrimeScriptTM RT Master Mix (Takara, Beijing, China). Quantitative real-time polymerase chain reaction (qRT-PCR) experiments were conducted using AceQ qPCR SYBR Green Master Mix (Vazyme, Nanjing, China). The relative messenger RNA (mRNA) expression levels of target genes were calculated through the 2−ΔΔCt method. Primers for qRT-PCR were as follows: NOTCH1 forward: 5′-GGACGTCAGACTT-GGCTCAG-3′; reverse: 5′-ACATCTTGGGACGCAT-CTGG-3′; NICD forward: 5′-TGCACACTATTCTGCCCCAG-3′; reverse: 5′-ACTTGAAGGCCTCCGGAATG-3′; c-Myc forward: 5′-TTCGGGTAGTGGAAAACCAG-3′; reverse: 5′-AGTAGAAATACGGCTGCACC-3′; hnRNPA1 forward: 5′-GCTCACGGACTGTGTGGTAA-3′; reverse: 5′-GCAAAGTATTGGCCTCCACC-3′; HES1 forward: 5′-CTGAGCACAGACCCAAGTGT-3′; reverse: 5′-GAGTGCGCACCTCGGTATTA-3′; and β-Actin forward: 5′-AGCGAGCATCCCCCAAAGTT-3′; reverse: 5′-GGGCACGAAGGCTCATCATT-3′.
Western blot
Total proteins extracted from purified B cells or cell lines were resolved by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE, Beyotime, Shanghai, China) and transferred to polyvinylidene fluoride (PVDF, Millipore, Billerica, Massachusetts, USA) membranes. Fast SDS-PAGE running buffer (Cwbio, Taizhou, China) and Tris-Glycine transfer buffer (Cwbio) were purchased from CoWin Biosciences. The membranes were blocked with QuickblockTM blocking buffer for Western blot (Beyotime) for 30 minutes followed by successive incubations with primary antibodies overnight at 4°C and corresponding secondary antibodies for 1 h. Protein expression levels were visualized using ECL Plus (Millipore) and Bio-Imaging System. The primary and secondary antibodies used in our experiments are NOTCH1 (#3608, CST, Danvers, Massachusetts, USA), cleaved NOTCH1 (#4147, CST), hnRNPA1 (#8443, CST), c-Myc (ab32072, Abcam, Boston, MA, USA), β-Actin (#3700, CST), GAPDH (#2118, CST), HRP-conjugated anti-mouse IgG (ZB-2305, ZSGB-BIO, Beijing, China), and HRP-conjugated anti-rabbit IgG (ZB-5301, ZSGB-BIO). Immunoblot bands were quantified through densitometry and ImageJ software (NIH, Bethesda, MD, USA). Relative folds were normalized to β-actin levels.
Cell count Kit-8 assay
Cell Count Kit-8 (CCK-8, Dojindo, Kumamoto, Japan) assay was applied to evaluate cell proliferation. A total of 1.0 × 104 cells/wells were seeded into 96-well plates with 100 μL corresponding medium (MEC-1: IMDM; JVM-3: RPMI 1640). 10 μL CCK-8 solution was added into specific wells at 0, 24, 48, and 72 hours followed by incubation in a 5% CO2 humidified incubator at 37°C for 4 h. Afterward, the plates were taken out and had their optical density value measured at wavelength 450 nm in Synergy H1 microplate reader (BioTek, Agilent, Santa Clara, CA, USA) immediately.
Cell apoptosis analysis
Cell apoptosis was tested by Annexin V-allophycocyanin (APC)/7-amino-actinomycin D (7–AAD) apoptosis kit (KeyGen Biotech, Nanjing, China). A total of 1.0 × 106 cells/tubes were stained with 5 μL Annexin V-APC and 5 μL 7–AAD in the dark at room temperature for 15 min. Afterward, apoptotic cells were measured with BD FACSCanto, and data were analyzed via FlowJo (X 10.0.7r2, Treestar, Ashland, OR, USA).
Bioinformatics analysis
GSE75122 in the GEO database was selected as the dataset for our research and analysis. The testing platform for this dataset was Agilent-014850 Whole Human Genome Microarray 4x44K G4112F. All the probe names were converted to gene names in line with the instructions of the platform. GEO2R (www.ncbi.nlm.nih.gov/geo/geo2r) was applied to analyze differentially expressed genes (DEGs). The standard of DEGs was defined as P < 0.05 and |log2 [fold change] | > 1. Heatmap was generated by supervised analysis. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was conducted in The Database for Annotation, Visualization, and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/, version 6.8).[13] The level of statistical significance in KEGG analysis was expressed as P < 0.05. Gene Set Enrichment Analysis (GSEA) was conducted via GSEA software (http://software.broadinstitute.org/gsea/downloads.jsp, UC San Diego and Broad Institute, USA). The statistical significance in GSEA analysis[14] was expressed as false discovery rate <0.05 and nominal P value < 0.05. Figures were drawn via R software (Version 3.5.3, The R Foundation for Statistical Computing, Vienna, Austria).
