STRUCTURED ABSTRACT
Objective:
To establish a liquid-biopsy assay to predict response to neoadjuvant therapy (NAT) in esophageal squamous cell carcinoma (ESCC) patients.
Summary Background Data:
Pretreatment prediction of resistance to NAT is of great significance for the selection of treatment options in ESCC patients. In this study, we comprehensively translated tissue-based microRNA (miRNA) and messenger RNA (mRNA) expression biomarkers into a liquid biopsy assay.
Methods:
We analyzed 186 clinical ESCC samples, which included 128 formalin-fixed paraffin-embedded and a matched subset of 58 serum samples, from 2 independent institutions. We performed quantitative reverse-transcription polymerase chain reaction, and developed a resistance-prediction model using the logistic regression analyses.
Results:
We first evaluated the potential of 4-miRNAs and 3-mRNAs panel, which robustly predicted resistance to NAT (area under the curve [AUC]: 0.85). Moreover, addition of tumor size to this panel increased predictive potential to establish a combination signature (AUC: 0.92). We successfully validated this signature performance in independent cohort, and our model was more accurate when the signature was combined with clinical predictors (AUC: 0.81) to establish a NAT resistance risk (NATRR) model. Finally, we successfully translated our NATRR model into a liquid biopsy assay (AUC: 0.78), and a multivariate regression analysis revealed this model as an independent predictor for response to NAT (odds ratio: 6.10; P < 0.01).
Conclusions:
We successfully developed a liquid biopsy-based assay that allows robust prediction of response to NAT in ESCC patients, and our assay provides fundamentals of developing precision-medicine.
Keywords: esophageal squamous cell carcinoma, neoadjuvant therapy, predictive biomarker, biomarker signature, chemo-resistance
Mini-Abstract
Pretreatment prediction of resistance to neoadjuvant therapy (NAT) is of great significance for the selection of treatment options in esophageal squamous cell carcinoma (ESCC) patients. In this study, we analyzed 186 clinical ESCC samples from multiple independent institutions, and successfully developed a liquid biopsy assay to allow robust prediction of response to NAT in ESCC patients.
INTRODUCTION
Esophageal cancer is the tenth most common cause of cancer and the sixth leading cause of cancer-related deaths, with 604,100 new cases and 544,076 cancer deaths in 2020 [1, 2]. There are two histologic subtypes of esophageal cancer – esophageal squamous cell carcinoma (ESCC) and adenocarcinoma (EAC). ESCC is one of the most aggressive and lethal subtypes, with a 5-year overall survival (OS) rate of less than 20% in United States [3, 4]. Surgery, which primarily involves esophagectomy and lymphadenectomy, remains the most attractive curative treatment in ESCC patients; however, in spite of the use of such treatment modalities, the survival outcomes have not significantly improved for decades [5]. Recent randomized controlled trials have indicated that addition of neoadjuvant therapy (NAT) might help improve the long-term survival in ESCC patients [6, 7], and accordingly the National Comprehensive Cancer Network (NCCN) Guidelines now recommend neoadjuvant chemotherapy (NAC) and neoadjuvant chemoradiotherapy (NACRT) in patients with advanced disease [8].
Nevertheless, the treatment outcomes of NAT have been heterogeneous and only 40–60% of ESCC patients with resected tumors demonstrate some histopathological response, while only 20–30% of patients achieve a complete pathological response [7, 9, 10]. In other words, non-responder patients harboring more than 50% of residual cancer after NAT, do not benefit from such therapies vis-à-vis upfront surgery. Unfortunately, the clinical challenge in such non-responsive patients is further exacerbated because these patients often miss out on the opportunity to undergo radical surgery due to their disease progression while receiving NAT, resulting in an overall worse prognosis. Furthermore, NAT in ESCC patients is often accompanies by significant undesirable effects, with more than 50% of patients experiencing grade 3 or 4 toxicities [11–13]. To make matters worse, some of the patients lose their ability to tolerate curative surgeries due to irreversible and severe adverse effects of NAT and succumb to death from the serious adverse effects of such treatments. As we usher into the era of precision oncology, it is imperative that patients are offered treatment that have higher likelihood of benefit and minimal toxicity; hence, prediction of therapeutic resistance to NAT prior to initiation of treatment is of great significance for appropriate selection of the subset of ESCC patients that stand to derive clinical benefit from such treatments.
In recent years, attempts have been made to develop various types of molecular biomarkers, including genes, microRNAs (miRNAs), DNA methylation, and somatic copy-number alterations, to predict response to cancer therapies and assess survival outcomes. In this context, a panel of tissue-based miRNA and messenger RNA (mRNAs) biomarkers that predicted the response to NACRT in ESCC patients were recently reported [14, 15]. While these biomarkers seem promising, their clinical significance remains unclear. Therefore, a systematic and comprehensive validation of these biomarkers in multiple and independent cohorts of ESCC patients might reveal the true predictive potential of these markers. Furthermore, if possible, the translation of these biomarkers into a blood-based liquid biopsy would be clinically attractive as it will obviate the need for availability of tissue, can overcome intratumor heterogeneity in cancer cells, and could offer a platform for non-invasive assay for predicting therapeutic response to NAT in ESCC patients.
Accordingly, in this study, we evaluated the feasibility of developing transcriptomic biomarkers (genes and miRNAs) for predicting response to NAT, as well as their translation into a liquid biopsy in ESCC patients. This transcriptomic signature was first systematically validated in two independent clinical cohorts, followed by its clinical performance in pre-treatment blood specimens. In conclusion, we have successfully established a liquid biopsy-based risk-prediction model for NAT in ESCC patients, which allows simple, facile and non-invasive pretreatment selection approach in patients suffering from this malignancy.
MATERIALS AND METHODS
Patient cohorts
This study included analysis of a total of 186 clinical specimens from ESSC patients, which involved 128 formalin-fixed paraffin-embedded (FFPE) specimens and 58 pretreatment serum samples to evaluate the predictive accuracy of the mRNA and miRNA biomarkers [14, 15]. For the initial training of biomarkers, we analyzed RNA derived from FFPE specimens from 43 ESCC patients enrolled at the Kumamoto University, Japan, between 2009 and 2015 (the clinical training cohort). For the clinical validation of the signature, we examined another independent cohort of 85 patients enrolled at Nagoya University, Japan, between 2008 and 2015 (the clinical validation cohort). Finally, for the performance evaluation of these tissue-based biomarkers in the blood specimens, we analyzed pretreatment serum specimens from 58 patients (the performance evaluation cohort), which included a subset of patients for which matched serum specimens were available from the clinical validation cohort. The clinicopathological characteristics of each clinical cohort are shown in Supplementary table S1.
All relevant clinical data were collected from a clinical database and/or electronic medical records at each enrolling institution. These data included patient demographics, comorbidities, and survival outcomes. All tumors were diagnosed histologically as ESCC and classified according to the Union for International Cancer Control (UICC) TNM classification of Malignant Tumors version 7. Exclusion criteria included the presence of distant metastasis or tumor histology other than ESCC. All the enrolled ESCC patients were evaluated by computed tomography (CT) and esophago-gastroduodenostomy before any cancer treatment and after NAT (if applicable). Follow-up was until patient death or January 2017 in the clinical training cohort and January 2018 in the clinical validation cohort. A written informed consent was obtained from each patient, and the study was conducted in compliance with the Declaration of Helsinki and was approved by the Institutional Review Boards of all participating institutions.
