Abstract
Periploca forrestii Schltr (P. forrestii) is an edible medicinal herb with various health benefits such as treating antirheumatoid arthritis (RA), reducing inflammation, and preventing tumor growth. The active ingredients in P. forrestii responsible for its protective effect against RA, however, remain unknown. In this study, the active ingredient of P. forrestii and its potential mechanism of action against RA were investigated by network pharmacology and enrichment analysis. The methods included predicting target genes of P. forrestii, constructing a protein interaction network, and performing gene-ontology (GO) and Kyoto-encyclopedia of genes and genomes (KEGG) enrichment analysis. We discovered targets of RA through retrieval of OMIM and GeneCards public databases. Cardiac glycosides (CGs) are considered the primarily active ingredients of P. forrestii, and the target genes of GCs were discovered to be overlapped with relevant targets of RA using the Venn diagram. After that, prediction of relevant targets of P. forrestii was accomplished with a network pharmacology-based approach. Through the Venn diagram, we discovered 99 genes shared in the target genes of P. forrestii and RA. Gene enrichment analysis showed that the mechanisms of CGs against RA are associated with 55 signaling pathways, including endocrine resistance, Epstein-Barr virus infection, bladder cancer, prostate cancer, and coronavirus disease (COVID-19) signaling pathways. Coexpression analysis indicated ADSL, ATIC, AR, CCND1, MDM2, and HSP90AA1 as the hub genes between putative targets of P. forrestii-derived CGs and known therapeutic targets of RA. In conclusion, we clarified the mechanism of action of P. forrestii against RA, which would provide a basis for further understanding the clinical application of P. forrestii.
1. Introduction
Rheumatoid arthritis (RA), one of the most common auto-immune diseases, is characterized by the destruction of articular cartilage (AC), synovial hyperplasia, and pannus formation [1]. RA also ranks among the top ten causes of disability worldwide. It has been shown that people with RA are considerably burdened by irreversible deformities and even dysfunction of the joints, which adversely affect the quality of life of these individuals [1]. However, the development of effective prophylactic and therapeutic treatments has been hampered by the complexity of RA's etiology as well as the lack of clarity on its pathogenesis, which is thought to involve both genetic and environmental components [2–4] identifying as anticancer, antidiarrheal, antidepressant, anti-inflammatory, analgesic, and thrombolytic agents [5–10]. RA is a complex disorder, and due to the various properties, a variety of TCMs have played a vital role in controlling RA symptoms, thus assisting anti-RA and nonsteroidal anti-inflammatory drugs [11–14].
Periploca forrestii Schltr (P. forrestii, also known as Hei-Long-Gu or Hei-Gu-Teng) is derived from the rhizomes and roots of a vining shrub of Periploca (Asclepiadaceae). For thousands of years, P. forrestii has been regarded as one of the most widely used Chinese herbal medicines (CHM) in Guizhou, China. It is historically believed to possess numerous medicinal properties, including activating the meridians, improving blood circulation, dispelling wind, and removing dampness, as well as being used to treat RA, bruises, and tumors [15]. Until now, the increased interest in CHM has led researchers to isolate and identify over 100 active compounds from P. forrestii. This means that CHM, P. forrestii, and its periplocin derivatives can exert anti-inflammatory activity by regulating the immune response. For example, in rats with adjuvant arthritis (AIA), P. forrestii has been shown to inhibit local joint inflammation and bone destruction [16]. Additionally, studies in pharmacology have indicated that cardiac glycosides (CGs) are the major active ingredients of P. forrestii which have wound healing and antitumor properties [17–19]. Despite the fact that P. forrestii has been extensively investigated for its therapeutic use in a variety of human diseases, our knowledge of its mechanism of action in treating RA is still restricted. Therefore, this study was undertaken in order to investigate the possible mechanisms of action of P. forrestii-derived CGs when these compounds are applied in RA treatment.
2. Materials and Methods
2.1. Chemical Structure of Compounds
Firstly, based on literature mining, we gathered information on the main active ingredients of P. forrestii-derived CGs. Next, the chemical structures of these gathered active ingredients were retrieved from the PubChem database. As an open-archive, the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) consists of compound, substance, and bioassay primary databases.