Statistical analysis
The cutoff value was determined by the use of X-tile,[15] a software that allowed users to move a cursor across the grid to test multiple divisions and accept the best P value. Data were analyzed by applying SPSS 23 software (IBM Corporation, Armonk, NY, USA) and presented as mean ± standard error of the mean. The student's t-test was applied to compare data between two groups. One-way analysis of variance was applied to compare data between multiple groups. P < 0.05 was defined as statistical significance. Graphs were made via GraphPad Prism 8.0 (GraphPad Software, Inc., La Jolla, CA, USA).
Results
Bioinformatics analysis of NOTCH1-mutated CLL cells
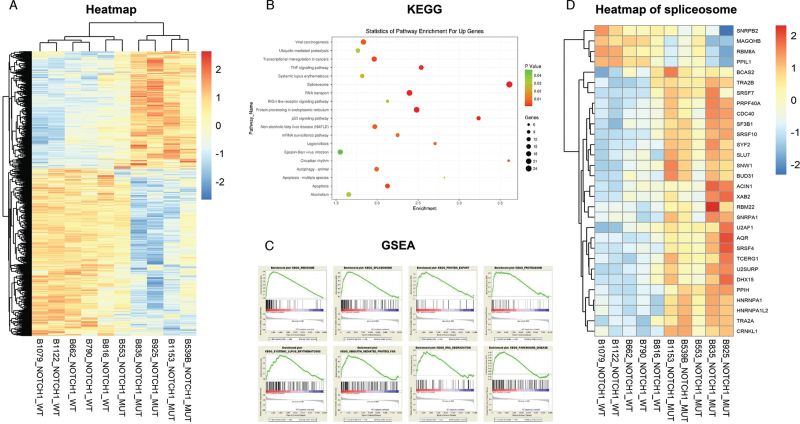
To explore different gene expression patterns between NOTCH1-mutated and NOTCH1-wt CLL cells, GSE75122 in the GEO database was analyzed and key signal pathways and downstream genes were identified. This dataset included five NOTCH1-mutated and five NOTCH1-wt CLL samples. Heatmap was generated by supervised analysis [Figure 1A]. A total of 2570 genes met the standard of DEGs with 1037 genes upregulated in NOTCH1-mutated CLL cells and 1533 downregulated. KEGG analysis indicated that highly expressed genes mainly contributed to spliceosome, protein processing in the endoplasmic reticulum, RNA transport, tumor protein 53 signaling pathway, tumor necrosis factor signaling pathway, apoptosis, etc. [Figure 1B]. GSEA revealed that genes involved in the ribosome, spliceosome, proteasome, protein export, ubiquitin-mediated proteolysis, RNA degradation, systemic lupus erythematosus, and Parkinson's disease were enriched in NOTCH1-mutated CLL cells [Figure 1C].
Figure 1.
Bio-informatics analysis of NOTCH1-mutated and NOTCH1-wt CLL cells In GSE75122. (A) The heatmap of five NOTCH1-mutated and five NOTCH1-wt CLL samples. (B) KEGG analysis of DEGs whose expressions were upregulated in NOTCH1-mutated CLL samples. (C) GSEA analysis of five NOTCH1-mutated and five NOTCH1-wt CLL samples. (D)The heatmap of genes involved in spliceosome. CLL: Chronic lymphocytic leukemia; DEGs: Differentially expressed genes; GSEA: Gene set enrichment analysis; KEGG: Kyoto Encyclopedia of Genes and Genomes.
Based on the results of KEGG and GSEA analysis, it was found that spliceosomes played an essential role in NOTCH1-mutated CLL cells. The heatmap of DEGs involved in spliceosome was presented in Figure 1D.
RNA splicing had been known to play a key part in various malignancies, but its functions in NOTCH1-mutated CLL cells remain unclear. Thus, hnRNPA1 was chosen for further examination due to its high expression in NOTCH1-mutated CLL cells and involvement in carcinogenesis in several solid tumors, hoping that it would bring insight into the role RNA splicing plays in these instances.