Neoadjuvant therapy and histologic evaluation
The NAT regimens were 5-fluorouracil (5-FU)- and cisplatin-based chemotherapy, as described previously [16–18]. Each NAT regimen was as follows; 3 cycles of DCF (docetaxel, 70 mg/m2/day, day 1; cisplatin, 70 mg/m2/day, day 1; 5-FU, 750mg/m2/day, days 1–5; every 3 weeks) [16], 2 cycles of FP (5-FU, 800mg/m2/day, days 1–5; cisplatin, 80mg/m2/day, day 1; every 3 weeks) [17], 2 cycles of SP (S-1, 40–80mg/m2/day, days 1–14; CDDP, 75mg/m2/day, day1; every 4 weeks) [18]. The radiotherapy was delivered with 6–10 MV photons to a total dose of 41.4 Gy in 23 fractions over 5 weeks [16]. After NAT, all patients underwent curative esophagectomy and lymphadenectomy within 3 to 5 weeks after completion of NAT.
After NAT and surgery, esophagectomy specimens were macroscopically assessed by pathologists, as previously described [19, 20]. Resected surgical specimens with no observable residual cancer cells at both the primary tumor site and the resected lymph nodes were defined as pathological complete response. Patients with 1–50% residual tumor were deemed to have partial response, and those with more than 50% residual tumors were considered non-response. Pathological complete and partial responders were categorized as responders of NAT in this study.
RNA extraction from FFPE and serum specimens
Total RNA extraction from FFPE and serum specimens was performed, as described previously [21–23]. The RNA was extracted from 10um-thick FFPE surgical specimens by microdissection for enrichment of cancer cells using an AllPrep DNA/RNA FFPE kit (Qiagen, Hilden, Germany). For serum samples, the RNA was extracted using the Qiagen miRNeasy Kit (Qiagen, Hilden, Germany). 200uL of serum was centrifuged at 3000g for 5 minutes to remove cell debris. Next, 200uL of the supernatant was lysed in 5 times the volume of QIAzol solution (Qiagen). Total RNA was subsequently enriched and purified following the manufacturer’s instructions. Extracted RNA from FFPE and serum specimens was reversely transcribed to complementary DNA (cDNA) before polymerase chain reaction (PCR) assays. For mRNA, synthesis of cDNA was performed using a high-capacity cDNA Reverse Transcription Kit (Thermo Fischer Scientific, Waltham, MA). For miRNA, a miRCURY LNA RT Kit (Qiagen) was used to synthesize cDNA.
Real-time quantitative reverse-transcription polymerase chain reaction assays
Real-time quantitative reverse-transcription polymerase chain reaction (qRT-PCR) assays was performed using a SensiFAST SYBR Lo-ROX Kit (Bioline, London, United Kingdom) and the QuantStudio 6/7 Flex RT-PCR System (Applied Biosystems, Foster City, CA). The miR-16–5p and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were used as an internal control for the normalization of miRNA and mRNA expression, respectively. The delta Ct method, where delta Ct is the difference in Ct values between the abundance of target transcripts and the internal control, was used for quantification. Normalized values were further log10 transformed [24–27]. The primers for mRNAs used in the present study were described in Supplementary table S2. The primers for miRNAs used in this study were purchased from Thermo Fisher Scientific (Catalog No: 4427975).
Statistical analysis
Two-sided Student’s t-test was used to analyze differences between continuous variables, and Fisher’s exact test was used to analyze categorical variables. Spearman’s rank correlation coefficient was indicated as R. The cut-off points for continuous variables were divided by the mean value in each clinical cohort. Survival curves were constructed using the Kaplan-Meir method and were compared with the log-rank test. Binary logistic regression model was used to train a classifier based on the expression of candidate mRNAs and miRNAs. Once the model was trained in the clinical training cohort, the same statistical model was applied in the clinical validation cohort and the performance evaluation cohort.
For all cohorts, receiver operating characteristics (ROC) curve and the area under the curve (AUC) values were used to evaluate the performance of the panel or signature for predicting the resistance of NAT. The factors acquired from univariate analysis (P < 0.10) were included in multivariate analysis with the binary logistic regression model. All P values were two-sided, and P < 0.05 was considered statistically significant. All statistical analyses were performed using EZR [28], which is a graphical user interface for R (R Foundation for Statistical Computing, Vienna, Austria, version 4.0.3) designed to add statistical functions and is frequently used in biostatistics.
RESULTS
ESCC patients with a lack of response to NAT exhibit worse survival outcomes
At the very outset, we compared the OS and recurrence-free survival (RFS) between responders and non-responders to NAT in the clinical training cohort. The 3-year OS and RFS rates were significantly worse in non-responders compared to responders (57.4% vs. 81.8%, P < 0.05; 59.6% vs. 86.4%, P = 0.04, respectively; Fig. 1A and B). We performed univariate logistic regression analyses by analyzing key clinical factors for predicting resistance to NAT in the clinical training cohort. This analysis revealed that larger tumor size was the singular significant predictive factor for predicting response to NAT in ESCC patients (odds ratio [OR]: 9.33; 95% confidence interval [CI]: 2.18–40.0; P < 0.01; Fig. 1C). The current treatment strategy, which includes NAT followed by surgery, is considered unacceptable for non-responders due to their worse prognosis, and our data revealed that only tumor size was able to predict non-responders in the clinical training cohort. These data highlight the need to develop more robust molecular biomarkers for predicting resistance to NAT in ESCC patients.
Figure 1: Clinical training phase for the combination signature for predicting resistance to NAT in ESCC patients.