2.2. Prediction of CGs Targets
Through the use of the pharmacophore mapping approach (PMA), we were able to identify potential therapeutic targets for these retrieved compounds in the PharmMapper (PM) database (http://www.lilab-ecust.cn/pharmmapper/) [20–22]. After that, the 3D chemical structures of these compounds were inputted into the PM database, and then twenty human targets with the highest matching scores were chosen for each of the compounds. Also, we removed probable CGs-related predicted targets that were repeated or nonstandard.
2.3. Analysis of Protein-Protein Interaction Network (PPIN) and Visualization
For predicted drug targets and known RA-related targets, an interaction network was selected with a score greater than or equal to 0.4 for the construction of a PPI network in a database, the Search Tool for Retrieving Interactive Genes (STRING). In light of the built interaction network, using the R package, we identified adjacent nodes with a greater number of connections to other nodes in our networks. For the purpose of constructing and visualizing interaction networks, Cytoscape software (Version 3.9.0) was used. The network displayed the active ingredients of CGs, and the nodes on the periphery of the network represented the key target genes of diseases and active components of CGs. As a result, the entire network demonstrates the link between drug-bioactive components-disease targets, while the mechanistic action of P. forrestii in RA treatment was explored by constructing this network.
2.4. SIN Targets Prediction for Treatment of RA
By searching for the term “rheumatoid arthritis” in the databases of GeneCards (https://www.genecards.org/) [23] and OMIM (https://www.omim.org/) [24], it was possible to retrieve therapeutic targets associated with RA. Moreover, using the Venn diagram, we were able to identify the overlapping genes between putative targets of P. forrestii and known therapeutic targets of RA.
2.5. Enrichment Analysis of Gene-Ontology (GO) and Kyoto-Encyclopedia of Genes and Genomes (KEGG) Pathway
A GO functional annotation was performed using the org.Hs.eg.db R package, while principal target genes of P. forrestii in RA treatment were converted into EntrezID for GO function enrichment analysis of the aforementioned genes (p < 0.05). Subsequently, we discovered key signaling pathways with overlapping targets and biological functions through KEGG pathway analysis, wherein p < 0.05.
3. Results
3.1. Target Genes that Associated with 10 Compounds and RA
As a result of a literature review, the 10 main chemical components of CGs obtained from P. forrestii were collected, and 654 corresponding target genes were determined using the pharmacophore model prediction tool. Afterward, we summarized and deduplicated the data in order to obtain 10 species with 279 target genes corresponding to the 10 main chemical components. Through GeneCards and the OMIM database, a total of 4821 target genes relating to RA were obtained. We performed a Venn diagram analysis on the target genes of CHM components and RA, resulting in 99 intersecting genes (Figure 1).
Figure 1.
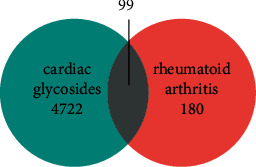
Using a Venn diagram analysis, 4722 target genes for the CHM component, GC, as well as 180 target genes for rheumatoid arthritis were identified, resulting in 99 overlapping genes.
3.2. Potential Signaling Pathways of CGs against RA
Based on these identified target genes, an analysis on a total of 29 GO term lists that were related to RA was performed (Figures 2(a) and 2(b)). These lists were combined with the top 20 results from a gene enrichment analysis of targets. As a result, this GO enrichment analysis showed biological processes and functions related to the activity of ligand activated transcription factors, the activity of nuclear-receptor, Hsp90 protein-binding, ubiquitin-like protein ligase-binding, RNA polymerase-II transcription initiation factor-binding, and so forth. There were also 55 signaling pathways that were identified in the KEGG analysis of P. forrestii (p < 0.05). Taken together, P. forrestii may treat RA by influencing endocrine resistance, Epstein-Barr virus infection, bladder cancer, prostate cancer, and COVID-19 signaling pathways, as well as other pathways (Figures 2(c) and 2(d). Table 1).
Figure 2.