Expression and prognostic value of hnRNPA1 in primary CLL
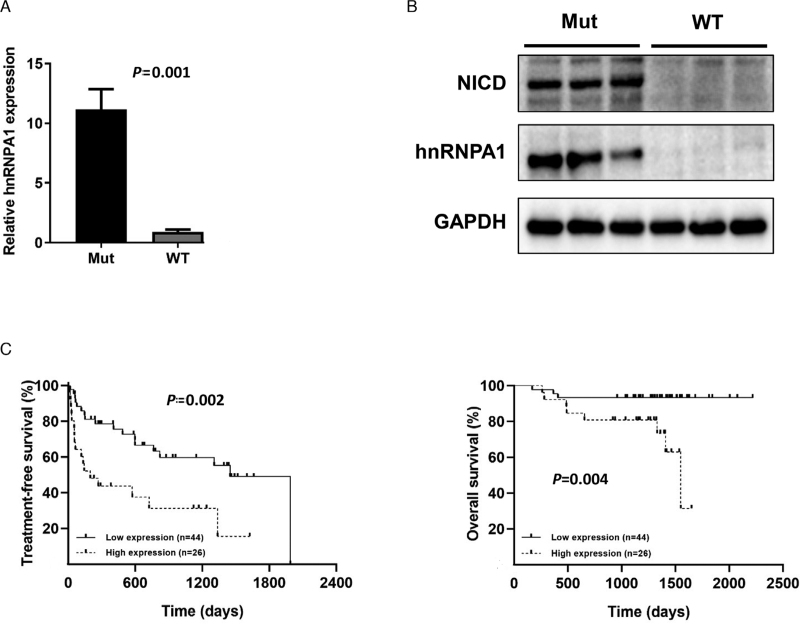
We first validated the results of bioinformatics analysis in our primary samples. The mRNA level of hnRNPA1 was determined in seven NOTCH1-mutated samples (six c.7541_7542delCT and one c.7443delC) and eight NOTCH1-wt CLL samples by qRT-PCR. The results revealed that hnRNPA1 was highly expressed in NOTCH1-mutated CLL samples [Figure 2A]. Increased NICD and hnRNPA1 protein levels in NOTCH1-mutated CLL cells were confirmed by Western blot [Figure 2B]. Afterward, another dataset GSE22762 was utilized to study the prognostic value of hnRNPA1 mRNA level in
Figure 2.
The expression of hnRNPA1 in CLL and its prognostic value. (A) The transcript level of hnRNPA1 in primary NOTCH1-mutated (n = 7) and NOTCH1-wt (n = 8). CLL samples was detected by qRT-PCR. (B) The protein level of NICD and hnRNPA1 in NOTCH1-mutated (n = 3) and NOTCH1-wt (n = 3) CLL samples was detected by Western blot. (C) Kaplan–Meier curves of TFS (left panel) and OS (right panel) for different mRNA level of hnRNPA1. CLL: Chronic lymphocytic leukemia; hnRNPA1: Heterogeneous nuclear ribonucleoprotein A1; mRNA: Messenger RNA; NICD: NOTCH1 intracellular domain; OS: Overall survival; qRT-PCR: Quantitative real-time polymerase chain reaction; TFS: Treatment-free survival.
CLL patients. This dataset contained the data of three platforms and GPL570 had larger amounts of cases than the other two platforms, hence GPL570 was chosen to conduct the survival analysis. Some missing data were deleted and 70 cases were reserved eventually. X-tile was applied to determine the cutoff value, according to which patients were divided into two groups, that is, high expression (exceeding cutoff value) and low expression (below or equal to cutoff value). Survival curves constructed through the Kaplan–Meier method revealed that patients with high mRNA level of hnRNPA1 had inferior TFS and OS [Figure 2C].
Effects of hnRNPA1 on proliferation and apoptosis of CLL
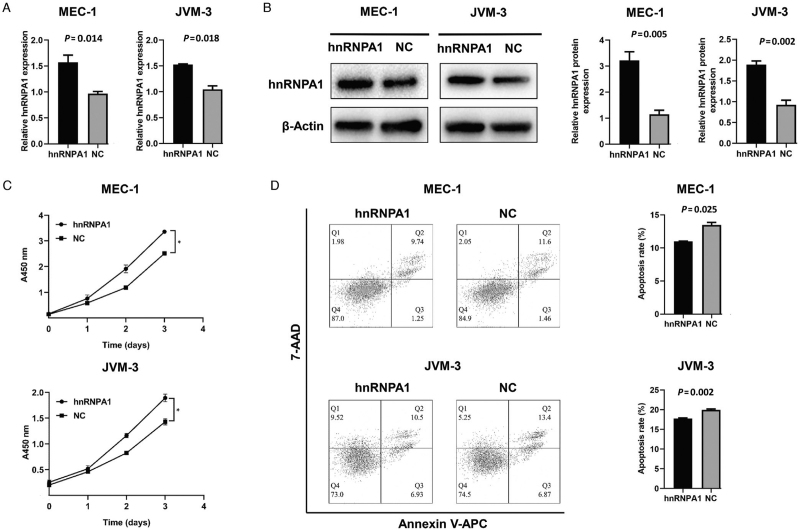
In vitro assays were conducted to investigate the effects of hnRNPA1 in the carcinogenesis of CLL. CLL-like cell lines MEC-1 and JVM-3 were selected to construct hnRNPA1-overexpressed cell lines. The transcript [Figure 3A] and protein level [Figure 3B] were confirmed through the corresponding methods as previously mentioned. Over-expressed hnRNPA1 led to a higher proliferation rate and a lower apoptosis rate of MEC-1 and JVM-3 [Figure 3C and 3D].
Figure 3.
