Abstract
Xiang pig (XP) is one of the best-known indigenous pig breeds in China, which is characterized by its small body size, strong disease resistance, high adaptability, favorite meat quality, small litter sizes, and early sexual maturity. However, the genomic evidence that links these unique traits of XP is still poorly understood. To identify the genomic signatures of selection in XP, we performed whole-genome resequencing on 25 unrelated individual XPs. We obtained 876.70 Gb of raw data from the genomic libraries. The LD analysis showed that the lowest level of linkage disequilibrium was observed in Xiang pig. Comparative genomic analysis between XPs and other breeds including Tibetan, Meishan, Duroc and Landrace revealed 3062, 1228, 907 and 1519 selected regions, respectively. The genes identified in selected regions of XPs were associated with growth and development processes (IGF1R, PROP1, TBX19, STAC3, RLF, SELENOM, MSTN), immunity and disease resistance (ZCCHC2, SERPINB2, ADGRE5, CYP7B1, STAT6, IL2, CD80, RHBDD3, PIK3IP1), environmental adaptation (NR2E1, SERPINB8, SERPINB10, SLC26A7, MYO1A, SDR9C7, UVSSA, EXPH5, VEGFC, PDE1A), reproduction (CCNB2, TRPM6, EYA3, CYP7B1, LIMK2, RSPO1, ADAM32, SPAG16), meat quality traits (DECR1, EWSR1), and early sexual maturity (TAC3). Through the absolute allele frequency difference (ΔAF) analysis, we explored two population-specific missense mutations occurred in NR6A1 and LTBP2 genes, which well explained that the vertebrae numbers of Xiang pigs were less than that of the European pig breeds. Our results indicated that Xiang pigs were less affected by artificial selection than the European and Meishan pig breeds. The selected candidate genes were mainly involved in growth and development, disease resistance, reproduction, meat quality, and early sexual maturity. This study provided a list of functional candidate genes, as well as a number of genetic variants, which would provide insight into the molecular basis for the unique traits of Xiang pig.
Subject terms: Computational biology and bioinformatics, Evolution, Genetics, Molecular biology
Introduction
Pigs (Sus scrofa) are important agriculture animals with an ancient domestication history and economic values and have become an important source of protein for human food. Moreover, the pig serves as a valuable biomedical model for some specific human diseases such as obesity and cardiovascular diseases1,2. Pigs from the original European and Asian wild boars were independently domesticated approximately 10,000 years ago3. From early domestication to modern breeding practices, pigs have undergone a series of natural and artificial selections in various environments which have resulted in high levels of phenotypic diversity in morphological, physiological and behavior traits. There are more than 730 pig breeds or lines worldwide, two thirds of which are distributed in China and Europe. These pig breeds show different characteristics with respect to size, color, body shape, ear carriage, behavior, prolificacy, as well as other traits. For example, European domestic Duroc and Yorkshire pigs are best known for their large body size, fast growth rate, larger feed conversion ratio, superior meat yield, and slaughter rate4. Both of them have been experienced very strong selection. Meishan pigs in East China are characterized by their high fecundity, wrinkled black skin, large drooping ears, and early maturity5. Tibetan pigs, a representative of Asian wild boar, have characteristics of small body size, low litter size, strong adaptability and body-resistance6. Whole genome-wide scans of diverse pig breeds can increase our understanding of the manner in which genomic variation has been shaped and how the advantageous characteristics have been evolved through artificial selection. With the development and application of next-generation sequencing techniques, increasing studies have been conducted to unveil the genetic mechanisms underlying the phenotypes of pigs5,6.
As a major center of early pig domestication, China possesses 88 indigenous pig breeds with unique characteristics7. Because of the diverse phenotypes among those pig breeds, the genetic for the phenotypic variations remain unknown, particularly for the indigenous Xiang pig breeds. Xiang pig (XP) is a representative indigenous mini-pig breed in China. Besides their small body size, this pig breed has characteristics of strong disease resistance, high adaptability, favorable meat quality, low litter sizes, and early sexual maturity8,9. The unique features of Xiang pig were evolved through long-time natural and artificial selections. Our previous studies have reported the copy number variations (CNVs) and structural variations (SVs) of Xiang pig by Porcine SNP60K BeadChip and resequencing approaches, respectively and identified a lot of genes invovled in growth and repoduction8,9. Based on RNA sequencing, we have also identified some genes which played an important role in estrous and litter size in female Xiang pigs10,11. However, the molecular mechanisms underlying the breed characteristics in Xiang pigs remain unclear.
In this study, we performed whole-genome resequencing on 25 unrelated individual XPs. Given the phenotypic and genomic differences between Xiang pig and Tibetan, Meishan, Duroc, and Yorkshire, we then separately conducted the first comparative genomic analysis between XPs and each of the pig breeds and sought to identify the genomic signatures of selection in XPs. We explored the genomic regions under selection in XPs using three complementary methods: Fst, θπ and the absolute allele frequency difference (ΔAF). We identified many heritable variants and a suite of potential candidate genes that are involved in crucial biological processes such as growth, disease resistance, reproduction, meat quality traits and early sexual maturity. These findings will provide useful information to improve our understanding of the molecular mechanisms underlying the unique traits of XPs.