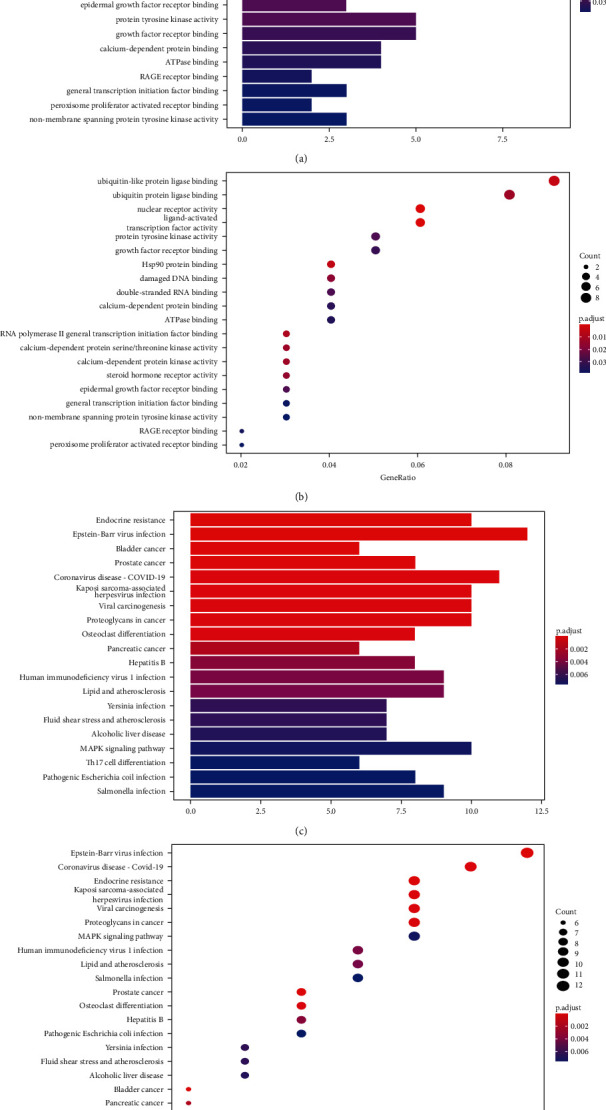
The top 20 gene-ontology (GO) categories of overlapping genes between RA and cardiac glycosides (CGs) were presented in a histogram and bubble diagram, respectively (a, b). The top 20 KEGG signaling pathways of overlapping genes were presented in a histogram and bubble diagram, respectively (c, d).
Table 1.
Top 10 target genes related to the occurrence and development of rheumatoid arthritis (RA) revealed by enrichment set analysis.
| Pathway ID | Description | Gene ID | p.adjust |
|---|---|---|---|
| hsa01522 | Endocrine resistance | MMP2/NOTCH1/HBEGF/E2F1/MAPK9/ESR1/ERBB2/CCND1/MDM2/ESR2 | 6.83E-06 |
| hsa05169 | Epstein-barr virus infection | B2M/CDK2/BTK/RAC1/E2F1/MAPK9/Table1/CCND1/IKBKG/EIF2AK2/MDM2/IRAK4 | 5.51E-05 |
| hsa05219 | Bladder cancer | MMP2/HBEGF/E2F1/ERBB2/CCND1/MDM2 | 1.83E-04 |
| hsa05215 | Prostate cancer | CDK2/E2F1/ERBB2/CCND1/AR/IKBKG/MDM2/HSP90AA1 | 2.30E-04 |
| hsa05171 | Coronavirus disease-COVID-19 | F13A1/C2/HBEGF/F2/MAPK9/IKBKG/EIF2AK2/IRAK4/FCGR2A/RPS3/IL6ST | 6.10E-04 |
| hsa05167 | Kaposi sarcoma-associated herpesvirus infection | IFNGR1/MAPKAPK2/HCK/RAC1/E2F1/MAPK9/CCND1/IKBKG/EIF2AK2/IL6ST | 6.30E-04 |
| hsa05203 | Viral carcinogenesis | CDK2/MAPKAPK2/RAC1/CCND1/DDB1/IKBKG/EIF2AK2/MDM2/HDAC6/IL6ST | 7.63E-04 |
| hsa05205 | Proteoglycans in cancer | MMP2/VAV3/FLNB/HBEGF/RAC1/PPP1CB/ESR1/ERBB2/CCND1/MDM2 | 7.63E-04 |
| hsa04380 | Osteoclast differentiation | IFNGR1/BTK/RAC1/MAPK9/NCF2/Table1/IKBKG/FCGR2A | 7.94E-04 |
| hsa05212 | Pancreatic cancer | RAC1/E2F1/MAPK9/ERBB2/CCND1/IKBKG | 2.03E-03 |
3.3. Hub Genes of CGs against RA
We used the online STRING database to construct a protein-protein interaction (PPI) network of overlapped targets with 4722 RA-related genes and 180 CGs-related genes (Figure 3(a)). Before it was used to visualize the overlapping genes between CGs and RA, the PPI network was constructed utilizing the STRING database and Cytoscape.