The characteristics of hnRNPA1-overexpressed CLL cells. (A) The transcript level of hnRNPA1 in hnRNPA1-overexpressed MEC-1 and JVM-3 was verified by qRT-PCR. (B) The protein level of hnRNPA1 in hnRNPA1-overexpressed MEC-1 and JVM-3 was verified by Western blot. (C) The proliferative ability of hnRNPA1-overexpressed MEC-1 as well as JVM-3 and corresponding negative control tested by CCK-8 assay. (D) The apoptosis of hnRNPA1-overexpressed MEC-1 as well as JVM-3 and corresponding negative control detected by Annexin V-APC/7-AAD apoptosis kit. Data are representative of three independent experiments. P < 0.05. CCK-8: Cell count kit-8; CLL: Chronic lymphocytic leukemia; hnRNPA1: Heterogeneous nuclear ribonucleoprotein A1; qRT-PCR: Quantitative real-time polymerase chain reaction.
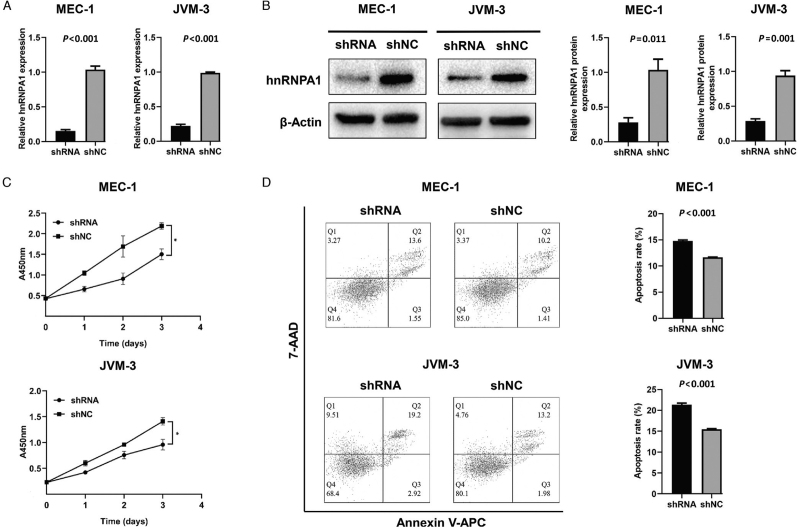
In addition, hnRNPA1-knockdown cell lines were also constructed. The transcript and protein levels were shown in Figure 4A and 4B, respectively. On the contrary, hnRNPA1-knockdown MEC-1 and JVM-3 indicated a lower proliferation rate [Figure 4C] and a higher apoptosis rate [Figure 4D].
Figure 4.
The characteristics of hnRNPA1-knockdown CLL cells. (A) The transcript level of hnRNPA1 in hnRNPA1-knockdown MEC-1 and JVM-3 was verified by qRT-PCR. (B) The protein level of hnRNPA1 in hnRNPA1-knockdown MEC-1 and JVM-3 was verified by Western blot. (C) The proliferative ability of hnRNPA1-knockdown MEC-1 as well as JVM-3 and corresponding negative control tested by CCK-8 assay. (D) The apoptosis of hnRNPA1-knockdown MEC-1 as well as JVM-3 and corresponding negative control detected by Annexin V-APC/7-AAD apoptosis kit. Data are representative of three independent experiments. ∗P < 0.05. CCK-8: Cell count kit-8; CLL: Chronic lymphocytic leukemia; hnRNPA1: Heterogeneous nuclear ribonucleoprotein A1; qRT-PCR: Quantitative real-time polymerase chain reaction.