Results
Characteristics of the genome datasets
Whole-genome resequencing yielded a total of 876.70 Gb raw paired-end reads from the 25 unrelated individual XPs. Within this data, 819.09 Gb (93.43%) of high-quality paired-end reads were obtained after strict quality control protocols. Subsequently, the high-quality clean reads were then aligned to the pig reference genome assembly (Sscrofa11.1), resulting in a mean mapping rate of 96.91%. The average sequencing depth for each sample was 12.98 × (from 11.60 × to 19.88 ×) (TableS1). To accurately detect genomic footprints, we obtained the publicly available whole genome data of 100 individuals pigs from NCBI database, including Tibetan pig (TT, n = 25), Meishan (MS, n = 25), Duroc (DU, n = 25) and Yorkshire (LW, n = 25). These downloaded datasets had a total of 3661 Gb raw reads. The average depth of the downloaded datasets was 12.93 × in the 100 publicly available pigs (TableS2).
Identification of heritable variants
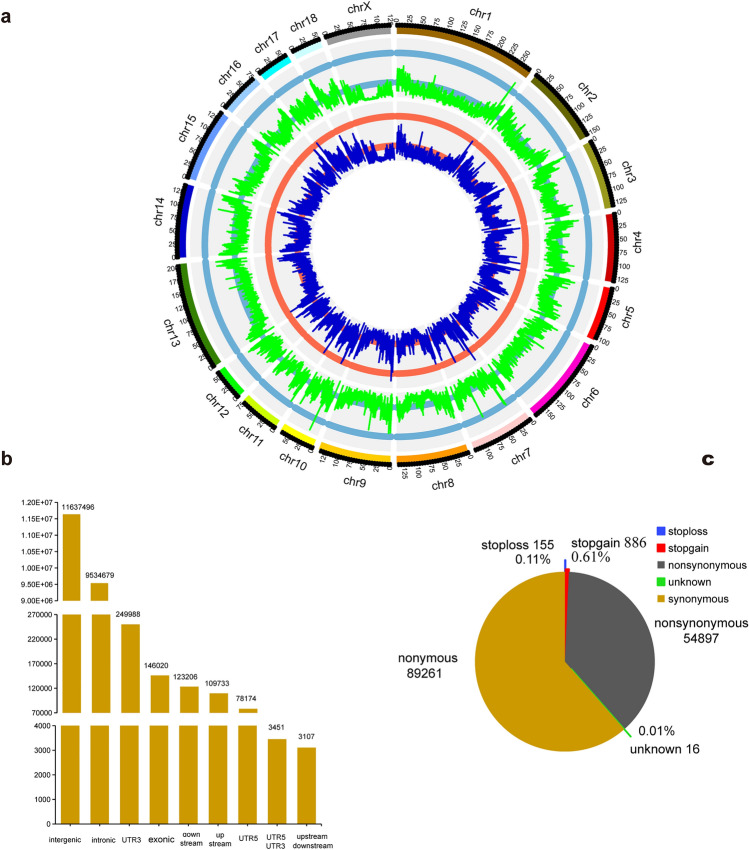
In total, 21,885,854 SNPs and 5,761,765 INDELs were obtained from Xiang pig genome (Table 1). Figure 1a showed the densities of SNPs and INDELs on each chromosome of XPs. We found that the maximum density of SNPs and INDELs was located on chromosomes 10 and 7, respectively. Of the total SNPs in XPs, 18,375,288 SNPs (83.96%) were identified in the dbSNP database, and 3,510,566 (16.04%) SNPs were newly found. Compared with other four pig breeds, 3,985,444 variants in our data set are Xiang-specific SNPs (Fig. S1).
Table 1.
Summary of the functional annotation statistics of SNPs in XP, TT, MS, DU and LW.
| SNP categories | XP | TT | MS | DU | LW |
|---|---|---|---|---|---|
| Upstream | 109,733 | 101,205 | 63,085 | 53,181 | 45,588 |
| UTR5 | 78,174 | 70,823 | 44,755 | 37,616 | 32,244 |
| Exonic | |||||
| Nonsynonymous | 54,897 | 47,693 | 32,950 | 28,676 | 22,030 |
| Synonymous | 89,261 | 78,034 | 53,931 | 43,824 | 37,320 |
| Stopgain | 886 | 769 | 520 | 397 | 298 |
| Stoploss | 155 | 151 | 85 | 98 | 70 |
| Unknown | 16 | 20 | 13 | 14 | 16 |
| Splicing | 805 | 725 | 469 | 421 | 379 |
| Intronic | 9,534,679 | 9,463,488 | 5,866,342 | 4,765,630 | 4,637,511 |
| Downstream | 123,206 | 242,955 | 152,657 | 125,375 | 117,879 |
| Upstream/downstream | 3107 | 3077 | 2017 | 1472 | 1324 |
| UTR3 | 249,988 | 119,609 | 74,922 | 62,124 | 56,394 |
| UTR5;UTR3 | 3451 | 2772 | 1536 | 1336 | 1224 |
| Intergenic | 11,637,496 | 11,770,026 | 7,300,453 | 6,046,060 | 5,739,841 |
| Numbers of total SNP | 21,885,854 | 21,901,347 | 13,593,735 | 11,166,224 | 10,692,118 |
XP, Xiang pig; TT, Tibetan; MS, Meishan; DU, Duroc; LW, Yorkshire.
Figure 1.
The distribution and annotation of genomic variants in XP. (a) Circos plot of distribution of variants density after filtration. The density of variants was calculated in each 100 kb step size. The circles from inside to outside display INDEL and SNP density per window, respectively. (b) SNP numbers in genomic annotation according to ANNOVAR. (c) The pie plot shows annotated SNPs at exonic regions.
