Abstract
Introduction:
Pulmonary fibrosis is an age-related, progressive, and fatal disease with a median survival of 3–5 years after diagnosis; idiopathic pulmonary fibrosis (IPF) is the most common type. It is characterized by fibroblast proliferation and accumulation of excessive extracellular matrix. Patients with IPF are at increased risk for lung cancer. Epigenetic mechanisms are involved in lung fibrosis and cancer, and DNA methylation is critical in disease pathogenesis and progression. Therefore, studies of DNA methylation contribute to better understanding of the underlying mechanisms of these two respiratory diseases, and can offer novel diagnostic and treatment options.
Areas covered:
This review discusses the latest advances in our understanding of epigenetic factors related to DNA methylation that impact development of lung cancer and pulmonary fibrosis, discusses the role of DNA methylation in promoting or inhibiting these diseases, and proposes its potential clinical significance in disease diagnosis and treatment.
Expert opinion:
DNA methylation plays a critical role in lung cancer and fibrosis pathogenesis. DNA methylation offers a new biomarker for disease diagnosis or monitoring, and provides a new therapeutic target for treatment.
Keywords: biomarkers, DNA methyl-binding protein, DNA methylation, DNA methyltransferases, epigenetic, idiopathic pulmonary fibrosis, lung cancer, pulmonary fibrosis
1. Introduction
Pulmonary fibrosis is an age-related progressive lung disease characterized by the overproduction of extracellular matrix proteins, resulting in lung tissue scarring and stiffness [1]. The most common aggressive form is idiopathic pulmonary fibrosis (IPF), which has limited treatment options, poor long-term prognosis, and an average life expectancy of only 3–5 years after diagnosis [2]. The exact etiology of IPF is unknown, but genetic factors, and environmental factors including smoking, chronic aspiration, viral infections, and advanced age have been implicated [3].
IPF is associated with a high-risk of lung cancer, and these two respiratory diseases share many similarities in pathological pathways and causative factors, including the high-risk factors, such as old age and smoking, and both have low survival rates [4–6] (Figure 1). The World Health Organization classifies lung cancer into two major histologic subtypes: non-small cell lung cancer (NSCLC), which accounts for approximately 85% of lung cancers, and small cell lung cancers (SCLC), accounting for the remaining 15%. NSCLC includes lung squamous cell carcinoma (LUSC), lung adenocarcinoma (LUAD), and large-cell carcinoma subtypes [7]. LUSC is the most frequent type of lung cancer in IPF patients, followed by LUAD [8]. Only a few cases of large-cell carcinoma and SCLC have been reported in IPF patients, possibly reflecting greater susceptibility of IPF to different histological subtypes of lung carcinoma [9]. Proteomics, epigenomics, and genetics have defined the etiology and heterogeneity of these complex diseases and have found multiple common genetic, molecular, and cellular signaling pathways linking lung fibrosis with lung cancer [10].
Figure 1. Idiopathic pulmonary fibrosis (IPF) and lung cancer have many similarities.
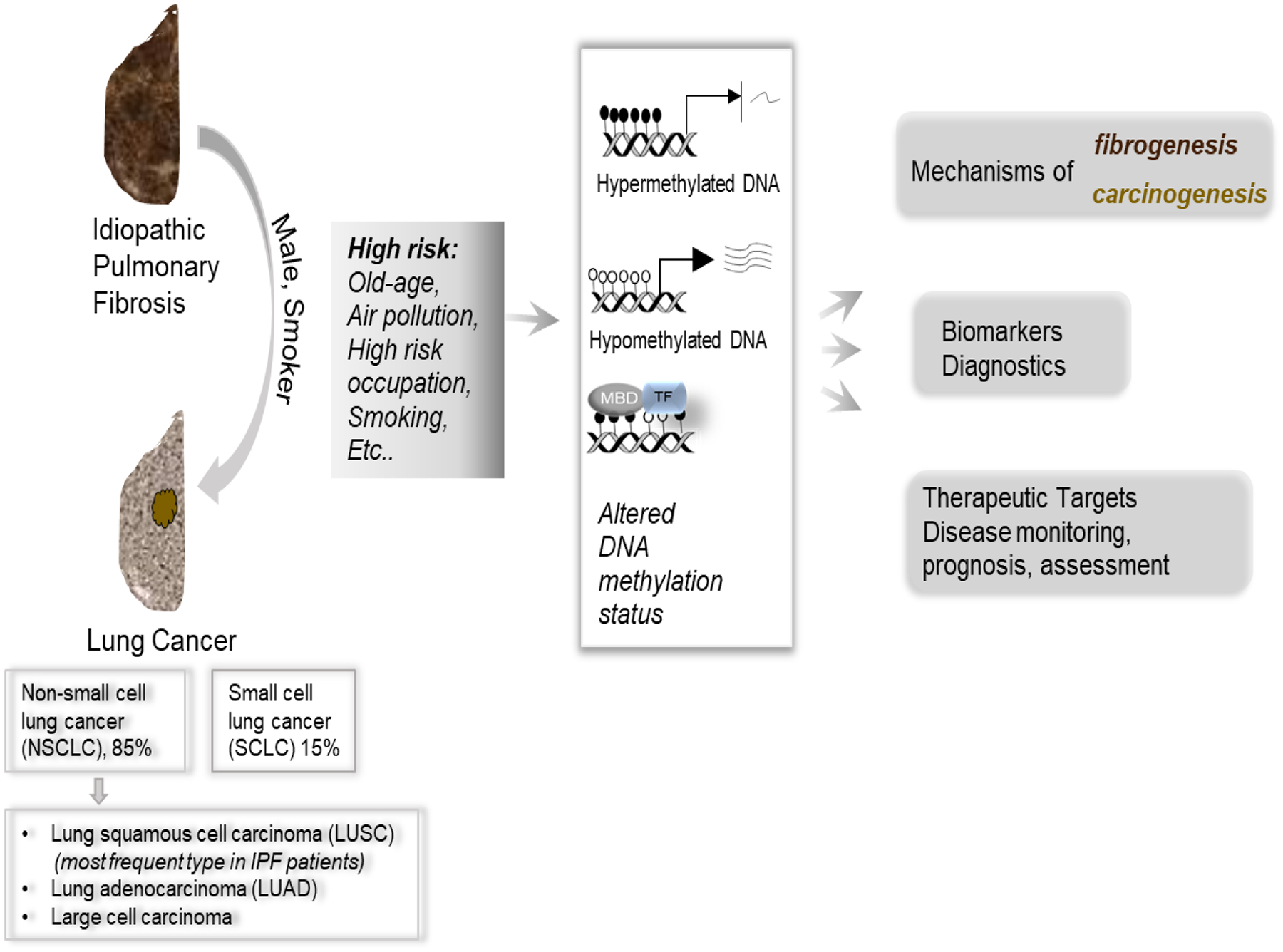
Lung cancer is classified into different subtypes, some of which are more frequently seen in IPF patients. There are many similarities between lung fibrosis and cancer. For example, high-risk factors for both diseases include aging, poor air, high-risk occupations, and smoking. IPF patients, especially with factors such as cigarette smoking and gender (male), are likely to develop lung cancer. These risk factors would alter DNA methylation status (hyper- or hypo-methylation), which would affect gene expression, and change the recruitment of methyl-binding proteins (MBD), and transcriptional factors (TF), which all contribute to the mechanisms underlying fibrogenesis and carcinogenesis. Altered DNA methylation can be targeted pharmacologically, used as diagnostic biomarkers, and used to monitor disease progression (see text for details).
In addition to these similarities, lung cancer and lung fibrosis share some common pathogenesis mechanisms and treatment methods, such as uncontrolled cell proliferation and aberrant activation of specific signaling pathways, and one anti-cancer agent, nintedanib, has been approved for the treatment of lung fibrosis[10]. Aberrant DNA methylation also contributes to pulmonary fibrosis and lung cancer development and progression [11,12]. For example, hypomethylation of oncogenes and hypermethylation of tumor suppressor genes are established pathogenic mechanisms in most tumors [13]. Similar findings have been reported in IPF, in which hypermethylation-silenced antifibrotic genes have been reported [14]. Genome-wide DNA methylation studies have revealed that differentially methylated cytosine phosphate guanine (CpG) islands overlap in lung cancer and IPF, suggesting common pathogenetic pathways between these two diseases, prompting us to explore DNA methylation as an epigenetic link between lung cancer and fibrosis [15]. In this review, we summarize the role of DNA methylation in lung cancer and fibrosis and its potential clinical applications as epigenetic markers and treatment targets.