NOTCH1 signaling and hnRNPA1 expression in CLL
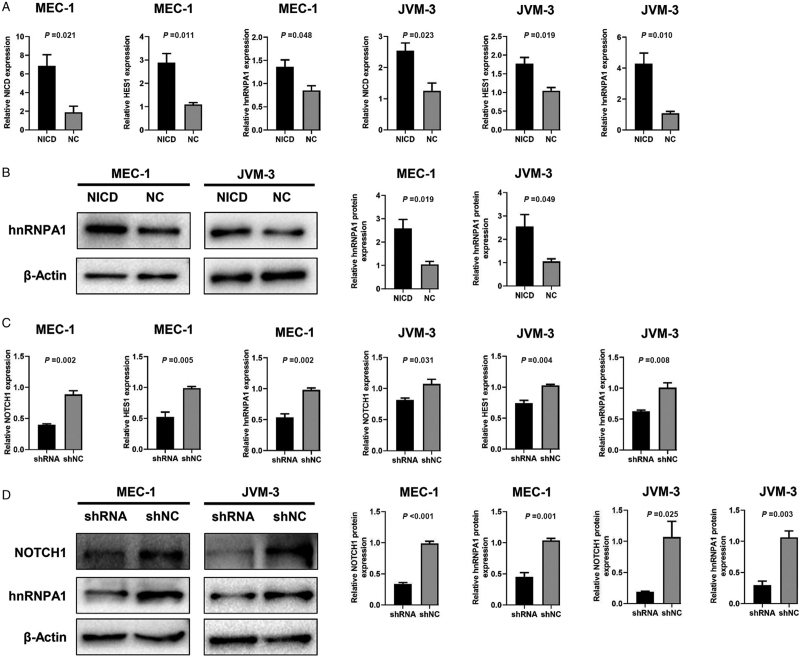
NOTCH1 mutation would cause impaired degradation of NICD and eventually cause continuous NOTCH1 signaling activation.[5] To evaluate the relationship between NOTCH1 signaling and hnRNPA1 expression in CLL, an adenovirus vector was applied to construct NICD-overexpressed cell lines. The transcript level of HES1 was an indicator for NOTCH1 signaling activation.[16] In NICD-overexpressed MEC-1 and JVM-3, the mRNA level of NICD, HES1, and hnRNPA1 was significantly higher than those in negative control cells [Figure 5A]. Similarly, the protein level of hnRNPA1 was upregulated in NICD-overexpressed cells [Figure 5B]. In addition, NOTCH1-knockdown MEC-1 and JVM-3 were constructed by using NOTCH1-shRNA to inhibit NOTCH1 signaling. Consequently, the mRNA level of NOTCH1, HES1, and hnRNPA1 was substantially downregulated in those cell lines [Figure 5C]. Similarly, the protein level of NOTCH1 and hnRNPA1 also decreased [Figure 5D].
Figure 5.
The relationships between NOTCH1 signaling and hnRNPA1 in CLL. (A) The transcript level of NICD, HES1, and hnRNPA1 in NICD-overexpressed MEC-1 and JVM-3 was tested by qRT-PCR. (B) The protein level of hnRNPA1 in NICD-overexpressed MEC-1 and JVM-3 was tested by Western blot. (C) The transcript level of NOTCH1, HES1, and hnRNPA1 in NOTCH1-knockdown MEC-1 and JVM-3 was tested by qRT-PCR. (D) The protein level of NOTCH1 and hnRNPA1 in NOTCH1-knockdown MEC-1 and JVM-3 was tested by Western blot. CLL: Chronic lymphocytic leukemia; hnRNPA1: Heterogeneous nuclear ribonucleoprotein A1; NICD: NOTCH1 intracellular domain; qRT-PCR: Quantitative real-time polymerase chain reaction.
c-Myc as a key regulator between NOTCH1 and hnRNPA1 in CLL
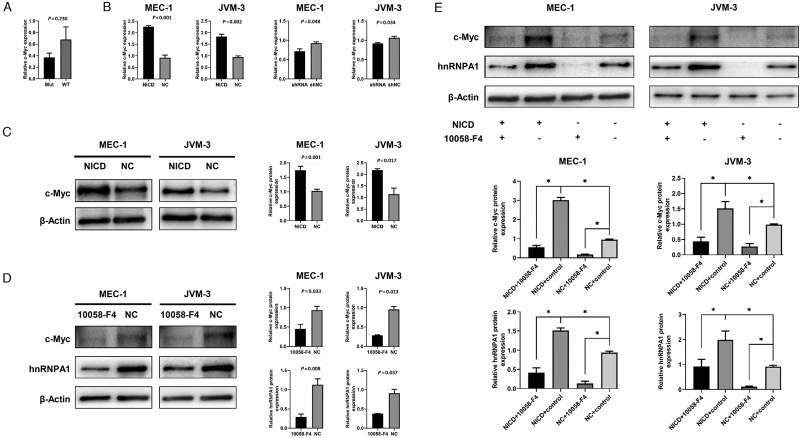
Previous research confirmed the direct NOTCH1 binding to the c-Myc promoter in CLL, which illustrated that c-Myc was a transcriptional target of NOTCH1.[8] In addition, a positive correlation was observed in the expression of c-Myc and hnRNPA1 in several solid tumors.[10,17] Thus, c-Myc was speculated as a vital regulator between NOTCH1 and hnRNPA1 in CLL. To verify this speculation, the transcript level of c-Myc between seven NOTCH1-mutated samples and eight NOTCH1-wt CLL samples were compared first [Figure 6A] and no difference between these two groups was found, which was consistent with the results of many previous researches.[8,18] Afterward, to explore the correlation of NOTCH1 signaling and c-Myc expression, the mRNA level of c-Myc in NICD-overexpressed and NOTCH1-knockdown cell lines was tested. The transcript level of c-Myc increased in NICD-overexpressed cells and declined in NOTCH1-knockdown cells [Figure 6B]. Similarly, NICD-overexpressed cells had higher c-Myc protein levels than the negative control [Figure 6C]. Furthermore, the relationship between c-Myc and hnRNPA1 expression was investigated and it was found that the protein level of c-Myc and hnRNPA1 simultaneously declined after treatment with 10058-F4 [Figure 6D]. Finally, a rescue experiment was conducted to verify the relationship among NOTCH1, c-Myc, and hnRNPA1. After 10058-F4 was added, the expression of c-Myc and hnRNPA1 was substantially downregulated in both NICD-overexpressed and corresponding negative control cell lines. The highest and lowest expression was observed in NICD-overexpressed cells treated with ethanol (as a control) and negative control cells treated with 10058-F4, respectively [Figure 6E].