Further functional annotation of these identified SNPs in the Xiang genome revealed that SNPs were partitioned into intergenic (11,637,496; 53.17%), intronic (9,534,679; 43.57%), exonic (145,215; 0.66%) and other gene regulatory regions (Table 1, Fig. 1b). In exonic regions, most variants resulted in synonymous (89,261) and nonsynonymous (54,897) mutations, followed by stop gain, stop loss, and unknown (< 1%) (Fig. 1c).
Population genetic structure and linkage disequilibrium
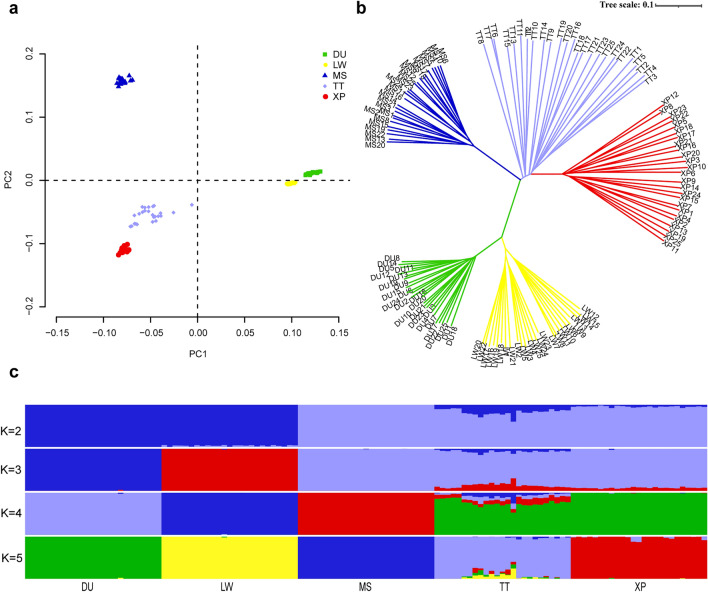
To assess the genetic structure among the pig breeds in this study, principal component analysis (PCA), neighbor-joining (NJ) trees, and ADMIXTURE were performed. The analysis of PCA (Fig. 2a) revealed that the whole pig population was divided into five groups: XP, TT, MS, DU and LW. The first component (PC1) separated the European-originated pig (DU and LW) from Chinese pig (XP, TT and MS). The second component separated XP and TT from MS. Meanwhile, DU and LW were divided by PC2. The branches of the phylogenetic tree (Fig. 2b) were consistent with the results of PCA, and successfully divided into five populations displaying genetically distinct clusters. Further population admixture analysis (Fig. 2c) clearly distinguished two breeds as expected based on their origin at K = 2. At K = 3, the European pig populations were separated, whereas the Chinese populations were still clustered together. At K = 4, MS were separated from Chinese pig populations. A separate ancestry for all four populations was visible at K = 5. With respect to the LD comparison among five populations (Fig. 3), we observed similar LD decay rates in Meishan and Duroc populations and the lowest level of LD in TT breed. XP had a lower level of LD compared to MS, DU and LW (Fig. 3).
Figure 2.
Population genetic analysis. (a) Plots of principal components 1 and 2 for the 125 individuals. (b) Neighbor-joining tree constructed from SNPs data among five pig populations. (c) Genetic structure analysis of samples using Admixture, with changing ancestral populations from K = 2 to K = 5.
Figure 3.
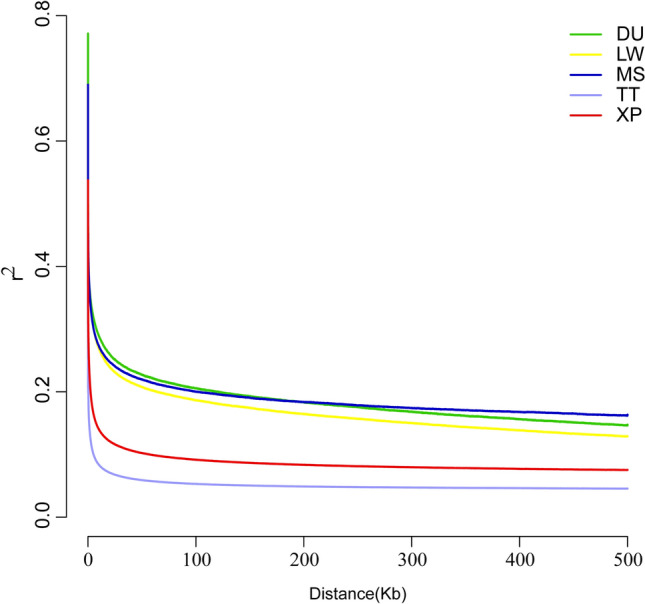
Decay of linkage disequilibrium in XP, TT, MS, DU and LW breeds.