2. DNA methylation
DNA methylation is a biological process that adds a methyl group to the 5’ carbon of the cytosine pyrimidine ring in CpG dinucleotides. Hypermethylation at promoter regions usually suppresses gene transcription, whereas hypomethylation underlies gene activation or overexpression, or causes abnormal expression of transposons and genetic instability [13]. DNA methylation is mainly modulated by DNA methyltransferases (DNMTs) and methyl-DNA-binding proteins (MBPs) (Figure 2).
Figure 2. Classification of DNA methyltransferases (DNMTs) and Methyl-binding Proteins (MBPs).
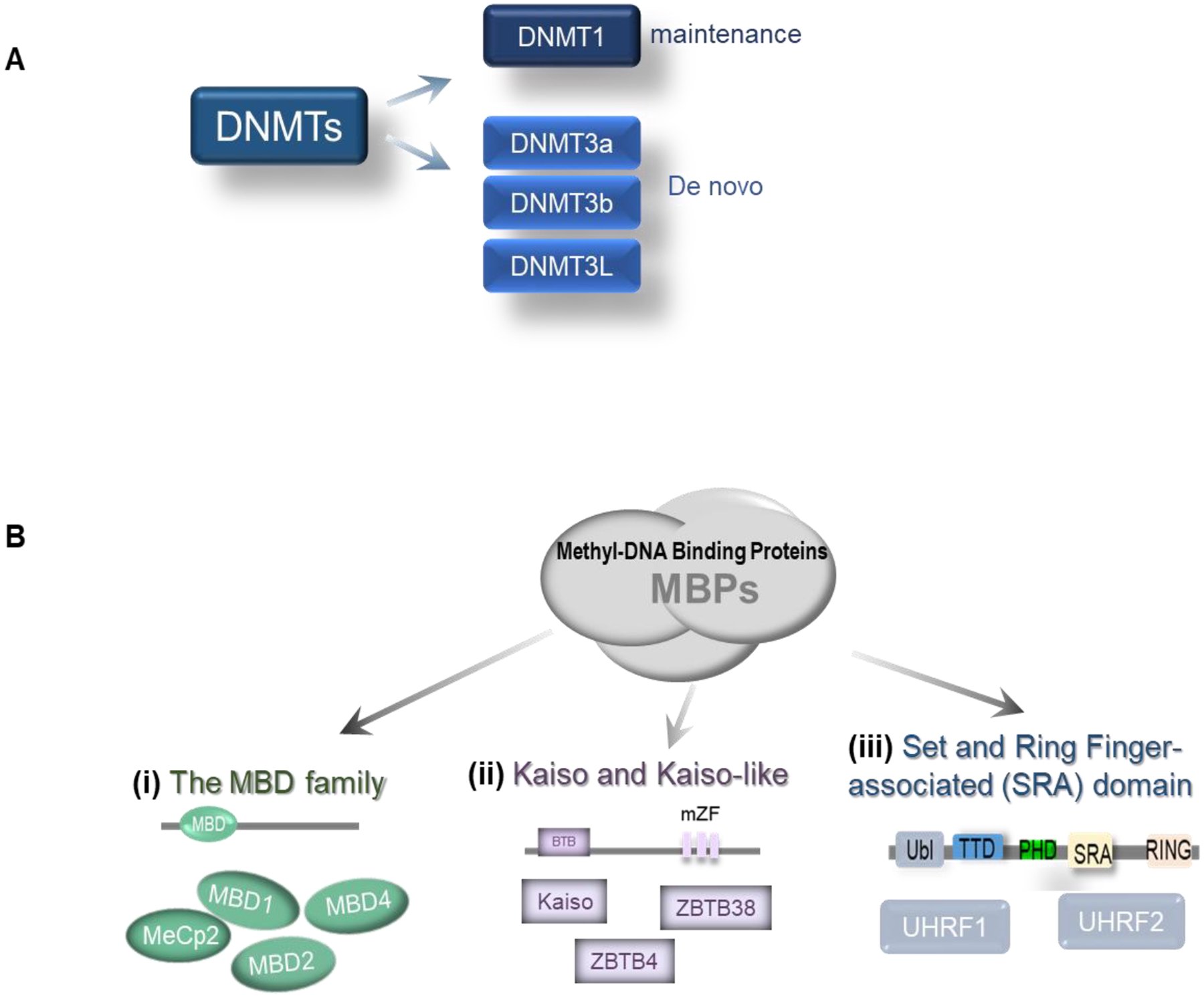
A. DNMTs are classified as maintenance or de novo DNA methyltransferases. B. Three major families of MBPs. (i). The MBD family (all with MBD domain), including MeCp2, MBD1, MBD2, and MBD4 preferentially bind methylated DNA. (ii). Kaiso- and Kaiso-like proteins, shown with the main BTB domain and methyl-CpG-binding zinc fingers (mZF). Including Kaiso, ZBTB4, and ZBTB38. (iii). Set and Ring Finger-associated (SRA) domain, two members of UHRF1 and UHRF2 (Ubiquitin-like with PHD and RING finger domains 1 and 2). Ubl, ubiquitin-like domain; TTD, tandem tudor domain; PHD, plant homeodomain finger domain; SRA, SET, and RING-associated domain.
All DNMTs share a common catalytic domain consisting of six conserved amino acid motifs in the carboxyl-terminus, and differ significantly at the N-terminus. According to their structure and function, DNMTs can be divided into two major groups in mammalian cells [16] (Figure 2A). The first group is represented by the maintenance methyltransferase DNMT1 (the largest methyltransferase with a molecular mass of 184 kDa) that binds to hemimethylated DNA and maintains DNA methylation after replication. In addition to catalyzing methyltransferase reactions, DNMT1 is a crucial element of the transcription suppression complex that interacts with DNMT1-associated proteins, such as E2F1 and HDAC [17]. The second group comprises de novo methyltransferases, including DNMT3a, DNMT3b, and DNMT3L. This group of enzymes does not require hemimethylated DNA to bind, and displays equal affinity for hemi- or non-methylated DNA. These enzymes cooperate with DNMT1 to extend methylation [18].
Inhibiting gene expression by DNA methylation requires the recruitment of specific proteins to methylated CpG sites and the recruitment of co-repressors, such as histone deacetylases (HDACs), to establish silencing complexes that inhibit gene expression [19]. These proteins are usually MBPs (Figure 2B). According to their structure and function, MBPs are divided into three different types in mammals: methyl-CpG-binding domain (MBD), Kaiso- and Kaiso-like proteins, and set and ring finger-associated (SRA) family proteins [20].
3. Changes of DNMTs in lung cancer and IPF
Highly expressed DNMTs have been reported to be involved in the development of lung cancer [21]. Increased DNMT levels have been suggested to reflect increased cell proliferation [21]. Several studies have illustrated the diagnostic potential of highly expressed DNMT1 in patients with NSCLC. These studies suggest that it may act as an independent prognostic factor for poor clinical prognosis [22,23]. Increased DNMT1 expression is consistent with the discovery of hypermethylated CpG islands in the promoter regions of tumor suppressor genes. Such hypermethylation results in silenced or decreased expression, leading to lung tumors [24].
A recent study has suggested that DNMT1 and the tyrosine kinase receptor KIT orchestrate lung tumorigenesis [25]. This study demonstrates a positive regulatory loop formed by DNMT1 and KIT. KIT overexpression in lung cancer leads to DNMT1 upregulation. Dual inhibition of DNMT1 and KIT expression synergistically inhibits lung tumor cell growth and metastasis, indicating that the regulatory and functional interplay between DNA methylation and tyrosine kinase signaling promotes tumorigenesis [25]. Similarly, overexpression of DNMT3a or DNMT3b has been reported in a variety of tumors [26,27]. Co-expression of DNMT1 and DNMT3a or DNMT3b in vivo leads to methylation spreading in the genome, suggesting cooperation between de novo and maintenance enzymes during DNA methylation [27].