Figure 3.
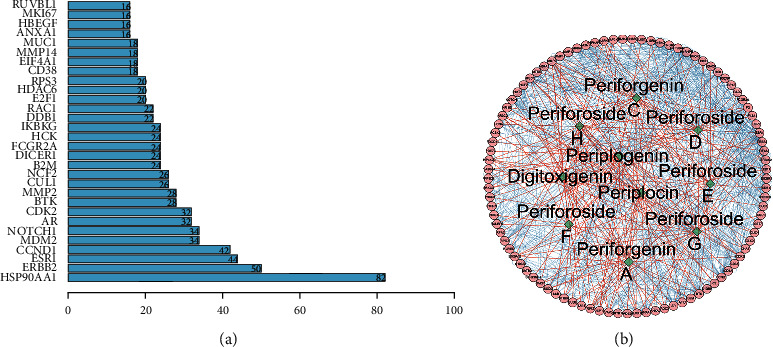
Key gene nodes in the rheumatic arthritis (RA)-related gene protein network (a). The interaction network of cardiac glycosides (CGs) and disease-related genes (b). Among them, the green diamonds represent the active components of CGs, whereas the pink circles represent key target genes, and the pink lines denote the relationship between each target gene.
3.4. Construction of Coexpression Network
Within the PPI network, the circle denotes the size of the degree value; a higher degree value corresponds to a transition from the color red to the color blue. The thickness of the edge represented the size of the combined score; a thicker edge suggests that the combined score has a bigger value. ADSL, ATIC, AR, CCND1, MDM2, and HSP90AA1 were among the key protein nodes, which were produced based on the credit scores of node interactions (Figure 4).
Figure 4.
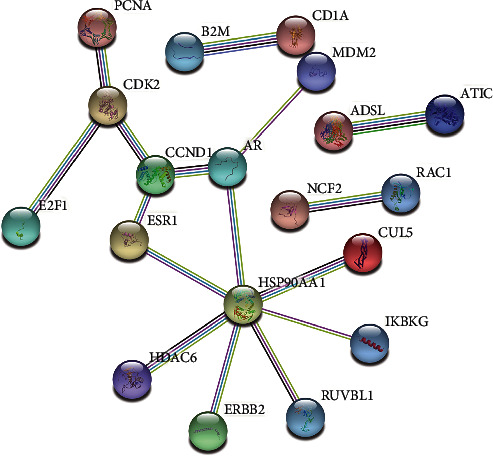
Coexpression network of related genes and target proteins.
4. Discussion
As a result of their chemical diversity and advantages such as simple availability and bioprocessing, scientists have always been interested in natural compounds [25–27]. P. forrestii has been reported to contain numerous bioactive compounds, including steroids, flavonoids, and phenylpropanoids [25]. These active molecules have previously shown a variety of functions, including antiproliferative, antioxidant, antinociceptive, and palliative effects [28–33]. In addition, TCMs containing these bioactive compounds have been demonstrated to be effective against COVID-19, showing their antivirus properties [34]. No doubt, P. forrestii and its extracts are capable of exhibiting these similar properties. Among the identified compounds in P. forrestii extracts, CGs are considered to be the main bioactive component. The CGs are mainly obtained from various plant and amphibian sources, such as members of the genus Bufo [35, 36]. It is widely accepted that heart disease and cancer treatment can benefit greatly from the application of these GCs, with some undergoing clinical trials (phase I and II) for solid tumor therapy [37–39].
In this study, the pharmacophore model was used to predict active compounds of CGs, and a network of “drug-active ingredient-disease” was built, in which we attempted an analysis of the CGs' mechanism of action. Besides, a coexpression network was established using the STRING database and used to identify key target genes. Furthermore, we performed function analysis of GO and KEGG pathway to reveal the related signaling pathways.