Identification of selection signatures in XPs
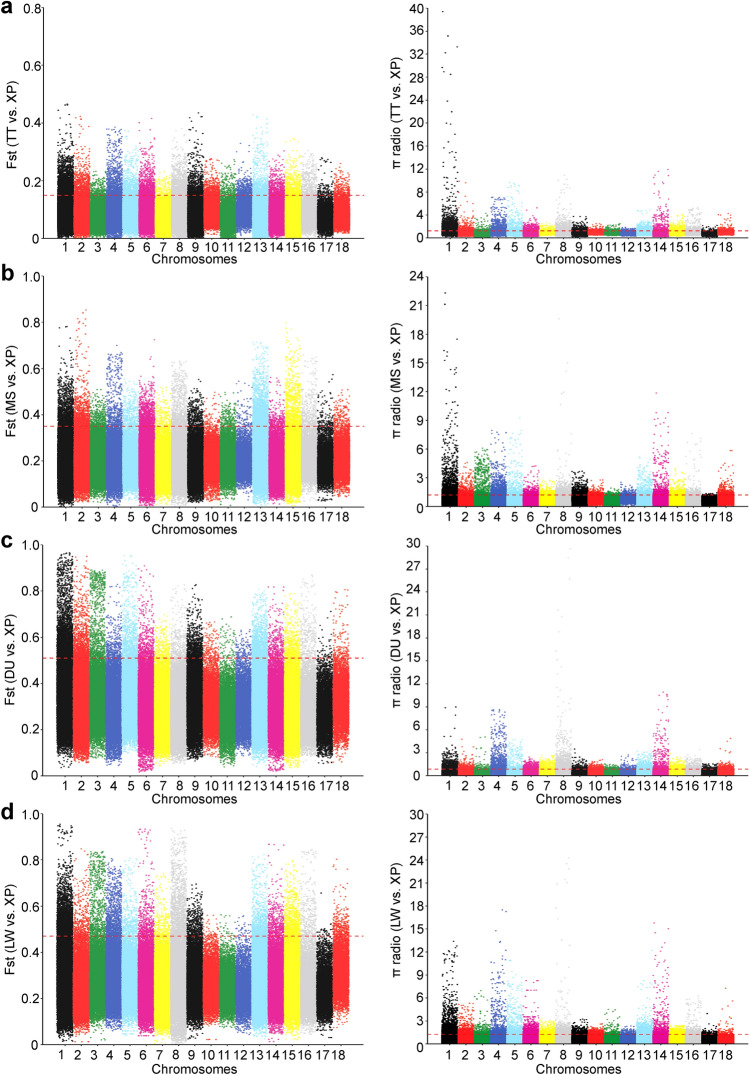
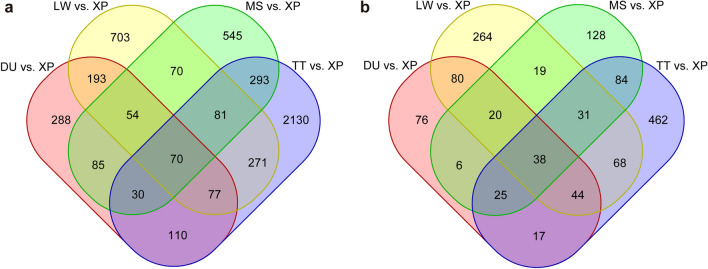
In order to detect the genome-wide selection signatures, we calculated pairwise Fst and θπ in 100-kb sliding windows with a step size of 10 kb across the autosomes between the XP and TT, MS, DU, and LW populations, respectively. For each comparison, the shared windows with both the Fst and θπ values in the top 5% were considered to be potentially positive selected regions. Based on the methods described above genome-wide screening revealed 3062 outlier windows as regions of selective sweeps with Fst > 0.15 and θπ > 1.23 in TT vs XP (Fig. 4a), 1228 outlier regions with Fst > 0.35 and θπ > 1.21 in MS vs XP (Fig. 4b), 907 outlier regions with Fst > 0.51 and θπ > 0.86 in DU vs XP (Fig. 4c), and 1519 outlier regions with Fst > 0.47 and θπ > 1.26 in LW vs XP (Fig. 4d). The numbers of the annotated genes within the selected regions were 769, 351, 306, and 564 in four comparative analyses (TT vs XP, MS vs XP, DU vs XP, and LW vs XP), respectively (Fig. 5b, Table S3). The numbers of unique and shared potential regions and candidate genes between four comparisons were shown in Fig. 5. We found that 38 candidate genes under selection were shared among all four comparisons (Fig. 5b, Table S4-1).
Figure 4.
Manhattan plots of genome-wide of Fst and θπ values across all 18 autosomes identified in XP population from four different comparisons. (a) TT vs XP. (b) MS vs XP. (c) DU vs XP. (d) LW vs XP. Dashed lines represented the threshod of top 5% Fst and θπ values.
Figure 5.
Venn diagrams represented the numbers of unique and shared (a) positively selected regions and (b) the candidate genes between four comparisons.