Although it is well established that DNA methylation contributes to the pathogenesis of IPF [28], how DNMT expression and activity influences IPF is not entirely understood. In one study [29], higher DNMT3a and DNMT3b expression were found in patients with IPF relative to levels in normal control subjects, but no significant difference was observed in DNMT1 expression, suggesting that de novo DNMTs may be upregulated in IPF. The most intense DNMT3a staining occurs in proliferative epithelial cells covering fibroblast foci [29], indicating active methylation changes in these cells. These data suggest that DNA methylation mediated by different DNMTs may contribute to IPF pathogenesis.
4. Changes in MBPs in lung cancer and IPF
Methylated CpG sites are usually recognized by MBPs; at least four (MeCP2, MBD1, MBD2, and MBD4) have been reported in mammals [30]. Some studies have demonstrated that MBPs, such as MeCP2, MBD1, and MBD2, can be recruited to different sites to bind methylated CpG islands [31,32]. For example, MeCP2 and MBD1/2 bind differently to methylated CpG islands in the E-cadherin promoter region [33]. MeCP2 acts as an oncogene in tumorigenesis by silencing genes through hypermethylation in many cancers, including lung cancer [34]. In squamous lung carcinoma tissues, MeCP2 interacts directly with DNMT1 and binds to hypermethylated promoters of tumor suppressor genes such as FHIT, p16INK4a, and RARβ, resulting in lung tumorigenesis and poor prognosis [35,36].
The DNA-binding domain of MBPs, SRA, can recognize 5-methylcytosine (5mC) in hemimethylated CpG dinucleotides [37]. There are two family members: UHRF1 and UHRF2 (Figure 2B). UHRF1 recruits DNMT1 to maintain epigenetic inheritance of DNA methylation. In lung cancer, UHRF1 controls the cell cycle through promoter methylation of CDKN2A and RASSF1 [38]. SRA domain-containing proteins are attractive therapeutic targets because of their role in silencing tumor suppressor genes. Targeting SRA domain conjugates with small molecules to reduce abnormal DNA methylation is a potential therapeutic option for cancer [37].
MBPs also contribute to the pathogenesis of IPF [29]. In addition to their usual repressive roles, some MBPs play active roles in gene expression. MeCP2 has been reported to be a transcriptional activator involved in various fibrotic diseases, including lung fibrosis [39–41]. Studies have shown that MeCP2 positively regulates expression of the enzyme ASH1L (absent, minor, or homeotic disc1-like), which methylates H3K4 (histone3 lysine4); H3K4 is associated with the upregulation of α-SMA, TGF-β1, and another myofibrotic gene expression in liver fibrosis [39]. In a murine model of lung fibrosis, MeCP2 showed enhanced affinity for the methylated site of the α-SMA gene promoter, and overexpression of MeCP2 lead to increased α-SMA expression [40]. In another study on human lung fibroblasts, treatment with the DNA demethylation agent 5’-azacytidine was reported to downregulate α-SMA expression and reduce MeCP2 association with α-SMA [41]. These studies suggest that MeCP2 is a critical activator of the α-SMA gene and is essential for regulating α-SMA gene expression in lung fibroblasts.
5. Genes altered by DNA methylation in lung cancer and IPF
5.1. DNA methylation in lung cancer
Many genes are silenced or deregulated in lung cancer through epigenetic modifications [42]. Emerging studies on DNA methylation have identified many hypermethylated genes in lung cancer [43,44]. For example, many tumor suppressor genes are silenced by promoter hypermethylation in NSCLC cells. These tumor suppressor genes play essential roles in normal cellular functions, such as p16 acting in cell cycle regulation [45], DAPK and caspase-8 in apoptosis [46], TIMP-3 in suppression of invasion [47], and MGMT in DNA repair [48]. When hypermethylated, these tumor suppressor genes display reduced expression, which may correlate with tumor development and recurrence [49]. In patients with early NSCLC, the promoter methylation status of a panel of apoptosis/survival genes is associated with TNM-stages and reduced overall survival [50].
Besides apoptosis-related genes, O6-Methylguanine-DNA methyltransferase (MGMT) is one of the most studied DNA repair proteins [48]. MGMT silencing is involved in the carcinogenic process induced by smoking [51]. Higher levels of methylation (>50%) in the MGMT promoter are associated with the formation of bulky DNA adducts, suggesting that MGMT promoter hypermethylation is a common event in lung cancer patients [52]. In addition to MGMT, CDKN2A is another widely studied protein critical for controlling cell cycle progression, cellular senescence, and lung cancer development [53]. In lung cancer, CDKN2A promoter regions are methylated at a 20–70% frequency and can be feasibly detected in exhaled respiratory condensate [53]. Therefore, the methylation status of CDKN2A may be a valuable biomarker for NSCLC diagnosis [54].
DNA methylation status has been proposed for use in cancer diagnosis [55], especially given the convenience of isolating cell-free DNA from body fluids [56]. To improve the accuracy of diagnosing early-stage lung cancer, an ultrasensitive MOB-qMSP approach was used to detect the promoter methylation status of eight lung cancer-specific genes in patients with stage IA or IB NSCLC [55]. In this study, with plasma from patients, detectable methylated CDO1 (cysteine dioxygenase 1), TAC1 (tachykinin precursor 1), SOX17 (the gene of sex-determining region Y-box 17), and HOXA7 (the seventh gene of cluster A of the homeobox genes) were significantly increased compared to the benign group [55]. The combined measurement of CDO1, SOX17, and HOXA7 showed the highest sensitivity and specificity (90% and 71%, respectively) [55]. This study indicates that highly sensitive DNA methylation tests can be used to diagnose early-stage lung cancers.
Although hypermethylated DNA usually silences tumor suppressor genes, hypomethylated or demethylated DNA promotes lung cancer development [57]. Activation of tumor-associated genes is one of the main mechanisms of carcinogenesis [58]. DNA hypomethylation activates proto-oncogenes that drive the growth of malignant cells [59]. A recent study found that restoring the expression of tumor suppressor secreted frizzled-related proteins by demethylation inhibited the invasiveness of NSCLC [60]. DNA methylation changes usually occur during the early stages of carcinogenesis [61]. Genome-wide methylation analysis of LUSC showed that low-methylation epigenotypes were associated with unfavorable outcomes [12].
In addition to DNA, some miRNAs can be dysregulated by methylation, allowing them to promote cancer invasion and migration. For example, hypermethylation reduces miR-1247 expression in lung cancer, whereas miR-1247 overexpression or demethylation by 5-Aza-cytidine treatment markedly inhibits tumor cell growth and migration, and cell cycle progression [62].
5.2. DNA methylation in IPF
DNA methylation is not only involved in oncogenesis but also in IPF pathogenesis. Compared to healthy controls, patients with IPF have differentially methylated CpGs in genes related to apoptosis, biosynthesis, morphogenesis, proliferation, and EMT [63]. In myofibroblasts, DNA methylation directly regulates the expression of α-SMA [40] and other fibrosis-related proteins, such as thymocyte differentiation antigen-1 (Thy-1) [14,64]. Expression of Thy-1 has been reported to attenuate interstitial pulmonary fibrosis by inhibiting myofibroblast differentiation and increasing lung fibroblast apoptosis [65]. Thy-1 promoter methylation silences the gene in lung fibroblasts, increases cell apoptosis resistance, and increases ECM deposition and lung scar formation [14]. Through detecting genome-wide DNA methylation and RNA expression by array hybridization, a study found that the DNA methylation status and RNA expression of 16 genes were significantly changed in IPF compared with the control group; among the 16 genes, previous literature has demonstrated that eight of those are highly associated with lung fibrosis [29]. Another study performed integrative genomic analyses of DNA methylation and gene expression in 94 IPF patients and 67 control subjects, identifying 738 differentially methylated regions associated with significant changes in gene expression [66]. In addition to studies on lung tissues, a recent study evaluated the CpG methylation status and differential gene expression in IPF fibroblasts and controls [67]. These independent genome-wide studies in IPF revealed that DNA methylation is involved in the pathogenesis of IPF and may offer novel therapeutic targets [11,29,66].