Figure 6.
The relationships among NOTCH1, c-Myc, and hnRNPA1 in CLL. (A) The transcript level of c-Myc in primary NOTCH1-mutated (n = 7) and NOTCH1-wt (n = 8) CLL samples was detected by qRT-PCR. (B) The transcript level of c-Myc in NICD-overexpressed and NOTCH1-knockdown MEC-1 as well as JVM-3 detected by qRT-PCR. (C) The protein level of c-Myc in NICD-overexpressed MEC-1 and JVM-3 was tested by Western blot. (D)The protein level of c-Myc and hnRNPA1 after treatment with c-Myc inhibitor 10058-F4 in MEC-1 and JVM-3 tested by Western blot. (E)The protein level of c-Myc and hnRNPA1 after treatment with c-Myc inhibitor 10058-F4 in NICD-overexpressed MEC-1 and JVM-3 tested by Western blot. P < 0.05. CLL: Chronic lymphocytic leukemia; hnRNPA1: Heterogeneous nuclear ribonucleoprotein A1; NICD: NOTCH1 intracellular domain; qRT-PCR: Quantitative real-time polymerase chain reaction.
Discussion
NOTCH1 mutation is one of the most essential molecular biological aberrations in CLL.[2] In the era of chemo-immunotherapy, patients with NOTCH1 mutation showed a poor response to treatment and had an adverse prognosis.[19] In addition, NOTCH1 and B cell receptor signals could support each other to promote the development of CLL. NOTCH1 mutation enhanced the expression of the B cell receptor signal by activating the NOTCH1 pathway, which in turn facilitated the translation and activation of the NOTCH1 signal. The efficacy of ibrutinib was negatively correlated with NOTCH1 pathway activation. Based on this, NOTCH1 mutation was associated with ibrutinib resistance.[6] Therefore, comprehending the changes caused by NOTCH1 mutation allows us to provide better treatment for those patients. After analyzing the gene expression profile of five NOTCH1-mutated and five NOTCH1-wt samples, we found that several genes involved in RNA splicing were upregulated in NOTCH1-mutated samples, one of which was hnRNPA1. In this research, we investigated the role of hnRNPA1 in NOTCH1-mutated CLL cells. We validated the higher mRNA and protein level of hnRNPA1 in our primary NOTCH1-mutated CLL samples, which were consistent with gene expression profile data. The functions of hnRNPA1 were explored by the CCK-8 assay and the cell apoptosis analysis in hnRNPA1-overexpressed and knockdown cell lines. These results illustrated that hnRNPA1 overexpression could promote proliferation and inhibit apoptosis in CLL cells, which might partly account for the poor prognosis of NOTCH1 mutation. However, the reasons why hnRNPA1 overexpression led to CLL development remain unknown. As stated in previous studies, hnRNPA1 could regulate the splicing of pyruvate kinase by promoting the formation of pyruvate kinase isozymes M2 rather than M1 to affect metabolism.[20] In that case, hnRNPA1 mediated metabolism changes might be an underlying mechanism for CLL development. Hence RNA sequencing or microarray is urgently needed to further study the role of hnRNPA1 in NOTCH1-mutated CLL cells.
NOTCH1 mutation leads to impaired NICD degradation, which eventually results in sustained NOTCH1 signaling activation.[5] In this study, the mRNA and protein level of hnRNPA1 increased after NOTCH1 signaling was over-activated by transfection with NICD-overexpressed adenovirus vector. On the contrary, the expression of hnRNPA1 declined after NOTCH1 signaling was inhibited by transfection with NOTCH1-shRNA lentiviral vector. These results indicated the positive correlation between NOTCH1 signaling and hnRNPA1 expression.
Previous research reported that c-Myc was a direct transcriptional target of the NOTCH1 activation complex in CLL confirmed by chromatin immunoprecipitation assay.[8] In addition, c-Myc could regulate the expression of hnRNPA1 in prostate cancer and gliomas.[17,21] However, the relationship among NOTCH1 signaling, c-Myc, and hnRNPA1 in CLL remains unclear. Consistent with earlier studies, we failed to find a higher mRNA level of c-Myc in NOTCH1-mutated samples, which might be due to the rapid turnover of c-Myc.[8,22] Activating NOTCH1 signaling by overexpressing NICD could upregulate both transcript and protein levels of c-Myc. Due to low c-Myc protein expression in CLL cells, we only examined mRNA level in NOTCH1-knockdown cells and proved that c-Myc mRNA expression declined after the NOTCH1 signaling was inhibited. These results confirmed the previously mentioned relationship between NOTCH1 signaling and c-Myc expression in CLL.