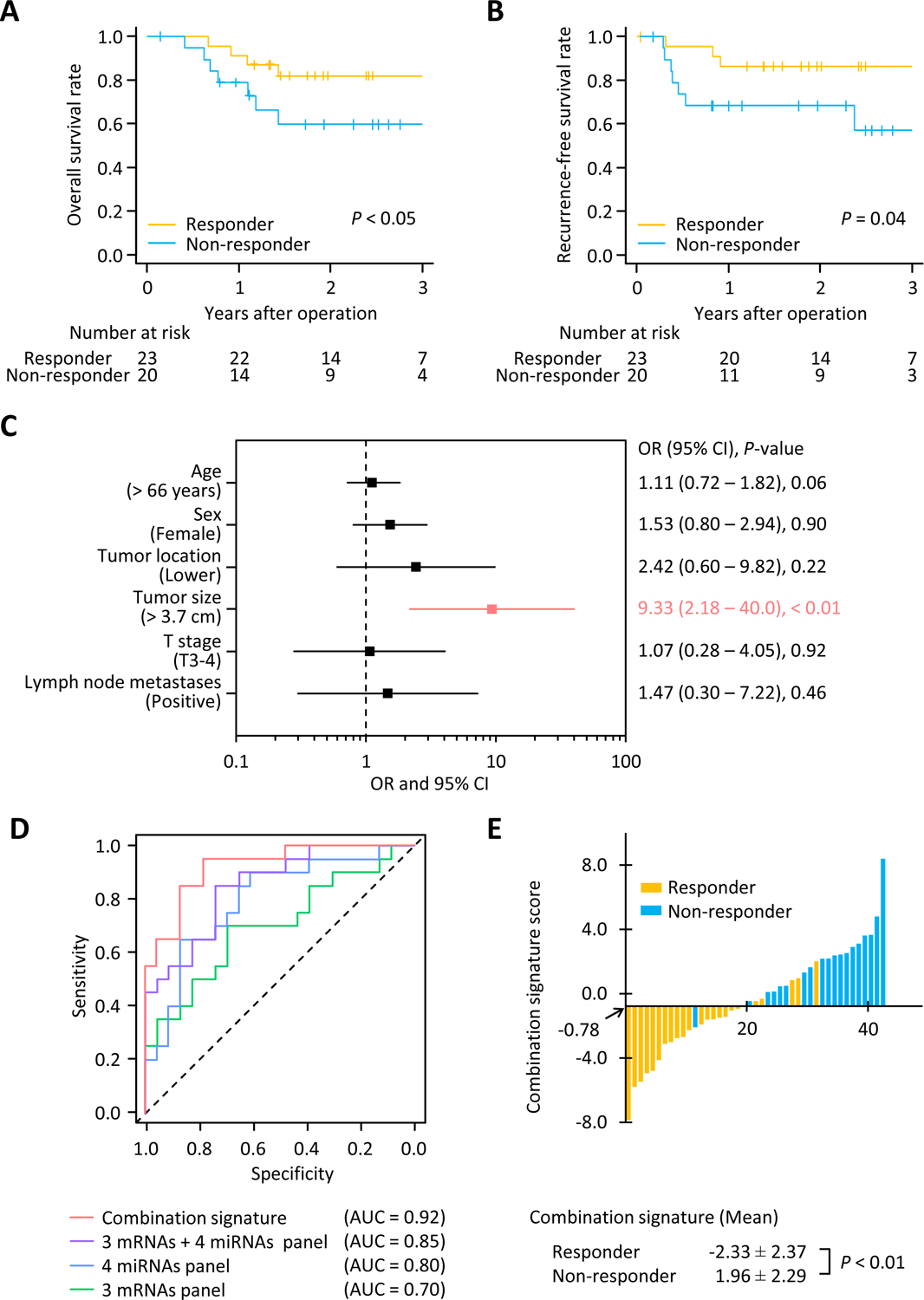
(A) Kaplan-Meier curves depicting the overall survival for responders (n = 23) or non-responders (n = 20) to NAT. (B) Kaplan-Meier curves exhibiting the recurrence-free survival in responders and non-responders to NAT. (C) Forest plot with odds ratio for each of the key clinical characteristics in univariate logistic regression analysis within the clinical training cohort (n = 43). (D) Receiver operating characteristics curve values for each signature or panels in the clinical training cohort (AUC: 0.70 for 3 mRNAs panel; 0.80 for 4 miRNAs panel; 0.85 for 3 mRNAs + 4 miRNAs panel; 0.92 for combination signature). (E) Waterfall plot representing the combination signature score in clinical training cohort. NAT, neoadjuvant therapy; OR, odds ratio; CI, confidence interval; AUC, area under the curve.
A combination signature predicts resistance to NAT in patients with ESCC
The previous studies reported a tissue-based panel of 4 miRNAs (miR-145-5p, miR-152, miR-193b-3p, and miR-376a-3p) and 3 genes (matrix metalloproteinase 1 [MMP1], LIM and calponin homology domain 1 [LIMCH1], and chromosome 1 open reading frame 226 [C1orf226]) for predicting the response of NAT in ESCC patients [14, 15]. However, these biomarkers were established in a single study and lacked a systematic and comprehensive validation in multiple independent cohorts, which would likely reveal their true clinical significance. Moreover, these panels required tissue specimens, while adaptation to a liquid biopsy assay is clinically more attractive, which would enable a noninvasive and facile assay for ESCC patients. To address these concerns, in this study, we evaluated the expression of these transcriptomic biomarkers in multiple, large, independent cohorts in our study, including their translation in a liquid biopsy assay.
Using logistic regression analysis, we first established a resistance-prediction model in the clinical training cohort. This risk-prediction panel had a reasonable accuracy even when using 3 mRNAs (AUC: 0.70; 95% CI: 0.54–0.86) or 4 miRNAs (AUC: 0.80; 95% CI: 0.66–0.93; Fig. 1D). However, intriguingly, when we combined all 3 mRNAs and 4 miRNAs together, the predictive potential of our panel was significantly superior to individual panels using either type of transcriptomic biomarkers (AUC: 0.85; 95% CI: 0.74–0.96; Fig. 1D). Furthermore, when we included additional clinicopathological features to this model, particularly, the tumor size, this resulted in an even higher predictive power compared to the transcriptomic panel alone (AUC: 0.92; 95% CI: 0.64–0.99; Fig. 1D and E). This combination signature was calculated based on the biomarker coefficients derived from the clinical training cohort as follows; Logit (P) = (−1.3548 × MMP1) + (1.6179 × LIMCH1) + (−0.4391 × C1orf226) + (2.1503 × miR-145–5p) + (5.0902 × miR-152) + (0.1270 × miR-193b-3p) + (−10.1033 × miR-376a-3p) + (0.6431 × Tumor size (cm)) – 15.4989. Herein, these data show we successfully established a signature that could robustly predict resistance to NAT in ESCC patients.
Successful validation of the risk prediction signature for NAT in an independent cohort of ESCC patients
We next performed the validation of predictive power for our combination signature in an independent cohort of ESCC patients (clinical validation cohort). In this phase, during univariate analyses, distal tumor location, larger tumor size, and the presence of lymphatic invasion were identified as significant predictive clinicopathological factors (OR: 2.83, 95% CI: 1.10–5.17, P = 0.03; OR: 2.84, 95% CI: 1.15–6.99, P = 0.02; OR: 3.27, 95% CI: 1.18–9.03, P = 0.01, respectively; Fig. 2A). Unfortunately, the predictive potential of any of these clinicopathological features on their own for their ability to predict resistance to NAT was insufficient (Fig. 2B). Interestingly, the combination signature established in the clinical training cohort was still superior to any of these clinicopathological factors (AUC: 0.78; 95% CI: 0.67–0.88; Fig. 2B). However, when we included significant clinicopathological predictors to this signature, it led to a further enhancement of the predictive power, and we defined this as NAT resistance risk (NATRR) model (AUC: 0.81; 95% CI: 0.70–0.89; Fig. 2B and C). This NATRR model derived from the clinical validation cohort was as follows; Logit (P) = (0.3827 × combination signature score) + (0.9833 × Tumor location) + (0.6612 × lymphatic invasion) – 0.8569.
Figure 2: Clinical validation phase of a NATRR model for predicting resistance of NAT in ESCC patients.