6. Clinical application
6.1. Epigenetic biomarkers
Recently, digital PCR has emerged as a sensitive tool for detecting epigenetic changes and point mutations [68–70]. Technological advances and increased use of high-throughput epigenetic screening approaches allow the identification of relevant epigenomic biomarkers. DNA methylation is one of the most widely studied biomarkers in cancer. Methylated biomarkers associated with lung fibrosis and lung cancer are shown in Table 1. Among them, RASSF1A (RAS association domain family protein 1A) is a candidate tumor suppressor that has been extensively studied in many human tumors [28]. It is frequently inactivated by methylation of its promoter region. Through analysis of bronchoalveolar lavage fluid (BALF) and venous blood, a recent study found that the positive rate of RASSF1A aberrant methylation in BALF was higher in the lung cancer group than in the control group [28]. Another study found that RASSF1A methylation status combined with either the RARB or L1RE1 panel could achieve satisfactory sensitivity and specificity in lung cancer diagnosis, especially for separating lung cancer and non-cancerous tissue [71]. SOX17 is another proposed biomarker whose promoter has a significantly higher frequency of methylation in primary and advanced NSCLC tumors and in corresponding plasma samples [72]. Methylation of the SOX17 promoter also has a statistically significant effect on survival time [72].
Table1.
Epigenetic biomarkers associated with DNA methylation in lung cancer and lung fibrosis.
| Disease | Gene | Result | Biospecimen | Ref | |
|---|---|---|---|---|---|
| NSCLC | SOX17 | SOX17 promoter is highly methylated in primary tumors and in corresponding plasma samples, both in operable and advanced NSCLC | Tumor tissue and plasma | [72] | |
| NSCLC | APC | APC methylation in tumor sample may be a useful marker for superior survival in NSCLC patients | Tumor tissue | [94] | |
| NSCLC | RAR-β | RAR-β methylation detected in lung tissue may be used as a predictive marker for NSCLC diagnosis | Tumor tissue | [94] | |
| Lung cancer | RASSF1A | The methylation analysis of the RASSF1A panel achieved a satisfactory sensitivity and specificity in lung cancer diagnosis, especially in an early stage | bronchoalveolar lavage fluid | [95] | |
| Lung cancer | TMEM196 | Multivariate models showed TMEM196 methylation is an independent prognostic marker in lung cancer | Tumor tissue | [96] | |
| Lung cancer | L1RE1 | L1RE1 in combination with either RARB or RASSF1 could function as biomarkers for separating lung cancer and non-cancerous tissue | Tumor tissue | [71] | |
| LUAD | PITX1 | PITX1 might serve as a potential biomarker for early detection and prognosis prediction of LUAD patients | Tumor tissue | [97] | |
| LUAD | HOXA9 | HOXA9 promoter methylation could potentially inform the clinical management of patients with early-stage lung adenocarcinoma. | Tumor tissue | [98] | |
| LUAD | SMAD3 | the histotype-specific regulation of tumor fibrosis in lung cancer is mediated through differential SMAD3 promoter methylation in TAFs. | Tumor tissue | [99] | |
| IPF | Thy-1 | Inhibiting DNMT1 with silencing RNA attenuated TGF-β1-induced DNMT activity and its downstream suppression of Thy-1 mRNA and protein expression, as well as inhibited α-SMA mRNA and Col1A1 mRNA and protein expression, with a decreased trend in Thy-1 promoter methylation | lung tissue | [64] | |
| IPF | PTGER2 | down-regulation of PTGER2 and consequent PGE2 resistance are both mediated by DNA hypermethylation | lung tissue | [100] | |
| IPF | P14 | IPF fibroblasts have reduced expression of the proapoptotic p14(ARF) attributable to promoter hypermethylation and indicate that epigenetic mechanisms may underlie their resistance to apoptosis. | fibroblasts | [101] | |
| IPF | CDKN2B,CARD10, MGMT | CDKN2B and CARD10 were hypermethylated and MGMT was hypomethylated in IPF cells. Aberrant methylation of CDKN2B, CARD10, and MGMT in IPF fibroblasts is associated with altered gene expression and increased fibroblast proliferation | lung tissue | [102] | |
| IPF | Cox-2 | COX-2 hypermethylated in IPF fibroblasts | lung tissue | [103] | |
| IPF | c8orf4 | The decreased capacity of fibrotic lung fibroblasts to up-regulate COX-2 expression and COX-2-derived PGE2 synthesis is due to an indirect epigenetic mechanism involving hypermethylation of the transcriptional regulator, c8orf4. | Lung tissue | [2] | |
In addition to their use for diagnostics, some biomarkers, such as circulating epigenetic markers, can be used to predict treatment response and aid in decision-making after surgery before adjuvant chemotherapy [73]. For example, dynamic measurements of DNA methylation levels in plasma before and after chemotherapy can be used to monitor treatment response [73]. Studies have shown that within 24 h after chemotherapy, increased APC/RASSF1A methylation levels are associated with better chemotherapy response in advanced lung cancer [74]. However, many challenges remain in finding highly sensitive and specific biomarkers in cell-free DNA obtained by “liquid biopsy.” Some obstacles, such as equipment, sample collection, contamination, and tumor heterogeneity, need to be addressed before circulating epigenetic markers can be applied in clinical practice.
6.2. Epigenetic treatment
Preclinical studies have demonstrated the feasibility of using demethylating agents to re-express epigenetically silent tumor/fibrotic-suppressor genes for clinical treatment. One study on lung cancer indicated that antroquinonol D, a demethylating agent, could decrease aberrant promoter methylation of CCND2 to induce CCND2 expression, resulting in cell cycle arrest and inhibition of cancer cell growth and migration [75]. Several studies have shown that DNA methylation is associated with cisplatin resistance. Cisplatin was the first metal-based chemotherapeutic drug, and its resistance causes therapeutic failure and tumor recurrence [76]. For example, hypermethylated and significantly downregulated HOXA11 expression was observed in a cisplatin-resistant LUAD cell line (A549/DDP) and LUAD tissues [77]. In another study that profiled DNA methylation and mRNA expression in cisplatin-resistant NSCLC cells using high-throughput microarrays, researchers found that a total of 372 genes were hypermethylated with reduced expression in cisplatin-resistant A549 cells, and 10 selected genes (S100P, GDA, WISP2, LOXL1, TIMP4, ICAM1, CLMP, HSP8, GAS1, and BMP2) were found to be associated with cisplatin resistance [78]. Another report found that the development of resistance to the cisplatin analog was associated with hypermethylation of ABCB1 in the human LUAD cell line A549 and clinical tumor samples [79].
The most extensively studied epigenetic drugs currently in clinical trials are DNMT inhibitors (DNMTi), including 5-Aza-cytidine and 5-Aza-2’-deoxycytidine, which increase tumor sensitivity to chemotherapy in lung cancer and several malignancies [80,81]. Low-dose 5-Aza-2’-deoxycytidine was shown to decrease the incidence of lung cancer by 30% in tobacco carcinogen-induced murine lung cancer, and a 50% decrease was achieved by combining 5-Aza-2’-deoxycytidine with HDACi sodium phenylbutyrate [82]. A clinical phase I study of 5-Aza-2’-deoxycytidine in combination with valproic acid in NSCLC patients suggested a route to a novel clinical strategy to treat lung cancer [83].
For lung cancer patients with pulmonary fibrosis, current research has focused on the TGF-β pathway, which is central to tissue fibrosis and tumorigenesis [84]. Abundant TGF-β in IPF lung tissues can induce dysregulated immune surveillance and provide a microenvironment favorable to cancer initiation and progression [84]. BMP endothelial cell precursor-derived regulator (BMPER) is a crucial regulator of fibroblast activation and is associated with IPF progression. BMPER binds directly to BMPs and regulates TGF-β/BMP signaling [85]. Although BMPER gene methylation status was not examined, BMPER was downregulated by the demethylation agent 5’-azacytidine in fibroblasts. This treatment also reduced lung fibrosis in mice in vivo [85]. 5-Aza-2’-deoxycytidine treatment attenuates hyperoxia-induced pulmonary fibrosis in neonatal rats by decreasing TGF-β1 expression and increasing p16 expression by reversing the hypermethylation of p16 [86]. In addition to TGF-β1, many other biomarkers, including CXCL10 and TET1, may be potential targets for epigenetic therapy of pulmonary fibrosis [87,88].