In light of our findings, enrichment analysis indicated that the treatment of RA with P. forrestii may be related to endocrine resistance, Epstein-Barr virus infection, bladder cancer, prostate cancer, and COVID-19 signaling pathways, suggesting that P. forrestii might treat RA by altering these signaling pathways. There is a long-established association between RA and cancer, based on studies that have demonstrated an increased lymphoma risk in patients with RA [40]. Though not all types of cancer are equally at risk, lung cancer and lymphoma incidences are higher among RA patients than in the general population as demonstrated by standardized incidence rates [41]. Therefore, CGs participate in a wide range of cancer-related pathways that are involved in RA treatment, which provides a novel perspective for the combination therapy. Not just cancer, but also COVID-19 was discovered to be connected with RA. According to the findings of Williamson and colleagues [42], people with RA, systemic lupus erythematosus, or psoriasis (combined) were more likely to die of COVID-19. Other researchers have also observed that some common treatment strategies like cytokine inhibition were effective in both COVID-19 and RA [43]. Furthermore, it was found that analysis of the GO function enrichment of core genes was primarily concentrated in the activity of ligand activated transcription factor, nuclear-receptor, Hsp90 protein-binding, ubiquitin-like protein ligase-binding, and RNA polymerase-II transcription initiation factor-binding. However, the current research on P. forrestii did not report on this specific aspect, which needs further investigation. Likewise, our coexpression analysis revealed that ADSL, ATIC, AR, CCND1, MDM2, and HSP90AA1 were the key genes that linked potential targets of P. forrestii to known therapeutic targets within RA.
Through these molecular mechanisms, CGs may play an important role in treating RA, thereby providing a theoretical foundation for future research and clinical application of these organic compounds. Also, based on their antitumor property, GCs have been increasingly studied, exhibiting cytotoxic effects against some tumor types, including prostate cancer [44, 45], melanoma [37], and lung cancer [46–48]. In view of this, the CGs' potential mechanism and potential in vivo application deserve further study. On the other hand, although CGs have shown some excellent safety profiles and efficacy, future clinical trials will need to confirm the long-term efficacy and safety of their extracts and chemical components due to the complexity of P. forrestii compounds.
5. Conclusion
In summary, a network of essential genes was established using the network pharmacology-based approach, connecting the putative therapeutic targets of P. forrestii to the known targets of RA. This network screened out six hub genes, including ADSL, ATIC, AR, CCND1, MDM2, and HSP90AA1. The significant biological processes, molecular functions, cellular components, and KEGG pathways were obtained by performing GO and KEGG pathway analysis. The hub genes and enrichment analysis results indicated that the primary active ingredients, GCs, appeared frequently in the application to treat RA, cancer, and COVID-19. The therapeutic effect of GCs in treating RA is mainly by regulating ligand activated transcription factors, activity of nuclear-receptor, Hsp90 protein-binding, ubiquitin-like protein ligase-binding, and RNA polymerase-II transcription initiation factor-binding. These pathways were most commonly observed in the host's immune inflammatory response, oxidative stress, cell repair, and cell migration. These findings provide a basis for understanding the further clinical application of P. forrestii and its ingredients, GCs, in the treatment of RA.
Acknowledgments
This work was supported by National Natural Science Foundation of China (82160869 and 81804127), Guizhou Province Science and Technology Plan Project (Qian Ke He Platform Talents (2016) 5650 and (2020) 2202), Qian Ke He Support (2021) general 006; Qian Ke He Basis (2020]1Y361), and National Key Research and Development Program: Research on Modernization of Traditional Chinese Medicine (2017YFC1703904).
Contributor Information
Hongyan Wu, Email: wu.hy@163.com.
Wukai Ma, Email: fsmymwk@163.com.
Data Availability
The data presented in this study are available on request from the corresponding author.
Disclosure
Qiuyi Wang and Xueming Yao should be considered co-first authors.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
Authors' Contributions
Yi Ling, Ying Huang, Changming Chen, Lei Hou, Yutao Yang, Hongyan Wu, and Wukai Ma provided important suggestions about designing and performing the overall research. Qiuyi Wang and Xueming Yao collected and analyzed the data, and wrote the paper. Qiuyi Wang and Xueming Yao contributed equally to this work.