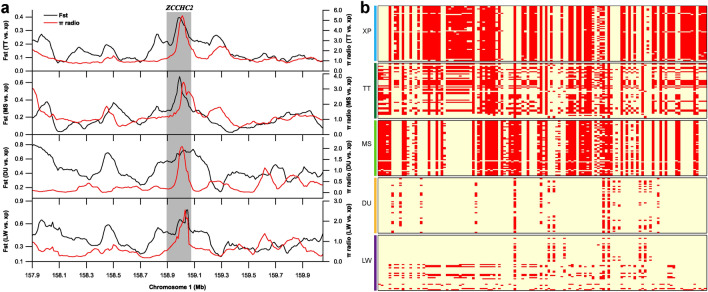
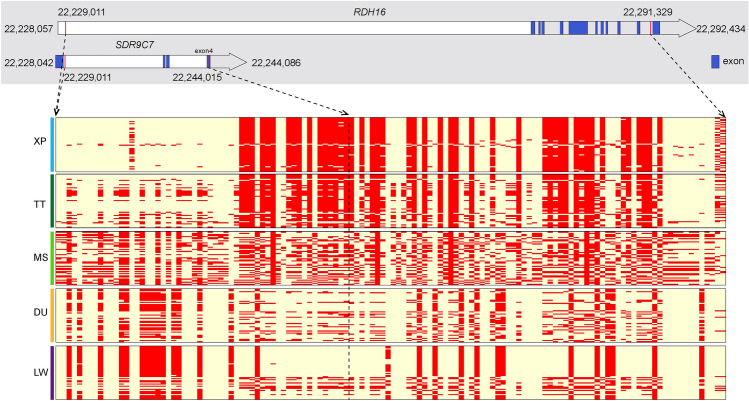
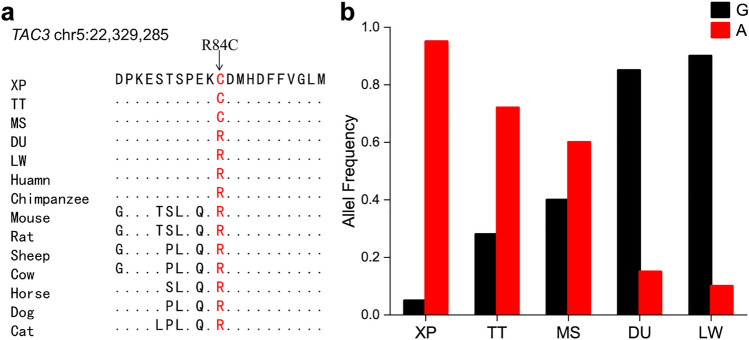
Among the 38 co-selection genes, we observed that ZCCHC2 on SSC1 showed strong selection signals. Both Fst and θπ ratios showed high values around the ZCCHC2 locus in all four comparisons (Fig. 6a). Moreover, XP had different haplotype patterns with other four pig breeds (Fig. 6b), suggesting a strong selection for XP in this region. In addition, one gene on SSC4, five genes on SSC5 and one gene on SSC15 were selected for further research based on their known biological function, including SLC26A7, SDR9C7, RDH16, TAC3, MYO1A, LYZ, PDE1A. We found that the haplotype patterns of the SDR9C7 and RDH16 genes in XPs were different from that of the other four populations (Fig. 7), which suggested a strong selection for XP in these two genes. To analyze the signs of selection in detail, we detected the SNPs with highly significant effects, which located in exons, UTRs and downstream/upstream of the seven genes mentioned above. We found 36 SNPs with different mutation frequencies between the XP and the other four pig breeds in SDR9C7, RDH16, TAC3, MYO1A, LYZ, and PDE1A (Table S5). The genotype pattern of these SNPs showed significant differences among the five pig breeds (Fig. S2). We also noted that twelve of these 36 variants were nonsynonymous mutations (Table S6). Of note, a novel missense variant/splice variant (c.G895A; chr5:22,354,739) in frequency of the derived allele “A” was 0.98 in XPs (Table S6). In TAC3, one nonsynonymous variants (G > A; p.R84C; chr5:22,329,285) was predicted as functional-altering variant (SIFT = 0) (Table S6). The p.R84C variant in XPs was significantly different from MS, DU and LW populations (Fig. 8).
Figure 6.
Characterization of selection signals around ZCCHC2 gene locus in XPs. (a) The Fst and θπ values around the ZCCHC2 locus. (b) Haplotype plots spanning ZCCHC2 gene among the five pig populations. The allele consistent with reference genome was indicated in lightyellow and derived allele in red color.
Figure 7.
Haplotype plots spanning SDR9C7 and RDH16 genes among the five pig populations. The allele consistent with reference genome was indicated in lightyellow and derived allele in red.
Figure 8.
Putative functional variant (p.R84C) in TAC3 gene. (a) Multispecies alignment of the protein sequences around the variant. The dots in the alignment indicated the amino acids which were identical with those in XP. (b) Distribution of the allele frequency of the variant in five pig populations.
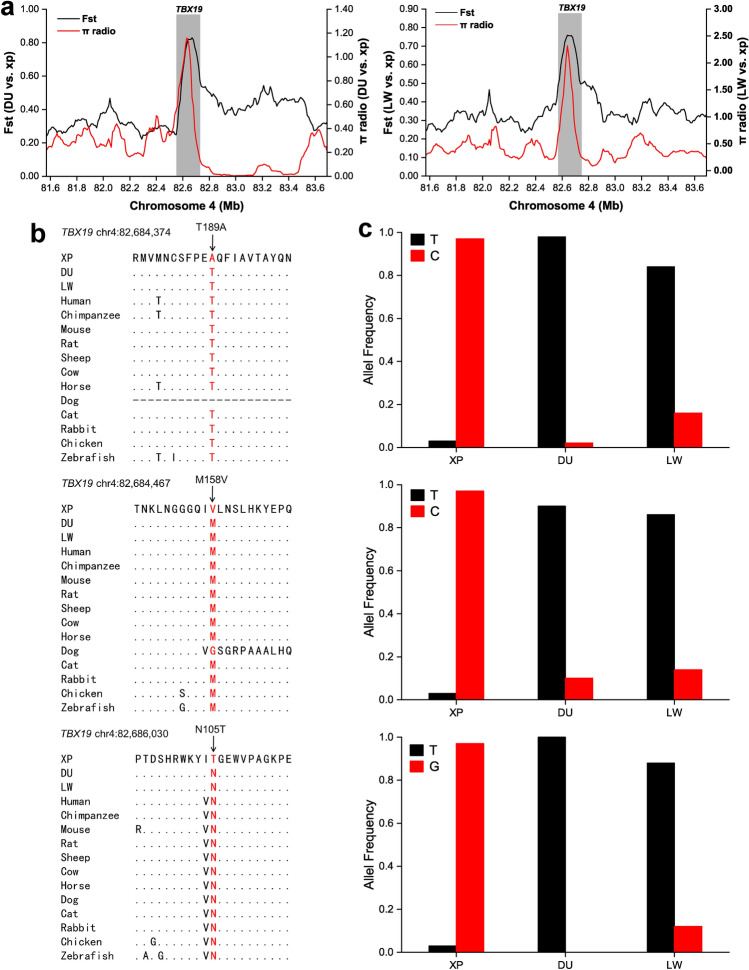
In addition, 274 and 120 genes were shared between two and three comparisons, respectively (Fig. 5b, Table S4). These genes were associated with multiple phenotype terms (Table 2), such as growth and development processes (IGF1R, PROP1, TBX19, STAC3, RLF, SELENOM, MSTN), immune response (SERPINB2, ADGRE5, STAT6, IL2, CD80, RHBDD3, PIK3IP1), environmental adaptation (NR2E1, SERPINB8, SERPINB10, UVSSA, EXPH5, VEGFC), reproduction (CCNB2, TRPM6, EYA3, CYP7B1, LIMK2, RSPO1, ADAM32, SPAG16), and meat quality traits (DECR1, EWSR1)6,12–41. In particularly, TBX19 on SSC4 showed high Fst and θπ values compared with neighboring regions in the DU vs XP and LW vs XP comparisons (Fig. 9a), and LIMK2 on SSC14 was also strongly selected in these two comparisons (Fig. S3b). EYA3 on SSC6 was strongly selected in the LW vs XP and MS vs XP comparisons (Fig. S3a). ADAM32 on SSC15 showed significant signals in the TT vs XP and MS vs XP comparisons (Fig. S3c). For TBX19, 364 highly differentiated SNPs in the upstream, downstream, UTR5, UTR3, exon, and intron regions were simultaneously distributed in both DU vs XP and LW vs XP comparisons (Table S7). Among 364 SNPs, 13 SNPs were observed in five exons and six of these were nonsynonymous variants (Table S8). Furthermore, we found that three nonsynonymous variants (p.N105T, p.M158V, p.T189A) were predicted as functional-altering variants (SIFT < 0.05) (Fig. 9c, Table S8), and they were highly conserved among multiple vertebrate species (Fig. 9b). Further analysis of LIMK2 gene showed that 115 homozygous SNPs had already been fixed in XPs (Table S9).