The DNA methylation machinery is a promising target for lung cancer and fibrosis treatment. Some of the drawbacks of using DNMT inhibitors, such as drug toxicity and reversal of their effects after drug withdrawal, may be overcome by combined therapy with other chemotherapeutic agents. Additionally, identifying novel DNA methylation-related biomarkers to select patients who would best respond to specific epigenetic therapeutic methods would provide better-targeted treatment.
7. Expert Opinion
Lung cancer and fibrosis are two distinct diseases that share multiple cellular and molecular mechanisms, including alterations in DNA methylation. In this review, we focus on traditional DNA methylation, excluding other forms of DNA modification, such as the recently identified DNA hydroxymethylation, which shows its importance as an epigenetic regulator of gene expression. DNA methylation and demethylation play pivotal roles in lung cancer and fibrosis. DNA methylation at a gene’s regulatory region can either directly regulate gene expression or recruit MBPs to areas that affect related regulatory complexes, activating or repressing gene expression according to cellular cues. Alterations in methylation patterns can be used for diagnosis or therapeutic targets. Although epigenetic biomarkers reveal substantial potential for clinical application, these studies are still in their infancy. DNA methylation signatures are stable and relatively easy to detect in tissues and body fluids [89,90]. Establishing such markers would be invaluable for the early diagnosis and prognosis of lung cancer and lung fibrosis, which would also aid in predicting treatment efficacy and tracking treatment efficiency or resistance. Efforts to identify and establish methylation biomarkers using plasma cell-free DNA have been reported in both lung cancer and fibrosis, and were able to differentiate between lung cancer, pulmonary fibrosis, and healthy subjects [91]. However, lung cancer transcriptome and methylation profiles in patients with IPF remain unclear.
Genome-wide DNA methylation studies in lung cancer and fibrosis provide a better understanding of the major players in both diseases. In comparison to DNA methylation, the proteins that are involved in DNA methylation, such as MBPs, methyltransferases, and demethylases, have been relatively underexplored. Changes in these proteins, like their expression and posttranslational modifications, could lead to altered DNA methylation patterns and chromatin structures, which would result in altered transcriptional regulation and underlying human diseases [31]. Emerging studies have indicated their importance in the maintenance of epigenetic homeostasis. Future studies should reveal their roles in preserving the normal epigenome and correcting disease outcomes.
DNA methylation is a major part of the epigenome, whereas lung cancer and lung fibrosis display different profiles. More extensive studies are needed to establish DNA methylation profiles that can be used to predict lung cancer or lung fibrosis and monitor disease progression and treatment efficiency. DNA methylation is a good candidate as an early biomarker to detect lung cancer or fibrosis and as a therapeutic target to restore the epigenome to physiological conditions. Epigenetic modifiers, such as DNMT inhibitors, are in clinical trials for lung cancer treatment, but have shown limited success to date. Some tests with histone deacetylation inhibitors have shown better outcomes [92]. Because many of these inhibitors are not gene-specific, the methylation status of other genes would be altered by treatment with these inhibitors, causing side effects that were not discovered in preclinical studies. Extensive studies and expanded markers are required to broadly monitor changes. Specific inhibitors would provide better-targeted treatment. With advances in biotechnology, techniques that could target the correct specific gene methylation status would have minimal side effects while providing maximum efficacy in restoring the native epigenome. In vivo studies of DNA methylation modifiers have primarily been performed in lung cancer, mainly in preclinical models of lung fibrosis.
Personalized therapy is of utmost importance due to the high variability among lung cancer and fibrosis patients. Drug resistance is partially caused by individual epigenetic heterogeneity. Different cancer or fibrosis subtypes would have varying sensitivities to a given treatment, probably due to epigenetic and genetic differences [93]. DNA methylation patterns are stable and relatively easy to detect, especially cfDNA, which could provide an early marker for diagnosis and treatment monitoring. Soon, integrating data from genomics, transcriptomics, and epigenomics to facilitate the discovery of relevant epigenetic therapeutic targets will be incorporated into the clinical approach to addressing chemoresistance and reversing immune escape in lung cancer and/or fibrosis treatment. A better understanding of pulmonary fibrosis pathogenesis and its association with cancer will aid the development of more advanced diagnostic and therapeutic strategies for both pulmonary fibrosis and lung cancer.
Article Highlights.
There are many similarities between lung cancer and fibrosis, including the same high-risk factors (old age and smoking), similar pathogenic pathways, and low survival rates.
Aberrant DNA methylation contributes to both diseases development and progression.
Increased DNA methyltransferase expression has been reported in lung cancer; however, their roles in IPF remain unclear.
Methyl-binding proteins are involved in lung cancer and fibrosis pathogenesis.
DNA hypermethylation silences or downregulates tumor suppressor and/or antifibrotic genes in both diseases. Hypomethylated genes promote lung cancer development, which may not be typical for IPF. However, differentially methylated regions associated with gene expression alterations have been reported in IPF.
A unique profile of DNA methylation changes occur at early stages in lung cancer or lung fibrosis, allowing DNA methylation to be used as a biomarker for early diagnosis, and to monitor treatment response.
DNA methylation is a promising therapeutic target for lung cancer and fibrosis treatment.
Acknowledgments
The authors thank Dr. Shan Zhu for editorial assistance.
Funding:
This work was partially supported by the National Natural Science Foundation of China (No. 81470256) (X.Z.), Natural Science Foundation of Hunan Province, China (No.2020JJ5802) (J.D.), and US NIH grants R01AG050567 and R01HL151702 (Y.Y.S).
Declaration of interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Reference:
- 1.Fang Y, Tian J, Fan Y, et al. Latest progress on the molecular mechanisms of idiopathic pulmonary fibrosis. Mol Biol Rep. 2020. Dec;47(12):9811–9820. [DOI] [PubMed] [Google Scholar]
- 2.Evans IC, Barnes JL, Garner IM, et al. Epigenetic regulation of cyclooxygenase-2 by methylation of c8orf4 in pulmonary fibrosis. Clinical science (London, England : 1979). 2016. Apr;130(8):575–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sack C, Raghu G. Idiopathic pulmonary fibrosis: unmasking cryptogenic environmental factors. Eur Respir J. 2019. Feb;53(2). [DOI] [PubMed] [Google Scholar]
- 4.Karampitsakos T, Tzilas V, Tringidou R, et al. Lung cancer in patients with idiopathic pulmonary fibrosis. Pulm Pharmacol Ther. 2017. Aug;45:1–10. [DOI] [PubMed] [Google Scholar]; * of interest. Reviewed the common pathology, clinical and therapetutic data involved in both diseases.