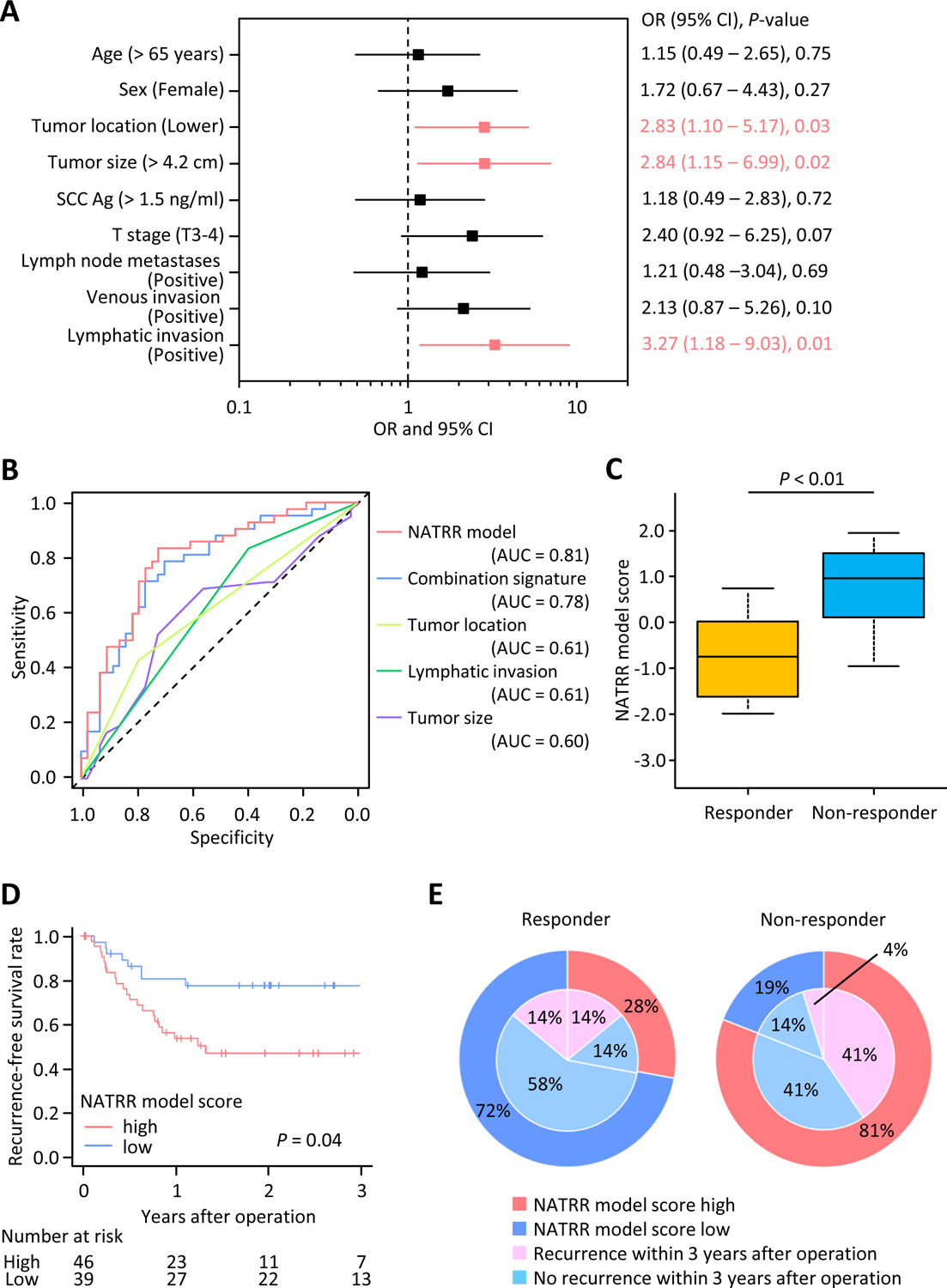
(A) Forest plot with odds ratios of each key clinicopathological characteristics in univariate logistic regression analysis for clinical validation cohort (n = 85). (B) Receiver operating characteristics curve values for each model, signature or characteristics in the clinical validation cohort. NATRR model includes the combination signature and significant key clinical predictors (tumor location and lymphatic invasion) and exhibited a superior predictive potential than the combination signature and other clinicopathological factors individually (AUC: 0.81). (C) Box plots representing the NATRR model score in the clinical validation cohort. (D) Kaplan-Meier curves of the recurrence-free survival in patients with NATRR model score high (n = 46) or low (n = 39). (E) Pie chart with NATRR model score and recurrence. Regardless of pathological responder or non-responders, patients with high NATRR model score possessed higher recurrence rates than those with low score. NAT, neoadjuvant therapy; SCC Ag, squamous cell carcinoma antigen; OR, odds ratio; CI, confidence interval; AUC, area under the curve.
We categorized all patients into high- and low- NATRR model scoring groups using cut-off thresholds derived from the Youden’s index [29], which was −0.16 (sensitivity: 0.85; specificity: 0.74). Among the key clinicopathological features, tumor location and lymphatic invasion, were significantly related to the NATRR model (P < 0.01; P < 0.01, respectively; Supplementary table S3). To evaluate the clinical applicability of our NATRR model, we examined the correlation between our model and tumor recurrence. The 3-year RFS rate was significantly worse in patients with high-risk scores than in those with low scores (47.9% vs. 74.9%, P = 0.04; Fig. 2D). Furthermore, half of pathological responders with high NATRR model score developed recurrence, which was significantly higher recurrence rate than those with low score. On the other hand, pathological non-responders with low scores exhibited lower recurrence rates. Regardless of pathological responders or non-responders, patients with high NATRR model scores experienced higher recurrence rate than those with low score (Fig. 2E). Collectively, we successfully validated our transcriptomic signature and the newly established a NATRR model to robustly predict response to NAT in ESCC patients. Furthermore, our NATRR model was able to predict not only response to NAT but also survival outcomes, such as RFS, in ESCC patients.
A NATRR model robustly predicts resistance to NAT in both early- and advanced-stage ESCC patients
To examine that our NATRR model can be applied to various patients and treatment options, we performed subgroup analyses in the patients within the clinical validation cohort. Our NATRR model yielded a remarkable accuracy for discriminating non-responders from responders to NAT in patients within each stage category, with an AUC of 0.80 in stage I & II patients and 0.79 in stage III & IV patients (Fig. 3A and B), which means a NATRR model can adapt to patients with various degrees of tumor progression. Furthermore, our model had sufficiently impressive predictive potential in each patient who received either NACRT or NAC (AUC: 0.88; AUC: 0.77, respectively; Fig. 3C and D). The predictive potential of our model didn’t depend on the regimen of chemotherapy and was equally relevant and exhibited high predictive power in all patients who received either FP or SP (AUC: 0.84; AUC:0.76, respectively; Fig. 3C and D). Furthermore, non-responders exhibited a significantly higher NATRR model score compared to responders in all subgroups (Fig. 3E). Accordingly, these findings indicate that our model can robustly predict response to NAT in a variety of patients and treatment strategies.
Figure 3: Subgroup analyses of the NATRR model for predicting the resistance to NAT in ESCC patients in clinical validation phase.