References
- 1.Sokka T., Kautiainen H., Pincus T., et al. Disparities in rheumatoid arthritis disease activity according to gross domestic product in 25 countries in the QUEST-RA database. Annals of the Rheumatic Diseases . 2009;68(11):1666–72. doi: 10.1136/ard.2009.109983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deighton C. M., Walker D. J., Griffiths I. D., Roberts D. F. The contribution of HLA to rheumatoid arthritis. Clinical Genetics . 2008;36(3):178–82. doi: 10.1111/j.1399-0004.1989.tb03185.x. [DOI] [PubMed] [Google Scholar]
- 3.Deane K. D., Demoruelle M. K., Kelmenson L. B., Kuhn K. A., Norris J. M., Holers V. M. Genetic and environmental risk factors for rheumatoid arthritis. Best Practice & Research Clinical Rheumatology . 2017;31(1):3–18. doi: 10.1016/j.berh.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang K., Yang S. M., Kim S. H., Han K. H., Park S. J., Shin J. I. Smoking and rheumatoid arthritis. International Journal of Molecular Sciences . 2014;15(12):22279–22295. doi: 10.3390/ijms151222279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmed S., Khan H., Aschner M., Mirzae H., Küpeli Akkol E., Capasso R. Anticancer potential of furanocoumarins: mechanistic and therapeutic aspects. International Journal of Molecular Sciences . 2020;21(16):p. 5622. doi: 10.3390/ijms21165622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan M. F., Kader F. B., Arman M., et al. Pharmacological insights and prediction of lead bioactive isolates of Dita bark through experimental and computer-aided mechanism. Biomedicine & Pharmacotherapy . 2020;131 doi: 10.1016/j.biopha.2020.110774.110774 [DOI] [PubMed] [Google Scholar]
- 7.Freitas M. A., Vasconcelos A., Gonçalves E. C. D., et al. Involvement of opioid system and TRPM8/TRPA1 channels in the antinociceptive effect of spirulina platensis. Biomolecules . 2021;11(4):p. 592. doi: 10.3390/biom11040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goni O., Khan M. F., Rahman M. M., et al. Pharmacological insights on the antidepressant, anxiolytic and aphrodisiac potentials of Aglaonema hookerianum Schott. Journal of Ethnopharmacology . 2021;268 doi: 10.1016/j.jep.2020.113664.113664 [DOI] [PubMed] [Google Scholar]
- 9.Akter A., Islam F., Bepary S., et al. CNS depressant activities of Averrhoa carambola leaves extract in thiopental-sodium model of Swiss albino mice: implication for neuro-modulatory properties. Biologia . 2022;77(5):1337–1346. doi: 10.1007/s11756-022-01057-z. [DOI] [Google Scholar]
- 10.Tagde P., Tagde P., Islam F., et al. The multifaceted role of curcumin in advanced nanocurcumin form in the treatment and management of chronic disorders. Molecules . 2021;26(23):p. 7109. doi: 10.3390/molecules26237109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hossain S., Urbi Z., Karuniawati H., et al. Andrographis paniculata (burm. F.) wall. Ex nees: an updated review of phytochemistry, antimicrobial pharmacology, and clinical safety and efficacy. Life (Basel) . 2021;11(4):p. 348. doi: 10.3390/life11040348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farooq U., Khan T., Shah S. A., et al. Isolation, characterization and neuroprotective activity of folecitin: an in vivo study. Life (Basel) . 2021;11(8):p. 825. doi: 10.3390/life11080825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakraborty A. J., Mitra S., Tallei T. E., et al. Bromelain a potential bioactive compound: a comprehensive overview from a pharmacological perspective. Life (Basel) . 2021;11(4):p. 317. doi: 10.3390/life11040317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moni J. N. R., Adnan M., Tareq A. M., et al. Therapeutic potentials of syzygium fruticosum fruit (seed) reflected into an array of pharmacological assays and prospective receptors-mediated pathways. Life (Basel) . 2021;11(2):p. 155. doi: 10.3390/life11020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang Y., Li B. Flora of China 63 . Beijing: Science Press; 1977. [Google Scholar]