- 5.Poletti V, Ravaglia C, Buccioli M, et al. Idiopathic pulmonary fibrosis: diagnosis and prognostic evaluation. Respiration. 2013;86(1):5–12. [DOI] [PubMed] [Google Scholar]
- 6.Tzouvelekis A, Spagnolo P, Bonella F, et al. Patients with IPF and lung cancer: diagnosis and management. The Lancet Respiratory medicine. 2018. Feb;6(2):86–88. [DOI] [PubMed] [Google Scholar]
- 7.Duruisseaux M, Esteller M. Lung cancer epigenetics: From knowledge to applications. Seminars in cancer biology. 2018. Aug;51:116–128. [DOI] [PubMed] [Google Scholar]
- 8.Lee T, Park JY, Lee HY, et al. Lung cancer in patients with idiopathic pulmonary fibrosis: clinical characteristics and impact on survival. Respiratory medicine. 2014. Oct;108(10):1549–55. [DOI] [PubMed] [Google Scholar]
- 9.Ballester B, Milara J, Cortijo J. Idiopathic Pulmonary Fibrosis and Lung Cancer: Mechanisms and Molecular Targets. Int J Mol Sci. 2019;20(3):593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kinoshita T, Goto T. Molecular Mechanisms of Pulmonary Fibrogenesis and Its Progression to Lung Cancer: A Review. Int J Mol Sci. 2019. Mar 22;20(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rabinovich EI, Kapetanaki MG, Steinfeld I, et al. Global methylation patterns in idiopathic pulmonary fibrosis. PLoS One. 2012;7(4):e33770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hata A, Nakajima T, Matsusaka K, et al. A low DNA methylation epigenotype in lung squamous cell carcinoma and its association with idiopathic pulmonary fibrosis and poorer prognosis. Int J Cancer. 2020. Jan 15;146(2):388–399. [DOI] [PubMed] [Google Scholar]
- 13.Das PM, Singal R. DNA methylation and cancer [Research Support, U.S. Gov’t, Non-P.H.S. Research Support, U.S. Gov’t, P.H.S. Review]. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2004. Nov 15;22(22):4632–42. [DOI] [PubMed] [Google Scholar]
- 14.Sanders YY, Pardo A, Selman M, et al. Thy-1 promoter hypermethylation: a novel epigenetic pathogenic mechanism in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2008. Nov;39(5):610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tzouvelekis A, Kaminski N. Epigenetics in idiopathic pulmonary fibrosis. Biochem Cell Biol. 2015. Apr;93(2):159–70. [DOI] [PubMed] [Google Scholar]
- 16.Gujar H, Weisenberger DJ, Liang G. The Roles of Human DNA Methyltransferases and Their Isoforms in Shaping the Epigenome [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Review]. Genes. 2019. Feb 23;10(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robertson KD, Ait-Si-Ali S, Yokochi T, et al. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nature genetics. 2000. Jul;25(3):338–42. [DOI] [PubMed] [Google Scholar]
- 18.Okano M, Bell DW, Haber DA, et al. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999. Oct 29;99(3):247–57. [DOI] [PubMed] [Google Scholar]
- 19.Choi WI, Jeon BN, Yoon JH, et al. The proto-oncoprotein FBI-1 interacts with MBD3 to recruit the Mi-2/NuRD-HDAC complex and BCoR and to silence p21WAF/CDKN1A by DNA methylation [Research Support, Non-U.S. Gov’t]. Nucleic acids research. 2013. Jul;41(13):6403–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Hu M, Lyu X, et al. DNA methylation regulated gene expression in organ fibrosis. Biochim Biophys Acta Mol Basis Dis. 2017. Sep;1863(9):2389–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang M, Xu W, Wang Q, et al. Potential of DNMT and its Epigenetic Regulation for Lung Cancer Therapy. Current genomics. 2009. Aug;10(5):336–52. [DOI] [PMC free article] [PubMed] [Google Scholar]; *of interest. Summarized the functions of DNMTs, their role in lung cancer, and related theraputic methods.
- 22.Wu XY, Chen HC, Li WW, et al. DNMT1 promotes cell proliferation via methylating hMLH1 and hMSH2 promoters in EGFR-mutated non-small cell lung cancer. Journal of biochemistry. 2020. Aug 1;168(2):151–157. [DOI] [PubMed] [Google Scholar]
- 23.Parbin S, Pradhan N, Das L, et al. DNA methylation regulates Microtubule-associated tumor suppressor 1 in human non-small cell lung carcinoma. Exp Cell Res. 2019. Jan 15;374(2):323–332. [DOI] [PubMed] [Google Scholar]
- 24.Liang G, Weisenberger DJ. DNA methylation aberrancies as a guide for surveillance and treatment of human cancers [Review Research Support, N.I.H., Extramural]. Epigenetics. 2017. Jun 3;12(6):416–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan F, Shen N, Pang J, et al. A regulatory circuit composed of DNA methyltransferases and receptor tyrosine kinases controls lung cancer cell aggressiveness. Oncogene. 2017. Dec 14;36(50):6919–6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weinberg DN, Papillon-Cavanagh S, Chen H, et al. The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature. 2019. Sep;573(7773):281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]; **of considerable interest. Studies connected chromatin regulatory pathway with DNA methylation.
- 27.Kim GD, Ni J, Kelesoglu N, et al. Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. EMBO J. 2002. Aug 1;21(15):4183–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang C, Yu W, Wang L, et al. DNA Methylation Analysis of the SHOX2 and RASSF1A Panel in Bronchoalveolar Lavage Fluid for Lung Cancer Diagnosis. Journal of Cancer. 2017;8(17):3585–3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanders YY, Ambalavanan N, Halloran B, et al. Altered DNA methylation profile in idiopathic pulmonary fibrosis [Comparative Study Research Support, N.I.H., Extramural]. American journal of respiratory and critical care medicine. 2012. Sep 15;186(6):525–35. [DOI] [PMC free article] [PubMed] [Google Scholar]; **of considerable interest. Performed DNA methylation array with IPF lung tissues; examined hyper- or hypo- DNA methylation status effects on RNA expression in these IPF lung tissues.
- 30.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998. Nov;18(11):6538–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du Q, Luu PL, Stirzaker C, et al. Methyl-CpG-binding domain proteins: readers of the epigenome. Epigenomics. 2015;7(6):1051–73. [DOI] [PubMed] [Google Scholar]
- 32.Gunther K, Rust M, Leers J, et al. Differential roles for MBD2 and MBD3 at methylated CpG islands, active promoters and binding to exon sequences. Nucleic acids research. 2013. Mar 1;41(5):3010–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koizume S, Tachibana K, Sekiya T, et al. Heterogeneity in the modification and involvement of chromatin components of the CpG island of the silenced human CDH1 gene in cancer cells. Nucleic acids research. 2002. Nov 1;30(21):4770–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parry L, Clarke AR. The Roles of the Methyl-CpG Binding Proteins in Cancer. Genes & cancer. 2011. Jun;2(6):618–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ozbayer C, Degirmenci I, Ustuner D, et al. miRSNPs of miR1274 and miR3202 Genes that Target MeCP2 and DNMT3b Are Associated with Lung Cancer Risk: A Study Conducted on MassARRAY Genotyping. Journal of environmental pathology, toxicology and oncology : official organ of the International Society for Environmental Toxicology and Cancer. 2016;35(3):223–236. [DOI] [PubMed] [Google Scholar]
- 36.Lin RK, Hsu HS, Chang JW, et al. Alteration of DNA methyltransferases contributes to 5’CpG methylation and poor prognosis in lung cancer [Research Support, Non-U.S. Gov’t]. Lung Cancer. 2007. Feb;55(2):205–13. [DOI] [PubMed] [Google Scholar]
- 37.Patnaik D, Esteve PO, Pradhan S. Targeting the SET and RING-associated (SRA) domain of ubiquitin-like, PHD and ring finger-containing 1 (UHRF1) for anti-cancer drug development [Review]. Oncotarget. 2018. May 25;9(40):26243–26258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daskalos A, Oleksiewicz U, Filia A, et al. UHRF1-mediated tumor suppressor gene inactivation in nonsmall cell lung cancer. Cancer. 2011. Mar 1;117(5):1027–37. [DOI] [PubMed] [Google Scholar]
- 39.Bian EB, Huang C, Wang H, et al. The role of methyl-CpG binding protein 2 in liver fibrosis. Toxicology. 2013. Jul 5;309:9–14. [DOI] [PubMed] [Google Scholar]
- 40.Hu B, Gharaee-Kermani M, Wu Z, et al. Essential role of MeCP2 in the regulation of myofibroblast differentiation during pulmonary fibrosis. Am J Pathol. 2011. Apr;178(4):1500–8. [DOI] [PMC free article] [PubMed] [Google Scholar]; *of interest. Studies showed how MeCP2 regulates alpha-smooth muscle actin expression in lung fibroblast.