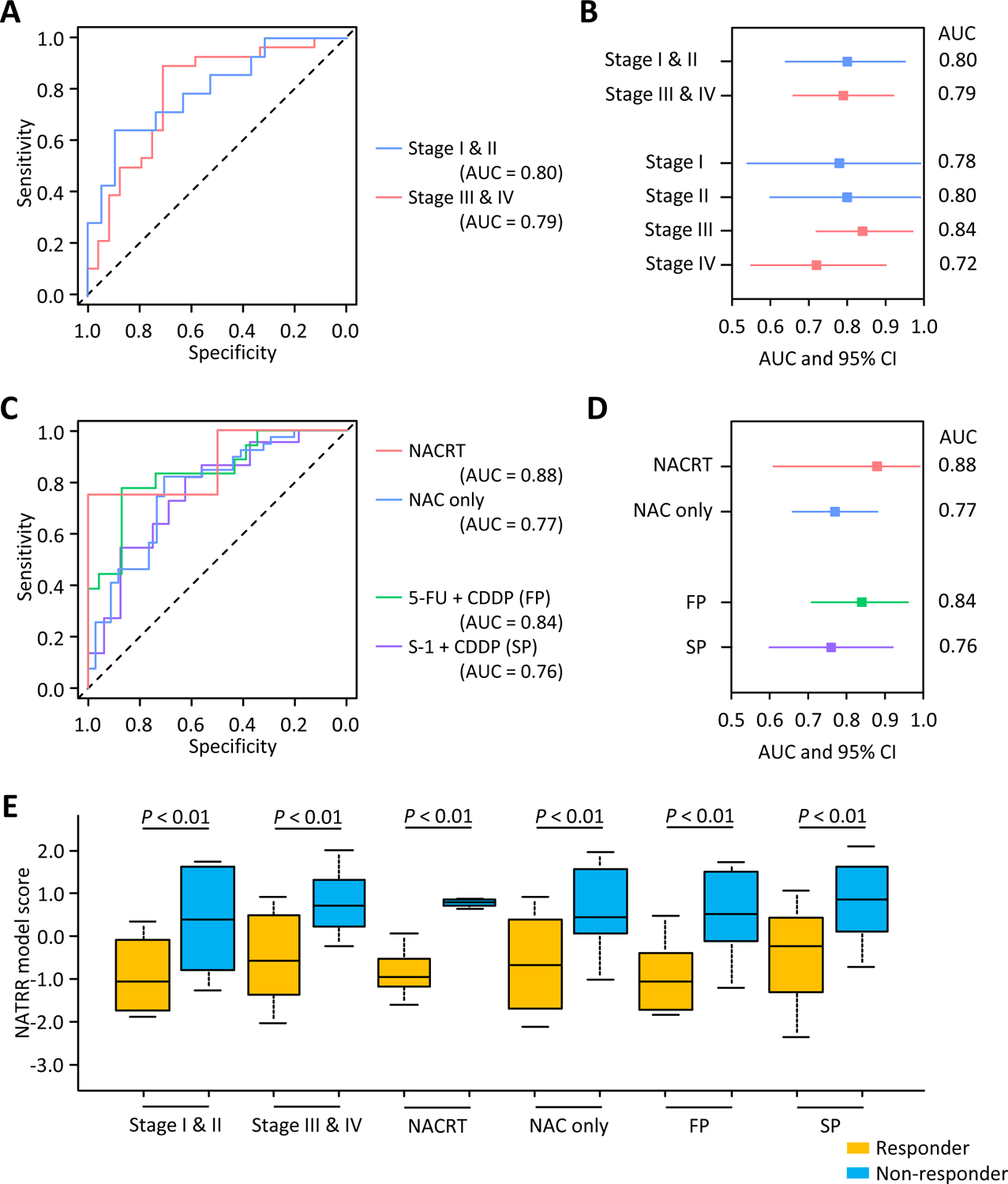
(A) Receiver operating characteristics curve values for the NATRR model in early- and advanced stage subgroups. NATRR model had sufficient potential in both early- and advanced-stage patients (AUC: 0.80; AUC: 0.79, respectively). (B) Forest plot with AUC of NATRR model in subgroup analyses focused on tumor progression. (C) Receiver operating characteristics curve values for the NATRR model in subgroup analyses focused on chemoradiotherapy and the regimen of chemotherapy. The NATRR model had sufficient potential in patients treated by chemoradiotherapy or chemotherapy only (AUC: 0.88; AUC: 0.77, respectively), and treated by FP or SP (AUC: 0.84; AUC:0.76, respectively). (D) Forest plot with AUC of NATRR model in subgroup analyses focused on chemoradiotherapy and regimen of chemotherapy. (E) Box plots representing the NATRR model score of subgroup analyses in the clinical validation cohort. AUC, area under the curve; CI, confidence interval; NACRT, neoadjuvant chemoradiotherapy; NAC, neoadjuvant chemotherapy; FP, 5-FU + CDDP; SP, S-1 + CDDP.
Successful translation of the NATRR model as a liquid biopsy assay in ESCC patients
To translate our NATRR model into a liquid biopsy assay, which would enable a noninvasive and facile assay for predicting resistance to NAT in ESCC patients, we analyzed a subset of 58 patients within the clinical validation cohort, from whom such matched pretreatment serum specimens were available (performance evaluation cohort). The NATRR model yielded an AUC value of 0.78 (95%CI: 0.67–0.90; sensitivity: 0.70; specificity: 0.71) for discriminating non-responders from responders to NAT (Fig. 4A and B). In line with the results from the clinical validation cohort, non-responders possessed a significantly higher NATRR model score compared to responders in the liquid biopsy specimens (Fig. 4C). When we compared AUC values among our NATRR model and key clinicopathological factors, our NATRR model revealed notably superior AUC values compared to any of the clinicopathological factors (Fig. 4D). In these key clinicopathological factors, only tumor location, was significantly related to the NATRR model (P < 0.01; Supplementary table S4). Furthermore, according to multivariate logistic regression analysis, which included factors acquired from univariate analysis (P < 0.10), our NATRR model emerged as a significant and independent predictor for resistance to NAT (OR: 6.10; 95%CI: 1.60–23.2; P < 0.01; Table 1).
Figure 4: Clinical performance evaluation phase of a NATRR model for predicting the resistance to NAT in ESCC patients in serum specimens.
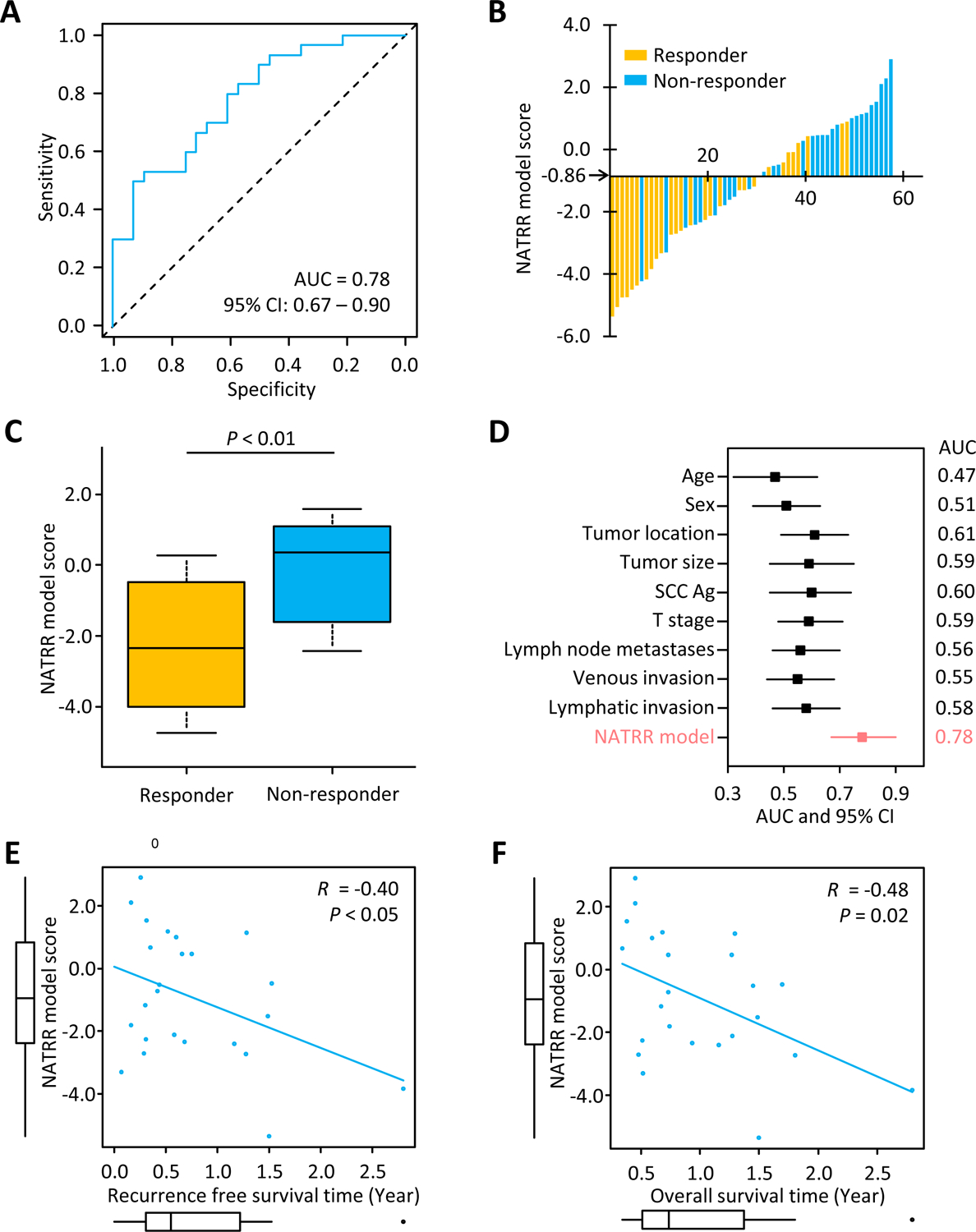
(A) Receiver operating characteristics curve values for NATRR model score in the clinical performance evaluation cohort (n = 58). (B) Waterfall plot represents the NATRR model score in the clinical performance evaluation cohort. (C) Box plots representing the NATRR model score in the performance evaluation cohort. (D) Forest plot with AUC of NATRR model and key clinicopathological factors to predict response to NAT in ESCC patients. (E) Pearson’s correlation of a NATRR model score and recurrence-free survival time in patients experienced recurrence (R = −0.40, P = 0.05). (F) Pearson’s correlation of a NATRR model score and overall survival time in patients experienced recurrence (R = −0.48, P = 0.02). AUC, area under the curve; CI, confidence interval; NAT, neoadjuvant therapy; SCC Ag, squamous cell carcinoma antigen.