Table 2.
A partial list of candidate genes with previous evidences that were shared among two or three comparisons datasets.
| Chr | Start | End | Candidate gene | Comparisons | Gene function | References |
|---|---|---|---|---|---|---|
| 1 | 74,271,042 | 74,292,681 | NR2E1 | TT vs XP/LW vs XP | Behavioral defense response | 12 |
| 1 | 112,907,047 | 112,942,219 | CCNB2 | TT vs XP/DU vs XP/LW vs XP | Litter size trait | 13 |
| 1 | 137,387,925 | 137,690,666 | IGF1R | TT vs XP/LW vs XP | Growth Traits | 14 |
| 1 | 157,820,645 | 157,836,904 | SERPINB8 | TT vs XP/LW vs XP | Environmental adaptation | 15 |
| 1 | 157,841,510 | 157,866,913 | SERPINB10 | TT vs XP/LW vs XP | Implicated in UV-induced stress | 16 |
| 1 | 157,872,212 | 157,886,753 | SERPINB2 | TT vs XP/LW vs XP | Immune response | 17 |
| 1 | 227,811,204 | 227,963,110 | TRPM6 | DU vs XP/MS vs XP | Litter size trait | 18 |
| 2 | 64,844,792 | 64,890,939 | ADGRE5 | LW vs XP/MS vs XP | An activation marker on T cells | 19 |
| 2 | 79,627,603 | 79,631,270 | PROP1 | TT vs XP/MS vs XP/LW vs XP | Growth traits | 20 |
| 4 | 46,730,461 | 46,767,427 | DECR1 | TT vs XP//MS vs XP/LW vs XP |
Carcass and meat quality, lipid composition |
21 |
| 4 | 69,616,035 | 69,806,496 | CYP7B1 | MS vs XP/DU vs XP/LW vs XP | Male reproductive behavior | 22 |
| 4 | 82,663,581 | 82,697,813 | TBX19 | DU vs XP/LW vs XP | Development and growth | 23 |
| 5 | 22,406,364 | 22,421,990 | STAT6 | MS vs XP/DU vs XP/LW vs XP | Immune response | 24 |
| 5 | 22,549,008 | 22,556,194 | STAC3 | DU vs XP/LW vs XP | Vertebrate skeletal muscle development | 25 |
| 6 | 85,095,409 | 85,185,799 | EYA3 | LW vs XP/MS vs XP | Male and female seasonal estrus | 26,27 |
| 6 | 93,665,138 | 93,685,197 | RSPO1 | TT vs XP/MS vs XP | Sex development | 28 |
| 6 | 95,900,086 | 95,994,044 | RLF | TT vs XP/MS vs XP/LW vs XP | Muscle development and growth | 29 |
| 8 | 573,421 | 601,944 | UVSSA | TT vs XP/DU vs XP/LW vs XP | Environmental adaptation | 30 |
| 8 | 101,640,944 | 101,645,609 | IL2 | TT vs XP/MS vs XP/DU vs XP | T-cell growth factor | 31 |
| 9 | 36,885,212 | 37,006,396 | EXPH5 | TT vs XP/DU vs XP/LW vs XP |
Adaptation to Arctic or Antarctic environments |
32 |
| 13 | 140,693,382 | 140,728,980 | CD80 | DU vs XP/LW vs XP | T cell activation | 33 |
| 14 | 46,400,026 | 46,408,366 | RHBDD3 | DU vs XP/LW vs XP | Suppress the production of IL-6 | 34 |
| 14 | 46,408,053 | 46,442,086 | EWSR1 | DU vs XP/LW vs XP | Intramuscular fat | 35 |
| 14 | 47,902,860 | 47,905,440 | SELENOM | DU vs XP/LW vs XP | Growth trait | 36 |
| 14 | 47,946,396 | 48,040,822 | LIMK2 | DU vs XP/LW vs XP | Male infertility | 37 |
| 14 | 48,040,881 | 48,051,660 | PIK3IP1 | DU vs XP/LW vs XP | Inhibition of T-cell activation | 38 |
| 15 | 39,037,893 | 39,133,041 | VEGFC | TT vs XP/MS vs XP | High-altitude adaptation | 6 |
| 15 | 47,388,300 | 47,520,236 | ADAM32 | TT vs XP/MS vs XP | Male reproduction | 39 |
| 15 | 94,620,269 | 94,628,546 | MSTN | DU vs XP/LW vs XP |
Inhibit of skeletal muscle development and growth |
40 |
| 15 | 115,809,180 | 116,760,790 | SPAG16 | TT vs XP/DU vs XP | Male reproductive function | 41 |
Figure 9.