- 41.Xiang Z, Zhou Q, Hu M, et al. MeCP2 epigenetically regulates alpha-smooth muscle actin in human lung fibroblasts. J Cell Biochem. 2020. Jul;121(7):3616–3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Darilmaz Yuce G, Ortac Ersoy E. [Lung cancer and epigenetic modifications]. Tuberkuloz ve toraks. 2016. Jun;64(2):163–70. [DOI] [PubMed] [Google Scholar]
- 43.Wang J, Zhao Y, Xu H, et al. Silencing NID2 by DNA Hypermethylation Promotes Lung Cancer. Pathology oncology research : POR. 2019. Mar 2. [DOI] [PubMed] [Google Scholar]
- 44.Wu J, He K, Zhang Y, et al. Inactivation of SMARCA2 by promoter hypermethylation drives lung cancer development. Gene. 2019. Mar 1;687:193–199. [DOI] [PubMed] [Google Scholar]
- 45.Ansari J, Shackelford RE, El-Osta H. Epigenetics in non-small cell lung cancer: from basics to therapeutics [Review]. Translational lung cancer research. 2016. Apr;5(2):155–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan X, Zhou R, Ma Z. Autophagy-Cell Survival and Death [Review]. Advances in experimental medicine and biology. 2019;1206:667–696. [DOI] [PubMed] [Google Scholar]
- 47.Gu C, Luo Y, Zhang S, et al. MAb NJ001 inhibits lung adenocarcinoma invasiveness by directly regulating TIMP-3 promoter activity via FOXP1 binding sites. Thorac Cancer. 2020. Sep;11(9):2630–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Christmann M, Kaina B. Epigenetic regulation of DNA repair genes and implications for tumor therapy [Research Support, Non-U.S. Gov’t Review]. Mutation research. 2019. Apr - Jun;780:15–28. [DOI] [PubMed] [Google Scholar]
- 49.Zhang W, Klinkebiel D, Barger CJ, et al. Global DNA Hypomethylation in Epithelial Ovarian Cancer: Passive Demethylation and Association with Genomic Instability. Cancers. 2020. Mar 24;12(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sutic M, Motzek A, Bubanovic G, et al. Promoter methylation status of ASC/TMS1/PYCARD is associated with decreased overall survival and TNM status in patients with early stage non-small cell lung cancer (NSCLC). Translational lung cancer research. 2019. Dec;8(6):1000–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]; *of interest. A preliminary study showing specific genes methylation status could be prognostic biomarkers in early stage lung cancer.
- 51.Povey AC, O’Donnell P, Barber P, et al. Smoking is associated with a decrease of O6-alkylguanine-DNA alkyltransferase activity in bronchial epithelial cells. Int J Cancer. 2006. Jul 15;119(2):463–6. [DOI] [PubMed] [Google Scholar]
- 52.Martinez-Ramirez OC, Perez-Morales R, Castro-Hernandez C, et al. Association of the Promoter Methylation and the rs12917 Polymorphism of MGMT with Formation of DNA Bulky Adducts and the Risk of Lung Cancer in Mexican Mestizo Population. DNA Cell Biol. 2019. Apr;38(4):307–313. [DOI] [PubMed] [Google Scholar]
- 53.Shi YX, Sheng DQ, Cheng L, et al. Current Landscape of Epigenetics in Lung Cancer: Focus on the Mechanism and Application. J Oncol. 2019;2019:8107318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiao P, Chen JR, Zhou F, et al. Methylation of P16 in exhaled breath condensate for diagnosis of non-small cell lung cancer. Lung Cancer. 2014. Jan;83(1):56–60. [DOI] [PubMed] [Google Scholar]
- 55.Chen C, Huang X, Yin W, et al. Ultrasensitive DNA hypermethylation detection using plasma for early detection of NSCLC: a study in Chinese patients with very small nodules. Clin Epigenetics. 2020. Mar 5;12(1):39. [DOI] [PMC free article] [PubMed] [Google Scholar]; **of considerable interest. Method to detect specific gene DNA methylation status with plasma samples in lung cancer patients.
- 56.Sanders YY. New Clue: Prediction from Cell-Free DNA. J Clin Med. 2020;9(7):2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vizoso M, Puig M, Carmona FJ, et al. Aberrant DNA methylation in non-small cell lung cancer-associated fibroblasts. Carcinogenesis. 2015. Dec;36(12):1453–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith J, Sen S, Weeks RJ, et al. Promoter DNA Hypermethylation and Paradoxical Gene Activation. Trends Cancer. 2020. May;6(5):392–406. [DOI] [PubMed] [Google Scholar]; *of interest. Updated current knowledge of promoter DNA methylation on gene expression, which not only associates with transcriptional repression, but also with high levels of gene expression; offered insights of the possible mechanisms.
- 59.Duthie SJ. Epigenetic modifications and human pathologies: cancer and CVD. Proc Nutr Soc. 2011. Feb;70(1):47–56. [DOI] [PubMed] [Google Scholar]
- 60.Zhang X, Rong X, Chen Y, et al. Methylation-mediated loss of SFRP2 enhances invasiveness of non-small cell lung cancer cells. Human & experimental toxicology. 2018. Feb;37(2):155–162. [DOI] [PubMed] [Google Scholar]
- 61.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005. Aug;6(8):597–610. [DOI] [PubMed] [Google Scholar]
- 62.Zhang J, Fu J, Pan Y, et al. Silencing of miR-1247 by DNA methylation promoted non-small-cell lung cancer cell invasion and migration by effects of STMN1. OncoTargets and therapy. 2016;9:7297–7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dowson C, O’Reilly S. DNA methylation in fibrosis [Review]. European journal of cell biology. 2016. Sep;95(9):323–30. [DOI] [PubMed] [Google Scholar]
- 64.Neveu WA, Mills ST, Staitieh BS, et al. TGF-beta1 epigenetically modifies Thy-1 expression in primary lung fibroblasts. Am J Physiol Cell Physiol. 2015. Nov 1;309(9):C616–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanders YY, Kumbla P, Hagood JS. Enhanced myofibroblastic differentiation and survival in Thy-1(−) lung fibroblasts. Am J Respir Cell Mol Biol. 2007. Feb;36(2):226–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang IV, Pedersen BS, Rabinovich E, et al. Relationship of DNA methylation and gene expression in idiopathic pulmonary fibrosis. American journal of respiratory and critical care medicine. 2014. Dec 1;190(11):1263–72. [DOI] [PMC free article] [PubMed] [Google Scholar]; **of considerable interest. Identified methylation marks that modify gene expression in IPF lungs.
- 67.Lee JU, Son JH, Shim EY, et al. Global DNA Methylation Pattern of Fibroblasts in Idiopathic Pulmonary Fibrosis. DNA Cell Biol. 2019. Sep;38(9):905–914. [DOI] [PubMed] [Google Scholar]
- 68.Foroni C, Zarovni N, Bianciardi L, et al. When Less Is More: Specific Capture and Analysis of Tumor Exosomes in Plasma Increases the Sensitivity of Liquid Biopsy for Comprehensive Detection of Multiple Androgen Receptor Phenotypes in Advanced Prostate Cancer Patients. Biomedicines. 2020. May 22;8(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sato H, Soh J, Aoe K, et al. Droplet digital PCR as a novel system for the detection of microRNA34b/c methylation in circulating DNA in malignant pleural mesothelioma. International journal of oncology. 2019. Jun;54(6):2139–2148. [DOI] [PubMed] [Google Scholar]
- 70.Frazzi R, Bizzarri V, Albertazzi L, et al. Droplet digital PCR is a sensitive tool for the detection of TP53 deletions and point mutations in chronic lymphocytic leukaemia. Br J Haematol. 2020. Apr;189(2):e49–e52. [DOI] [PubMed] [Google Scholar]
- 71.Walter RFH, Rozynek P, Casjens S, et al. Methylation of L1RE1, RARB, and RASSF1 function as possible biomarkers for the differential diagnosis of lung cancer. PLoS One. 2018;13(5):e0195716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Balgkouranidou I, Chimonidou M, Milaki G, et al. SOX17 promoter methylation in plasma circulating tumor DNA of patients with non-small cell lung cancer. Clinical chemistry and laboratory medicine. 2016. Aug 1;54(8):1385–93. [DOI] [PubMed] [Google Scholar]; *of interest. Evaluated DNA methylation status in circulating tumor DNA in plasma of lung cancer patients.