- 16.Liu Y., Li M., He Q., Yang X., Ruan F., Sun G. Periploca forrestii saponin ameliorates murine CFA-induced arthritis by suppressing cytokine production. Mediators of Inflammation . 2016;2016:1–11. doi: 10.1155/2016/7941684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y., Liu Y. B., Yu S. S., et al. Cytotoxic cardenolides from the stems of Periploca forrestii. Steroids . 2012;77(5):375–381. doi: 10.1016/j.steroids.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 18.Chen L., Li J., Ke X., et al. Chemical profiling and the potential active constituents responsible for wound healing in Periploca forrestii Schltr. Journal of Ethnopharmacology . 2018;224:230–241. doi: 10.1016/j.jep.2018.04.023. [DOI] [PubMed] [Google Scholar]
- 19.Chen L., Tang S., Li X., et al. A review on traditional usages, chemical constituents and pharmacological activities of periploca forrestii schltr. Journal of Ethnopharmacology . 2021;271 doi: 10.1016/j.jep.2021.113892.113892 [DOI] [PubMed] [Google Scholar]
- 20.Liu X., Ouyang S., Yu B., et al. PharmMapper server: a web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Research . 2010;38(suppl_2):W609–W614. doi: 10.1093/nar/gkq300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X., Pan C., Gong J., Liu X., Li H. Enhancing the enrichment of pharmacophore-based target prediction for the polypharmacological profiles of drugs. Journal of Chemical Information and Modeling . 2016;56(6):1175–1183. doi: 10.1021/acs.jcim.5b00690. [DOI] [PubMed] [Google Scholar]
- 22.Wang X., Shen Y., Wang S., et al. PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Research . 2017;45(W1):W356–W360. doi: 10.1093/nar/gkx374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen C., Peng H., Huang X., et al. Genome-wide profiling of DNA methylation and gene expression in esophageal squamous cell carcinoma. Oncotarget . 2016;7(4):4507–4521. doi: 10.18632/oncotarget.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKusick V. A. Mendelian inheritance in man and its online version, OMIM. The American Journal of Human Genetics . 2007;80(4):588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Demain A. L., Vaishnav P. Natural products for cancer chemotherapy. Microbial Biotechnology . 2011;4(6):687–699. doi: 10.1111/j.1751-7915.2010.00221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uddin Chy M. N., Adnan M., Chowdhury M. R., et al. Central and peripheral pain intervention by Ophiorrhizarugosa leaves: potential underlying mechanisms and insight into the role of pain modulators. Journal of Ethnopharmacology . 2021;276 doi: 10.1016/j.jep.2021.114182.114182 [DOI] [PubMed] [Google Scholar]
- 27.Bari M. S., Khandokar L., Haque E., et al. Ethnomedicinal uses, phytochemistry, and biological activities of plants of the genus Gynura. Journal of Ethnopharmacology . 2021;271 doi: 10.1016/j.jep.2021.113834.113834 [DOI] [PubMed] [Google Scholar]
- 28.Fernández J., Silván B., Entrialgo-Cadierno R., et al. Antiproliferative and palliative activity of flavonoids in colorectal cancer. Biomedicine & Pharmacotherapy . 2021;143 doi: 10.1016/j.biopha.2021.112241.112241 [DOI] [PubMed] [Google Scholar]
- 29.Rahman M. M., Reza A. A., Khan M. A., et al. Unfolding the apoptotic mechanism of antioxidant enriched-leaves of Tabebuia pallida (lindl.) miers in EAC cells and mouse model. Journal of Ethnopharmacology . 2021;278 doi: 10.1016/j.jep.2021.114297.114297 [DOI] [PubMed] [Google Scholar]
- 30.Rahman M., Majumder S., Akter F., Islam F., Shahriar M., Alam J. Pre-clinical investigation of analgesic, anti-diarrheal and CNS depressant effect of Pterocarpus indicus in Swiss albino mice. Jordan Journal of Pharmaceutical Sciences . 2021;14(1) [Google Scholar]
- 31.Akter A., Begh M. Z. A., Islam F., Afroz T., Hossain M. S., Faysal M. Phytochemical screening and evaluation of thrombolytic, analgesic and antidiarrhoeal activity of the leaves of cucumis sativus linn.(cucurbitaceae) of methanolic extracts. Journal of Pharmaceutical Sciences and Research . 2020;12(3):448–451. [Google Scholar]
- 32.Islam F., Azad A., Faysal M., Islam S., Sugandha N. J., Saha S. Phytochemical investigation and comparative anthelmintic activity of between methanol and acetone extract of limonia acidissima l (fruit peel) Fruit peel) Pharmacologyonline . 2019;2:241–246. [Google Scholar]