Table 1.
Univariate and multivariate analysis of factors contributing to resistance of neoadjuvant therapy in patients with ESCC
| Univariate | Multivariate | |||
|---|---|---|---|---|
| OR (95% CI) | P value | OR (95% CI) | P value | |
| Age, > 66 versus ≤ 66 years | 1.14 (0.41 – 3.21) | 0.80 | ||
| Sex, female versus male | 1.07 (0.35 – 3.32) | 0.91 | ||
| Tumor location, upper versus lower | 2.80 (0.88 – 8.91) | 0.08 | 1.82 (0.50 – 6.64) | 0.36 |
| Tumor size, > 4.2 versus ≤ 4.2 cm | 2.70 (0.93 – 7.82) | 0.07 | 2.58 (0.80 – 8.38) | 0.11 |
| SCC Ag, > 1.5 versus ≤ 1.5 ng/ml | 1.38 (0.48 – 3.96) | 0.55 | ||
| T stage, T3–4 versus T1–2 | 2.59 (0.80 – 8.36) | 0.11 | ||
| Lymph node metastases, positive versus negative | 2.13 (0.66 – 6.62) | 0.19 | ||
| Venous invasion, positive versus negative | 1.74 (0.56 – 5.39) | 0.34 | ||
| Lymphatic invasion, positive versus negative | 2.06 (0.69 – 6.21) | 0.20 | ||
| NATRR model, high versus low | 6.86 (1.91 – 24.6) | < 0.01 | 6.10 (1.60 – 23.2) | < 0.01 |
OR, odds ratio; CI, confidence interval; SCC Ag, squamous cell carcinoma antigen
Finally, using Spearman’s correlation test, we examined the associations between a NATRR model score and survival times. A NATRR model score confirmed a negative correlation with RFS time (R = −0.40, P < 0.05; Fig. 4E) and OS time (R = −0.48, P = 0.02; Fig. 4F). This indicates that patients with high NATRR model score might have risks of early recurrence and early death and highlights the potential clinical significance of our liquid biopsy model in ESCC patient. Accordingly, we successfully established a robust liquid biopsy model to predict response to NAT in ESCC patients.
DISCUSSION
Current well-designed randomized clinical trials demonstrated that NAC and NACRT prior to surgery improved the survival outcomes of patients with ESCC, and, NAT in particular, is becoming the standard treatment in patients with advanced ESCC. Accordingly, non-responders, who accounts for more than half of all patients with ESCC, not only did not benefit from these therapies compared to upfront surgery, but also suffered from undesirable toxicity and a missed opportunity for a curative operation. Therefore, identification of patients that will or will not benefit from such therapies is of great clinical significance, which will allow tailored development of individualized treatment strategies to maximize therapeutic efficacy in each patient. Advances in molecular biology technologies in recent years have led to the rapid development of precision-medicine in cancer, which allows novel multimodality treatment strategies for patients with cancer. With regards to ESCC, in spite of the fact that molecular biomarkers to predict response to NAT have been reported previously [14, 15, 30–38], none of these have yet been utilized in actual clinical practice. For the clinical application of molecular biomarkers, the need for the availability of accurate, noninvasive, and easy-to-use biomarkers are widely acknowledged. In particular, remarkable progresses have recently been made in the realm of liquid biopsy biomarkers, and such a platform is deemed awfully attractive. In this study, we undertook a systematic series of experiments to delineate a panel of molecular biomarkers that were successfully developed in a liquid biopsy assay for predicting resistance to NAT in ESCC patients.
In this study, we demonstrated a NATRR model using 7 molecular biomarkers [14, 15], and 3 clinicopathological factors, which included tumor size, tumor location, and lymphatic invasion. Several previous studies have identified molecular biomarkers, including mRNAs and miRNAs, to predict the response to NAT in ESCC patients [14, 15, 30–38]. However, most of these studies were performed in FFPE tissues and were based upon single institution cohorts and lacked independent validation of such marker panels. Our study had several unique advantages compared to most previous studies. First, we comprehensively validated our NATRR model in multiple independent clinical cohorts from different institutions, which ensured generalizability of our model. Second, we translated our model into a liquid biopsy assay, which offers a distinct advantage in terms of its easier clinical implementation and noninvasive nature. A liquid biopsy assay is attractive and can be easily applied in routine clinical practice. Clinicians and surgeons always collect and evaluate other biological analytes in blood prior to initiation of any cancer treatment; hence, availability of a predictive assay in a liquid biopsy sample will obviate the need for additional biospecimen collection and easier implementation of such an assay in clinical practice. Third, by translating into a liquid biopsy, we can overcome the molecular intratumoral heterogeneity. In clinical practice, ESCC patients are invariably biopsied endoscopically, however, the major limitation of tumor biopsy is its inability to detect intratumoral heterogeneity, which characterizes most of advanced ESCCs [39–41]. Endoscopic biopsy from one part of a solitary tumor may miss the molecular intratumoral heterogeneity. A liquid biopsy is superior to endoscopic biopsy in terms of this point. Fourth, we used a multi-omic transcriptomic biomarkers (both mRNAs and miRNAs) in this study, which were able to offer a superior predictive accuracy compared to individual types of markers.