- 73.Tomasetti M, Amati M, Neuzil J, et al. Circulating epigenetic biomarkers in lung malignancies: From early diagnosis to therapy. Lung Cancer. 2017. May;107:65–72. [DOI] [PubMed] [Google Scholar]
- 74.Wang H, Zhang B, Chen D, et al. Real-time monitoring efficiency and toxicity of chemotherapy in patients with advanced lung cancer. Clin Epigenetics. 2015;7:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hung CS, Wang SC, Yen YT, et al. Hypermethylation of CCND2 in Lung and Breast Cancer Is a Potential Biomarker and Drug Target. Int J Mol Sci. 2018. Oct 10;19(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Galluzzi L, Vitale I, Michels J, et al. Systems biology of cisplatin resistance: past, present and future. Cell death & disease. 2014. May 29;5(5):e1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Y, Yuan Y, Li Y, et al. An inverse interaction between HOXA11 and HOXA11-AS is associated with cisplatin resistance in lung adenocarcinoma. Epigenetics. 2019. Oct;14(10):949–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang YW, Zheng Y, Wang JZ, et al. Integrated analysis of DNA methylation and mRNA expression profiling reveals candidate genes associated with cisplatin resistance in non-small cell lung cancer. Epigenetics. 2014. Jun;9(6):896–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li A, Song J, Lai Q, et al. Hypermethylation of ATP-binding cassette B1 (ABCB1) multidrug resistance 1 (MDR1) is associated with cisplatin resistance in the A549 lung adenocarcinoma cell line. International journal of experimental pathology. 2016. Dec;97(6):412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fuller M, Klein M, Schmidt E, et al. 5-azacytidine enhances efficacy of multiple chemotherapy drugs in AML and lung cancer with modulation of CpG methylation. International journal of oncology. 2015. Mar;46(3):1192–204. [DOI] [PubMed] [Google Scholar]
- 81.Hu Q, Yu L, Chen R, et al. 5-aza-2’-deoxycytidine improves the sensitivity of endometrial cancer cells to progesterone therapy. Int J Gynecol Cancer. 2012. Jul;22(6):951–9. [DOI] [PubMed] [Google Scholar]
- 82.Belinsky SA, Klinge DM, Stidley CA, et al. Inhibition of DNA methylation and histone deacetylation prevents murine lung cancer. Cancer research. 2003. Nov 1;63(21):7089–93. [PubMed] [Google Scholar]
- 83.Chu BF, Karpenko MJ, Liu Z, et al. Phase I study of 5-aza-2’-deoxycytidine in combination with valproic acid in non-small-cell lung cancer. Cancer Chemother Pharmacol. 2013. Jan;71(1):115–21. [DOI] [PubMed] [Google Scholar]
- 84.Saito A, Horie M, Micke P, et al. The Role of TGF-beta Signaling in Lung Cancer Associated with Idiopathic Pulmonary Fibrosis. Int J Mol Sci. 2018. Nov 15;19(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huan C, Yang T, Liang J, et al. Methylation-mediated BMPER expression in fibroblast activation in vitro and lung fibrosis in mice in vivo. Scientific reports. 2015. Oct 7;5:14910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhao SM, Wu HM, Cao ML, et al. 5-aza-2’-deoxycytidine, a DNA methylation inhibitor, attenuates hyperoxia-induced lung fibrosis via re-expression of P16 in neonatal rats. Pediatric research. 2018. Mar;83(3):723–730. [DOI] [PubMed] [Google Scholar]
- 87.Zhang S, Liu H, Liu Y, et al. miR-30a as Potential Therapeutics by Targeting TET1 through Regulation of Drp-1 Promoter Hydroxymethylation in Idiopathic Pulmonary Fibrosis. Int J Mol Sci. 2017. Mar 15;18(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Coward WR, Brand OJ, Pasini A, et al. Interplay between EZH2 and G9a Regulates CXCL10 Gene Repression in Idiopathic Pulmonary Fibrosis. Am J Respir Cell Mol Biol. 2018. Apr;58(4):449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Konecny M, Markus J, Waczulikova I, et al. The value of SHOX2 methylation test in peripheral blood samples used for the differential diagnosis of lung cancer and other lung disorders. Neoplasma. 2016;63(2):246–53. [DOI] [PubMed] [Google Scholar]
- 90.Diaz-Lagares A, Mendez-Gonzalez J, Hervas D, et al. A Novel Epigenetic Signature for Early Diagnosis in Lung Cancer. Clin Cancer Res. 2016. Jul 1;22(13):3361–71. [DOI] [PubMed] [Google Scholar]; *of interest. Demonstrated methods to use hypermethylation DNA as minimally invasive epigenetic markers for early lung cancer diganosis.
- 91.Wielscher M, Vierlinger K, Kegler U, et al. Diagnostic Performance of Plasma DNA Methylation Profiles in Lung Cancer, Pulmonary Fibrosis and COPD. EBioMedicine. 2015. Aug;2(8):929–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Forde PM, Brahmer JR, Kelly RJ. New strategies in lung cancer: epigenetic therapy for non-small cell lung cancer. Clin Cancer Res. 2014. May 1;20(9):2244–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Toh TB, Lim JJ, Chow EK. Epigenetics in cancer stem cells. Mol Cancer. 2017. Feb 1;16(1):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Feng H, Zhang Z, Qing X, et al. Promoter methylation of APC and RAR-beta genes as prognostic markers in non-small cell lung cancer (NSCLC). Experimental and molecular pathology. 2016. Feb;100(1):109–13. [DOI] [PubMed] [Google Scholar]
- 95.Ren M, Wang C, Sheng D, et al. Methylation analysis of SHOX2 and RASSF1A in bronchoalveolar lavage fluid for early lung cancer diagnosis. Annals of diagnostic pathology. 2017. Apr;27:57–61. [DOI] [PubMed] [Google Scholar]
- 96.Liu WB, Han F, Huang YS, et al. TMEM196 hypermethylation as a novel diagnostic and prognostic biomarker for lung cancer. Molecular carcinogenesis. 2019. Apr;58(4):474–487. [DOI] [PubMed] [Google Scholar]
- 97.Song X, Zhao C, Jiang L, et al. High PITX1 expression in lung adenocarcinoma patients is associated with DNA methylation and poor prognosis. Pathology, research and practice. 2018. Dec;214(12):2046–2053. [DOI] [PubMed] [Google Scholar]
- 98.Lissa D, Ishigame T, Noro R, et al. HOXA9 methylation and blood vessel invasion in FFPE tissues for prognostic stratification of stage I lung adenocarcinoma patients. Lung Cancer. 2018. Aug;122:151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ikemori R, Gabasa M, Duch P, et al. Epigenetic SMAD3 repression in tumor-associated fibroblasts impairs fibrosis and response to the antifibrotic drug nintedanib in lung squamous cell carcinoma. Cancer research. 2019. Nov 6. [DOI] [PubMed] [Google Scholar]; **of condiserable interest. Study links smoking, anatomic location of adenocarcinoma and squamous cell carcinoma, and the differential SMAD2 promoter methylation, SMAD3/SMAD2 balance, fibrosis, and response to antifibrotic drug.
- 100.Huang SK, Fisher AS, Scruggs AM, et al. Hypermethylation of PTGER2 confers prostaglandin E2 resistance in fibrotic fibroblasts from humans and mice. Am J Pathol. 2010. Nov;177(5):2245–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cisneros J, Hagood J, Checa M, et al. Hypermethylation-mediated silencing of p14(ARF) in fibroblasts from idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2012. Aug 15;303(4):L295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang SK, Scruggs AM, McEachin RC, et al. Lung fibroblasts from patients with idiopathic pulmonary fibrosis exhibit genome-wide differences in DNA methylation compared to fibroblasts from nonfibrotic lung. PLoS One. 2014;9(9):e107055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Coward WR, Feghali-Bostwick CA, Jenkins G, et al. A central role for G9a and EZH2 in the epigenetic silencing of cyclooxygenase-2 in idiopathic pulmonary fibrosis [Research Support, Non-U.S. Gov’t]. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2014. Jul;28(7):3183–96. [DOI] [PMC free article] [PubMed] [Google Scholar]