It is known that c-Myc is a vital transcription factor that could regulate several target genes expression.[23] In the present research, inhibiting c-Myc by adding c-Myc inhibitor 10058-F4, which could interfere with c-Myc-Max interaction, could downregulate hnRNPA1 expression. Moreover, in NICD-overexpressed cells, the protein level of hnRNPA1 decreased after 10058-F4 was applied.
All these results demonstrated that c-Myc was a key regulator between NOTCH1 and hnRNPA1 in CLL.
Nevertheless, there are still certain limitations in our research. Wang et al[24] found that the untranslated part in NICD caused by NOTCH1 mutation exerted some unique functions beyond recognition and degradation. Therefore, applying full-length NICD to simulate NOTCH1 mutation status might cause a certain level of deviation. In this study, we only utilized one method to activate or inhibit NOTCH1 signaling. In further research, we could add ethylene diamine tetra-acetic acid and gamma-secretase inhibitor to activate and inhibit NOTCH1 signaling, respectively. Meanwhile, primary CLL cells could better reflect the real status and are more suitable for research compared with cell lines.[8]
RNA splicing has aroused increasing attention in recent years. With the accumulation of knowledge, the relationship between RNA splicing and tumor development is being gradually revealed.[25–27] Our research aimed to explore the molecular mechanisms of NOTCH1 mutation which focuses on splicing factors. In summary, we found that overexpression of c-Myc-related hnRNPA1 could promote proliferation and inhibit apoptosis in NOTCH1-mutated CLL cells. Nevertheless, further researches are needed to study which genes hnRNPA1 could change and what functions they could exert to better understand NOTCH1 mutation in CLL from the perspective of RNA splicing.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 81700193, 81970146, 82170186, and 81720108002), National Major Science and Technology Projects of China (No. 2018ZX09734-007), China Postdoctoral Science Foundation (No. 2021M691336), and Jiangsu Postdoctoral Science Foundation (No. 2021K083A).
Conflicts of interest
None.
Supplementary Material
Footnotes
How to cite this article: Zou Y, Tang H, Miao Y, Zhu H, Wang L, Fan L, Fu J, Xu W, Li J, Xia Y. Overexpression of c-Myc-dependent heterogeneous nuclear ribonucleoprotein A1 promotes proliferation and inhibits apoptosis in NOTCH1-mutated chronic lymphocytic leukemia cells. Chin Med J 2022;135:920–929. doi: 10.1097/CM9.0000000000002037
Supplemental digital content is available for this article.
References
- 1.Fernandez-Martinez JL, deAndres-Galiana EJ, Sonis ST. Genomic data integration in chronic lymphocytic leukemia. J Gene Med 2017; 19: doi: 10.1002/jgm.2936. [DOI] [PubMed] [Google Scholar]
- 2.Fabbri G, Dalla-Favera R. The molecular pathogenesis of chronic lymphocytic leukaemia. Nat Rev Cancer 2016; 16:145–162. doi: 10.1038/nrc.2016.8. [DOI] [PubMed] [Google Scholar]
- 3.Nadeu F, Delgado J, Royo C, Baumann T, Stankovic T, Pinyol M, et al. Clinical impact of clonal and subclonal TP53, SF3B1, BIRC3, NOTCH1, and ATM mutations in chronic lymphocytic leukaemia. Blood 2016; 127:2122–2130. doi: 10.1182/blood-2015-07-659144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D’Agaro T, Bittolo T, Bravin V, Dal Bo M, Pozzo F, Bulian P, et al. NOTCH1 mutational status in chronic lymphocytic leukaemia: clinical relevance of subclonal mutations and mutation types. Br J Haematol 2018; 182:597–602. doi: 10.1111/bjh.14843. [DOI] [PubMed] [Google Scholar]
- 5.Rosati E, Baldoni S, De Falco F, Del Papa B, Dorillo E, Rompietti C, et al. NOTCH1 aberrations in chronic lymphocytic leukemia. Front Oncol 2018; 8:229.doi: 10.3389/fonc.2018.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arruga F, Bracciama V, Vitale N, Vaisitti T, Gizzi K, Yeomans A, et al. Bidirectional linkage between the B-cell receptor and NOTCH1 in chronic lymphocytic leukemia and in Richter's syndrome: therapeutic implications. Leukemia 2020; 34:462–477. doi: 10.1038/s41375-019-0571-0. [DOI] [PubMed] [Google Scholar]
- 7.Arruga F, Gizdic B, Bologna C, Cignetto S, Buonincontri R, Serra S, et al. Mutations in NOTCH1 PEST domain orchestrate CCL19–driven homing of chronic lymphocytic leukemia cells by modulating the tumor suppressor gene DUSP22. Leukemia 2017; 31:1882–1893. doi: 10.1038/leu.2016.383. [DOI] [PubMed] [Google Scholar]