In the present study, for the definition of NAT response, we adopted pathological response instead of the Response Evaluation Criteria in Solid Tumors (RECIST) (version 1.1) system [42]. The RECIST system is the gold standard for evaluating chemotherapy response and is widely used in clinical practice. However, we did not have available data on RECIST classification in all patients of our cohort, and current systematic review and several previous studies for ESCC suggested that the RECIST system does not reflect actual treatment effects [43–45]. Despite the additional analyses using the RECIST data may be more helpful and beneficial, we used only pathological response to definite NAT response. For the definition of non-responders in this study, we defined pathological non-responders, with more than 50% residual cancer, as non-responders. The NCCN guidelines recommend a 3-category system, which divides all patients into pathological complete responders, partial responders (with 1–50% residual cancer), and non-responders (with more than 50% residual cancer) [46, 47]. However, due to the limited sample size in our cohorts, separating all patients into 3 categories were not possible. Furthermore, even in everyday clinical practice, identification of non-responders is more important, rather than all three subgroups. Therefore, we grouped pathological complete and partial responders as responders, and pathological non-responder as non-responder in our study.
Several limitations of our study must be acknowledged. First, our biomarker selection was based on previous data from a single institution, which could potentially introduce some inadvertent bias in the selection of patients and treatment approaches. Second, ours was a retrospective cohort study with a limited number of patients. To further confirm the predictive potential of our model, the results need to be validated in future large prospective clinical trials. Third, there were some minor inconsistencies in treatment strategies in each cohort. In clinical training cohort, none of the patients underwent radiation therapy, and the regimen of chemotherapy also differed between the two cohorts. While our final model exhibited remarkable predictive potential in patients who received each treatment option in subgroup analyses (Fig. 3C and D), to further ensure its predictive ability, the results need to be validated in future well-designed prospective clinical trials with larger patient cohorts. Finally, our liquid biopsy-based NATRR model included mRNAs, which can be unstable in the blood. Recent studies have demonstrated that mRNAs encapsulated within extracellular vesicles, such as exosomes and microparticles, are stable due to their protection by the cellular membrane and circulating RNAs include mRNA from these vesicles [48, 49]. In this study, we did not have information about which vesicles mRNAs are derived from, however, information of these extracellular vesicles may be helpful in the development of better methods. Thus, future analyses about these extracellular vesicles may be able to bring greater potentials of our liquid biopsy-based model.
In conclusion, we have successfully identified and developed a liquid biopsy-based model that allows robust prediction of resistance to NAT in patients with ESCC. Our findings could help in the selection of the best treatment strategy in patients with ESCC. Furthermore, our study provides a proof-of-concept precision-medicine assay, for its further validation in future prospective clinical trials.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs Rachana Garg, Kota Nakamura, Souvick Roy, Yogi Pratama, Viktor Hlavac, and Yuki Sunagawa for their thoughtful discussions and advice during the course of this project.
Funding:
This work was supported by, CA184792, CA187956, CA227602, CA072851 and CA202797 grants from the National Cancer Institute, National Institutes of Health.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed by the authors.
REFERENCES
- 1.Sung H, Ferlay J, Siegel RL et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021; 71: 209–249. [DOI] [PubMed] [Google Scholar]
- 2.Arnold M, Abnet CC, Neale RE et al. Global Burden of 5 Major Types of Gastrointestinal Cancer. Gastroenterology 2020; 159: 335–349.e315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Then EO, Lopez M, Saleem S et al. Esophageal Cancer: An Updated Surveillance Epidemiology and End Results Database Analysis. World J Oncol 2020; 11: 55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Jiang Y, Wu C et al. Comparison of clinicopathologic features and survival between eastern and western population with esophageal squamous cell carcinoma. J Thorac Dis 2015; 7: 1780–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herskovic A, Russell W, Liptay M et al. Esophageal carcinoma advances in treatment results for locally advanced disease: review. Ann Oncol 2012; 23: 1095–1103. [DOI] [PubMed] [Google Scholar]
- 6.Sjoquist KM, Burmeister BH, Smithers BM et al. Survival after neoadjuvant chemotherapy or chemoradiotherapy for resectable oesophageal carcinoma: an updated meta-analysis. Lancet Oncol 2011; 12: 681–692. [DOI] [PubMed] [Google Scholar]
- 7.van Hagen P, Hulshof MC, van Lanschot JJ et al. Preoperative chemoradiotherapy for esophageal or junctional cancer. N Engl J Med 2012; 366: 2074–2084. [DOI] [PubMed] [Google Scholar]
- 8.Ajani JA, D’Amico TA, Bentrem DJ et al. Esophageal and Esophagogastric Junction Cancers, Version 2.2019, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2019; 17: 855–883. [DOI] [PubMed] [Google Scholar]
- 9.Noble F, Lloyd MA, Turkington R et al. Multicentre cohort study to define and validate pathological assessment of response to neoadjuvant therapy in oesophagogastric adenocarcinoma. Br J Surg 2017; 104: 1816–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blum Murphy M, Xiao L, Patel VR et al. Pathological complete response in patients with esophageal cancer after the trimodality approach: The association with baseline variables and survival-The University of Texas MD Anderson Cancer Center experience. Cancer 2017; 123: 4106–4113. [DOI] [PubMed] [Google Scholar]
- 11.Zheng Y, Li Y, Liu X et al. Reevaluation of Neoadjuvant Chemotherapy for Esophageal Squamous Cell Carcinoma: A Meta-Analysis of Randomized Controlled Trials Over the Past 20 Years. Medicine (Baltimore) 2015; 94: e1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumagai K, Rouvelas I, Tsai JA et al. Meta-analysis of postoperative morbidity and perioperative mortality in patients receiving neoadjuvant chemotherapy or chemoradiotherapy for resectable oesophageal and gastro-oesophageal junctional cancers. Br J Surg 2014; 101: 321–338. [DOI] [PubMed] [Google Scholar]
- 13.Mariette C, Dahan L, Mornex F et al. Surgery alone versus chemoradiotherapy followed by surgery for stage I and II esophageal cancer: final analysis of randomized controlled phase III trial FFCD 9901. J Clin Oncol 2014; 32: 2416–2422. [DOI] [PubMed] [Google Scholar]
- 14.Wen J, Yang H, Liu MZ et al. Gene expression analysis of pretreatment biopsies predicts the pathological response of esophageal squamous cell carcinomas to neo-chemoradiotherapy. Ann Oncol 2014; 25: 1769–1774. [DOI] [PubMed] [Google Scholar]
- 15.Wen J, Luo K, Liu H et al. MiRNA Expression Analysis of Pretreatment Biopsies Predicts the Pathological Response of Esophageal Squamous Cell Carcinomas to Neoadjuvant Chemoradiotherapy. Ann Surg 2016; 263: 942–948. [DOI] [PubMed] [Google Scholar]
- 16.Nakamura K, Kato K, Igaki H et al. Three-arm phase III trial comparing cisplatin plus 5-FU (CF) versus docetaxel, cisplatin plus 5-FU (DCF) versus radiotherapy with CF (CF-RT) as preoperative therapy for locally advanced esophageal cancer (JCOG1109, NExT study). Jpn J Clin Oncol 2013; 43: 752–755. [DOI] [PubMed] [Google Scholar]