Characterization of selection signals around TBX19 gene locus in XPs, and three putative functional variants (p.N105T, p.M158V, p.T189A). (a) The Fst and θπ values around the TBX19 locus. (b) Cross-species alignment of the protein sequences around three functional variants (p.N105T, p.M158V, p.T189A) in TBX19 gene. The dots in the alignment indicated the amino acids which were identical with those in XPs, and the dashes indicated the missing data. (c) Distribution of the allele frequency of the three variants in three populations (XP, DU, and LW).
The absolute allele frequency difference (ΔAF) for each SNP was calculated in each comparison. As result, we identified 5885, 62,332, 855,864 and 483,774 SNPs with the criteria of AFXiang > 80% and AF non-Xiang < 20%, which located within 1466, 4937, 13,518, and 10,220 genes from comparisons between XP and TT, MS, DU, and LW, respectively (Table S10).
Next, we evaluated the overlap between the candidate genes in Fst-θπ analysis and those detected genes in the absolute allele frequency difference (ΔAF) analysis in each comparison. In total, we obtained 174, 132, 186, and 308 genes overlapped among the results from Fst-θπ and ΔAF analysis in the TT vs XP, MS vs XP, DU vs XP, and LW vs XP comparisons, respectively (Table S11). Many of these overlapped genes have already been mentioned above, such as ZCCHC2 and TBX19. Especially the ZCCHC2 gene, it was commonly identified by Fst-θπ and ΔAF analysis in all four comparisons. These results supported by Fst, θπ and ΔAF methods further indicated the reliability of the highlighted genes in this study.
Considering the impact of nonsynonymous SNPs (nsSNPs) on protein function, we further investigated the genes detected only by ΔAF analysis that contain missense variants. In total, we explored 143 missense variants with marked allele frequency differences distributed in 89 functional genes that might affect domestication or phenotypic traits of XPs among all four comparisons (Table S12). For example, we identified two missense mutations with the highest ΔAF (ΔAF = 1) occurring in NR6A1 and LTBP2 in both DU vs XP and LW vs XP comparisons. The first was a c.T485C (p.L162P, rs326780270) substitution in NR6A1 (nuclear receptor subfamily 6 group A member 1), which encodes an orphan nuclear receptor and has been considered a strong candidate for affecting the numbers of vertebrae in swine42. The other was a c.T2732C (p.M911T, rs328662847) substitution in LTBP2 (latent transforming growth factor beta binding protein 2) that are associated with thoracic vertebrae numbers in the pig population43. Our results indicated that both of the two genes might play important roles in the vertebrae development of Xiang pigs. Moreover, we detected other important missense variants with marked allele frequency differences occurring in the functional genes. All of the strongly differentiated missense variants identified from the four comparisons were presented in Table S12.