- 33.Fahad F. I., Barua N., Islam M. S., et al. Investigation of the pharmacological properties of lepidagathis hyalina nees through experimental approaches. Life (Basel) . 2021;11(3):p. 180. doi: 10.3390/life11030180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tallei T. E., Fatimawali, Niode N. J., et al. A comprehensive review of the potential use of green tea polyphenols in the management of COVID-19. Evidence-based Complementary and Alternative Medicine . 2021;2021:1–13. doi: 10.1155/2021/7170736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makarevich I. F., Kovalev S. V. Cardiac glycosides from Strophanthus kombe. Chemistry of Natural Compounds . 2006;42(2):189–93. doi: 10.1007/s10600-006-0075-9. [DOI] [Google Scholar]
- 36.Jian‐Xin C., Shi‐De L. Two novel types of cardiac glycosides from Parepigynum funingense and the possible biogenesis. Chinese Journal of Chemistry . 2005;23(7):905–912. doi: 10.1002/cjoc.200590905. [DOI] [Google Scholar]
- 37.Menger L., Vacchelli E., Kepp O., et al. Trial watch: cardiac glycosides and cancer therapy. OncoImmunology . 2013;2(2) doi: 10.4161/onci.23082.e23082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hou Y., Shang C., Meng T., Lou W. Anticancer potential of cardiac glycosides and steroid-azole hybrids. Steroids . 2021;171 doi: 10.1016/j.steroids.2021.108852.108852 [DOI] [PubMed] [Google Scholar]
- 39.Mekhail T., Kaur H., Ganapathi R., Budd G. T., Elson P., Bukowski R. M. Phase 1 trial of Anvirzel in patients with refractory solid tumors. Investigational New Drugs . 2006;24(5):423–427. doi: 10.1007/s10637-006-7772-x. [DOI] [PubMed] [Google Scholar]
- 40.Isomaki H. A., Hakulinen T., Joutsenlahti U. Excess risk of lymphomas, leukemia and myeloma in patients with rheumatoid arthritis. Journal of Chronic Diseases . 1978;31(11):691–696. doi: 10.1016/0021-9681(78)90071-1. [DOI] [PubMed] [Google Scholar]
- 41.Simon T. A., Thompson A., Gandhi K. K., Hochberg M. C., Suissa S. Incidence of malignancy in adult patients with rheumatoid arthritis: a meta-analysis. Arthritis Research and Therapy . 2015;17(1):p. 212. doi: 10.1186/s13075-015-0728-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williamson E. J., Walker A. J., Bhaskaran K., et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature . 2020;584(7821):430–436. doi: 10.1038/s41586-020-2521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schett G., Manger B., Simon D., Caporali R. COVID-19 revisiting inflammatory pathways of arthritis. Nature Reviews Rheumatology . 2020;16(8):465–470. doi: 10.1038/s41584-020-0451-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeh J. Y., Huang W. J., Kan S. F., Wang P. S. Inhibitory effects of digitalis on the proliferation of androgen dependent and independent prostate cancer cells. The Journal of Urology . 2001;166(5):1937–1942. doi: 10.1097/00005392-200111000-00087. [DOI] [PubMed] [Google Scholar]
- 45.Platz E. A., Yegnasubramanian S., Liu J. O., et al. A novel two-stage, transdisciplinary study identifies digoxin as a possible drug for prostate cancer treatment. Cancer Discovery . 2011;1(1):68–77. doi: 10.1158/2159-8274.cd-10-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaushik V., Yakisich J. S., Azad N., et al. Anti-tumor effects of cardiac glycosides on human lung cancer cells and lung tumorspheres. Journal of Cellular Physiology . 2017;232(9):2497–2507. doi: 10.1002/jcp.25611. [DOI] [PubMed] [Google Scholar]
- 47.Elbaz H. A., Stueckle T. A., Wang H. Y. L., et al. Digitoxin and a synthetic monosaccharide analog inhibit cell viability in lung cancer cells. Toxicology and Applied Pharmacology . 2012;258(1):51–60. doi: 10.1016/j.taap.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ayogu J. I., Odoh A. S. Prospects and therapeutic applications of cardiac glycosides in cancer remediation. ACS Combinatorial Science . 2020;22(11):543–553. doi: 10.1021/acscombsci.0c00082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.