- 8.Pozzo F, Bittolo T, Vendramini E, Bomben R, Bulian P, Rossi FM, et al. NOTCH1-mutated chronic lymphocytic leukemia cells are characterized by a MYC-related overexpression of nucleophosmin 1 and ribosome-associated components. Leukemia 2017; 31:2407–2415. doi: 10.1038/leu.2017.90. [DOI] [PubMed] [Google Scholar]
- 9.Pozzo F, Bittolo T, Arruga F, Bulian P, Macor P, Tissino E, et al. NOTCH1 mutations associate with low CD20 level in chronic lymphocytic leukemia: evidence for a NOTCH1 mutation-driven epigenetic dysregulation. Leukemia 2016; 30:182–189. doi: 10.1038/leu.2015.182. [DOI] [PubMed] [Google Scholar]
- 10.Yao A, Xiang Y, Si YR, Fan LJ, Li JP, Li H, et al. PKM2 promotes glucose metabolism through a let-7a-5p/Stat3/hnRNP-A1 regulatory feedback loop in breast cancer cells. J Cell Biochem 2019; 120:6542–6554. doi: 10.1002/jcb.27947. [DOI] [PubMed] [Google Scholar]
- 11.Yang H, Zhu R, Zhao X, Liu L, Zhou Z, Zhao L, et al. Sirtuin-mediated deacetylation of hnRNP A1 suppresses glycolysis and growth in hepatocellular carcinoma. Oncogene 2019; 38:4915–4931. doi: 10.1038/s41388-019-0764-z. [DOI] [PubMed] [Google Scholar]
- 12.Huang MJ, Cheng YC, Liu CR, Lin S, Liu HE. A small-molecule c-Myc inhibitor, 10058-F4, induces cell-cycle arrest, apoptosis, and myeloid differentiation of human acute myeloid leukemia. Exp Hematol 2006; 34:1480–1489. doi: 10.1016/j.exphem.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 13.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 14.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005; 102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res 2004; 10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 16.Palomero T, Ferrando A. Oncogenic NOTCH1 control of MYC and PI3K: challenges and opportunities for anti-NOTCH1 therapy in T-cell acute lymphoblastic leukemias and lymphomas. Clin Cancer Res 2008; 14:5314–5317. doi: 10.1158/1078-0432.CCR-07-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luan W, Wang Y, Chen X, Shi Y, Wang J, Zhang J, et al. PKM2 promotes glucose metabolism and cell growth in gliomas through a mechanism involving a let-7a/c-Myc/hnRNPA1 feedback loop. Oncotarget 2015; 6:13006–13018. doi: 10.18632/oncotarget.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 2011; 475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou Y, Fan L, Xia Y, Miao Y, Wu W, Cao L, et al. NOTCH1 mutation and its prognostic significance in Chinese chronic lymphocytic leukemia: a retrospective study of 317 cases. Cancer Med 2018; 7:1689–1696. doi: 10.1002/cam4.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamada M, Suematsu M, Saya H. Pyruvate kinase M2: multiple faces for conferring benefits on cancer cells. Clin Cancer Res 2012; 18:5554–5561. doi: 10.1158/1078-0432.CCR-12-0859. [DOI] [PubMed] [Google Scholar]
- 21.Nadiminty N, Tummala R, Liu C, Lou W, Evans CP, Gao AC. NF-kappaB2/p52:c-Myc:hnRNPA1 pathway regulates expression of androgen receptor splice variants and enzalutamide sensitivity in prostate cancer. Mol Cancer Ther 2015; 14:1884–1895. doi: 10.1158/1535-7163.MCT-14-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones TR, Cole MD. Rapid cytoplasmic turnover of c-myc mRNA: requirement of the 3’ untranslated sequences. Mol Cell Biol 1987; 7:4513–4521. doi: 10.1128/mcb.7.12.4513-4521.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, metabolism, and cancer. Cancer Discov 2015; 5:1024–1039. doi: 10.1158/2159-8290.CD-15-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang S, Ge M, Cui J, Qiao Z, Chen X, Wu S, et al. Diminished interaction between mutant NOTCH1 and the NuRD corepressor complex upregulates CCL17 in chronic lymphocytic leukemia. Leukemia 2019; 33:2951–2956. doi: 10.1038/s41375-019-0526-5. [DOI] [PubMed] [Google Scholar]
- 25.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 2008; 40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 26.Vu NT, Park MA, Shultz MD, Bulut GB, Ladd AC, Chalfant CE. Caspase-9b interacts directly with cIAP1 to drive agonist-independent activation of NF-κB and lung tumorigenesis. Cancer Res 2016; 76:2977–2989. doi: 10.1158/0008-5472.CAN-15-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sveen A, Kilpinen S, Ruusulehto A, Lothe RA, Skotheim RI. Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene 2016; 35:2413–2427. doi: 10.1038/onc.2015.318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.