- 17.Ando N, Kato H, Igaki H et al. A randomized trial comparing postoperative adjuvant chemotherapy with cisplatin and 5-fluorouracil versus preoperative chemotherapy for localized advanced squamous cell carcinoma of the thoracic esophagus (JCOG9907). Ann Surg Oncol 2012; 19: 68–74. [DOI] [PubMed] [Google Scholar]
- 18.Tahara M, Fuse N, Mizusawa J et al. Phase I/II trial of chemoradiotherapy with concurrent S-1 and cisplatin for clinical stage II/III esophageal carcinoma (JCOG 0604). Cancer Sci 2015; 106: 1414–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryan R, Gibbons D, Hyland JM et al. Pathological response following long-course neoadjuvant chemoradiotherapy for locally advanced rectal cancer. Histopathology 2005; 47: 141–146. [DOI] [PubMed] [Google Scholar]
- 20.Gu Y, Swisher SG, Ajani JA et al. The number of lymph nodes with metastasis predicts survival in patients with esophageal or esophagogastric junction adenocarcinoma who receive preoperative chemoradiation. Cancer 2006; 106: 1017–1025. [DOI] [PubMed] [Google Scholar]
- 21.Wada Y, Shimada M, Murano T et al. A Liquid Biopsy Assay for Noninvasive Identification of Lymph Node Metastases in T1 Colorectal Cancer. Gastroenterology 2021; 161: 151–162.e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishiwada S, Sho M, Cui Y et al. A gene expression signature for predicting response to neoadjuvant chemoradiotherapy in pancreatic ductal adenocarcinoma. Int J Cancer 2021; 148: 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishiwada S, Sho M, Banwait JK et al. A MicroRNA Signature Identifies Pancreatic Ductal Adenocarcinoma Patients at Risk for Lymph Node Metastases. Gastroenterology 2020; 159: 562–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 25.Matsuyama T, Kandimalla R, Ishikawa T et al. A novel mesenchymal-associated transcriptomic signature for risk-stratification and therapeutic response prediction in colorectal cancer. Int J Cancer 2020; 147: 3250–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kandimalla R, Ozawa T, Gao F et al. Gene Expression Signature in Surgical Tissues and Endoscopic Biopsies Identifies High-Risk T1 Colorectal Cancers. Gastroenterology 2019; 156: 2338–2341.e2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ozawa T, Kandimalla R, Gao F et al. A MicroRNA Signature Associated With Metastasis of T1 Colorectal Cancers to Lymph Nodes. Gastroenterology 2018; 154: 844–848.e847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanda Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant 2013; 48: 452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Youden WJ. Index for rating diagnostic tests. Cancer 1950; 3: 32–35. [DOI] [PubMed] [Google Scholar]
- 30.Lavery A, Turkington RC. Transcriptomic biomarkers for predicting response to neoadjuvant treatment in oesophageal cancer. Gastroenterol Rep (Oxf) 2020; 8: 411–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bollschweiler E, Hölscher AH, Herbold T et al. Molecular Markers for the Prediction of Minor Response to Neoadjuvant Chemoradiation in Esophageal Cancer: Results of the Prospective Cologne Esophageal Response Prediction (CERP) Study. Ann Surg 2016; 264: 839–846. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka K, Miyata H, Yamasaki M et al. Circulating miR-200c levels significantly predict response to chemotherapy and prognosis of patients undergoing neoadjuvant chemotherapy for esophageal cancer. Ann Surg Oncol 2013; 20 Suppl 3: S607–615. [DOI] [PubMed] [Google Scholar]
- 33.Yamasaki M, Makino T, Masuzawa T et al. Role of multidrug resistance protein 2 (MRP2) in chemoresistance and clinical outcome in oesophageal squamous cell carcinoma. Br J Cancer 2011; 104: 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujishima H, Fumoto S, Shibata T et al. A 17-molecule set as a predictor of complete response to neoadjuvant chemotherapy with docetaxel, cisplatin, and 5-fluorouracil in esophageal cancer. PLoS One 2017; 12: e0188098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Motoori M, Takemasa I, Yamasaki M et al. Prediction of the response to chemotherapy in advanced esophageal cancer by gene expression profiling of biopsy samples. Int J Oncol 2010; 37: 1113–1120. [DOI] [PubMed] [Google Scholar]
- 36.Pühringer-Oppermann F, Sarbia M, Ott N, Brücher BL. The predictive value of genes of the TGF-beta1 pathway in multimodally treated squamous cell carcinoma of the esophagus. Int J Colorectal Dis 2010; 25: 515–521. [DOI] [PubMed] [Google Scholar]
- 37.Slotta-Huspenina J, Drecoll E, Feith M et al. MicroRNA expression profiling for the prediction of resistance to neoadjuvant radiochemotherapy in squamous cell carcinoma of the esophagus. J Transl Med 2018; 16: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugimura K, Miyata H, Tanaka K et al. Let-7 expression is a significant determinant of response to chemotherapy through the regulation of IL-6/STAT3 pathway in esophageal squamous cell carcinoma. Clin Cancer Res 2012; 18: 5144–5153. [DOI] [PubMed] [Google Scholar]
- 39.Lin L, Lin DC. Biological Significance of Tumor Heterogeneity in Esophageal Squamous Cell Carcinoma. Cancers (Basel) 2019; 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li R, Li P, Xing W, Qiu H. Heterogeneous genomic aberrations in esophageal squamous cell carcinoma: a review. Am J Transl Res 2020; 12: 1553–1568. [PMC free article] [PubMed] [Google Scholar]
- 41.Cao W, Wu W, Yan M et al. Multiple region whole-exome sequencing reveals dramatically evolving intratumor genomic heterogeneity in esophageal squamous cell carcinoma. Oncogenesis 2015; 4: e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eisenhauer EA, Therasse P, Bogaerts J et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009; 45: 228–247. [DOI] [PubMed] [Google Scholar]
- 43.de Gouw D, Klarenbeek BR, Driessen M et al. Detecting Pathological Complete Response in Esophageal Cancer after Neoadjuvant Therapy Based on Imaging Techniques: A Diagnostic Systematic Review and Meta-Analysis. J Thorac Oncol 2019; 14: 1156–1171. [DOI] [PubMed] [Google Scholar]
- 44.Odawara S, Kitajima K, Katsuura T et al. Tumor response to neoadjuvant chemotherapy in patients with esophageal cancer assessed with CT and FDG-PET/CT - RECIST 1.1 vs. PERCIST 1.0. Eur J Radiol 2018; 101: 65–71. [DOI] [PubMed] [Google Scholar]
- 45.Yanagawa M, Tatsumi M, Miyata H et al. Evaluation of response to neoadjuvant chemotherapy for esophageal cancer: PET response criteria in solid tumors versus response evaluation criteria in solid tumors. J Nucl Med 2012; 53: 872–880. [DOI] [PubMed] [Google Scholar]
- 46.Al-Batran SE, Homann N, Pauligk C et al. Perioperative chemotherapy with fluorouracil plus leucovorin, oxaliplatin, and docetaxel versus fluorouracil or capecitabine plus cisplatin and epirubicin for locally advanced, resectable gastric or gastro-oesophageal junction adenocarcinoma (FLOT4): a randomised, phase 2/3 trial. Lancet 2019; 393: 1948–1957. [DOI] [PubMed] [Google Scholar]
- 47.Ychou M, Boige V, Pignon JP et al. Perioperative chemotherapy compared with surgery alone for resectable gastroesophageal adenocarcinoma: an FNCLCC and FFCD multicenter phase III trial. J Clin Oncol 2011; 29: 1715–1721. [DOI] [PubMed] [Google Scholar]
- 48.Lässer C. Identification and analysis of circulating exosomal microRNA in human body fluids. Methods Mol Biol 2013; 1024: 109–128. [DOI] [PubMed] [Google Scholar]
- 49.Tovar-Camargo OA, Toden S, Goel A. Exosomal microRNA Biomarkers: Emerging Frontiers in Colorectal and Other Human Cancers. Expert Rev Mol Diagn 2016; 16: 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