GO terms and KEGG pathway enrichment analyses
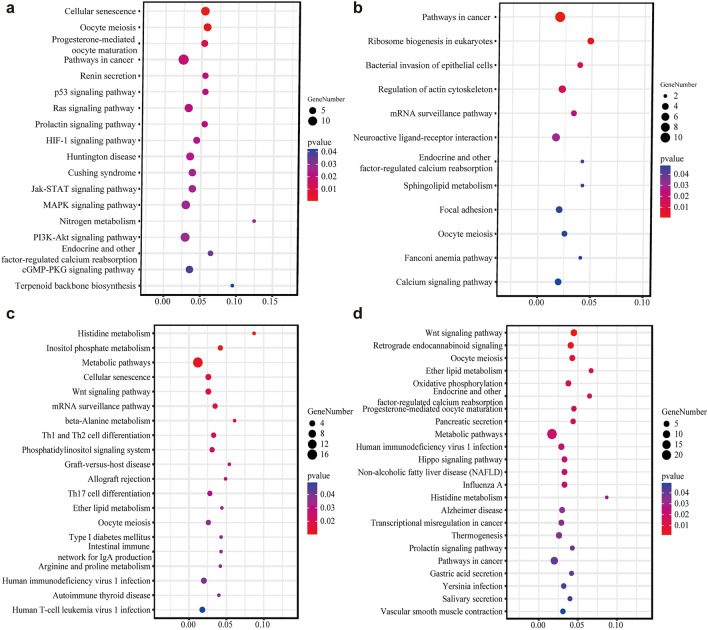
Subsequently, we searched for significantly overrepresented (P-value < 0.05) GO terms and KEGG pathways related to the candidate genes with gene symbols identified from both Fst and θπ statistics in each comparison. In the TT vs XP comparison, 427 candidate genes under selection were used for the GO and KEGG analysis, which resulted in 224 significant GO terms and 18 statistically significant KEGG terms. The top three most significant KEGG terms included cellular senescence, oocyte meiosis, and progesterone-mediated oocyte maturation terms (Fig. 10a). Among the 224 significant GO categories, we detected the terms involved in adaption, including “positive regulation of bone resorption”, “locomotory behavior”, “actin cytoskeleton”, “skin development”, “sensory perception of sound”, “actin cytoskeleton”, “actin cytoskeleton reorganization” (Table S13-1). In the MS vs XP comparison, 199 selected genes resulted in 187 significantly enriched GO terms (Table S13-2) and 12 KEGG pathways (Fig. 10b). Among the significantly enriched GO categories, the highest counts were associated with the immune system. The 12 KEGG enrichment pathways were significantly enriched in pathways involving cancer, bacterial invasion of epithelial cells, Fanconi anemia pathway, calcium signaling pathway, and other important biological processes. For the comparison between XPs and DUs, 180 selected genes resulted in 161 significant GO terms (Table S13-3) and 20 KEGG enrichment terms (Fig. 10c). The top three most significant terms were associated with the nucleus (GO:0005634; p-value = 1.72E−08), nucleoplasm (GO:0005654; p-value = 3.65E−06), and cytoplasm (GO:0005737; p-value = 1.08E−05) terms. In addition, some of these significantly enriched terms were related to morphogenesis and development. Disease-resistance related processes displayed the greatest KEGG enrichments, such as Th1 and Th2 cell differentiation, graft-versus-host disease, Th17 cell differentiation, and intestinal immune network for IgA production. Six enriched terms were related to metabolism. In the comparison group of XPs and LWs, 307 selected genes were significantly enriched in 128 GO terms (Table S13-4) and 23 KEGG path (Fig. 10d). Of which, eight GO categories were associated with muscle development and growth, including sarcomere, actin filament binding, actin cytoskeleton, actin monomer binding, actin binding, filamentous actin, negative regulation of cell growth, and regulation of growth. Moreover, two GO categories were involved in male reproduction, including spermatogenesis, sperm axoneme assembly and fertilization. The Wnt signaling pathway displayed the greatest KEGG enrichment. The remaining enriched pathways were primarily related to metabolism, secretion and disease (Fig. 10d).
Figure 10.
The significant KEGG pathway enrichment of the candidate genes under selection in XPs. (a) TT vs XP, (b) MS vs XP (c) DU vs XP, and (d) LW vs XP. Note: KEGG pathways were analyzed via KEGG database (https://www.kegg.jp/kegg/kegg1.html).
Discussion
It is well known that pig is one of the first domesticated animals. During the process of domestication and breeding, natural and artificial selection caused many changes of pig in body conformation, production performance, immune function, and adaptability to the environment44. The distinct imprints have been left in the genomes of domestic pigs. In recent years, selection signatures have been used to elucidate the genetic basis of many complex traits in agricultural animals5,6. In the present study, we performed the whole-genome resequencing of 25 unrelated individual Xiang pigs, and conducted population structure and LD analysis for the Xiang pig and other four breeds (i.e., Tibetan, Meishan, Duroc and Yorkshire). Our analyses revealed the population structure was mostly correlated with the geographic distribution. Xiang pigs displayed lower LD decay, indicating that Xiang pigs are less affected by artificial selection than the European and Meishan pig breeds. In addition, we explored the selection signatures that help provide potential genomic evidence linking the domestication of Xiang pigs with their breed characteristics.
To screen genome-wide regions under positive selection in Xiang pigs, we first selected windows with the top 5% of Fst and θπ values and identified 38 genes in the intersection of all four comparisons. Of those, seven genes (ZCCHC2, SLC26A7, SDR9C7, RDH16, TAC3, MYO1A, LYZ, and PDE1A) were involved in important biological processes. Except for PDE1A, other seven selection signatures were also confirmed by ΔAF analysis. ZCCHC2, as a member of the zinc finger CCHC-type (ZCCHC) superfamily protein, has nucleic acid–binding properties. Several literature have implicated the ZCCHC2 is correlated with diverse diseases. For example, ZCCHC2 participates in the regulation of retinoblastoma tumorigenesis by suppressing the activity of c-Myc45. A SNP in ZCCHC2 is associated with insect bite hypersensitivity in Exmoor ponies46 Besides, ZCCHC2 has previously been identified as important gene contributing to the benign (BEN) phenotypes of SIV/HIV infections, suggesting its role in the host defense against virus infections47. Intriguingly, ZCCHC2 showed high Fst and θπ scores in all four comparisons, implicating that ZCCHC2 gene could have been preferentially selected in XP. Our findings supported that ZCCHC2 likely played roles in disease resistance process of XP. SLC26A7 on SSC4 is one of the sulfate/anion transporter gene families and involves in the regulation of intracellular pH through chloride channels. It has been identified as a promising candidate gene for plateau adaptability in Tibetan pigs48. Interestingly, we found that four neighbor protein-coding genes (SDR9C7, RDH16, TAC3 and MYO1A) on SSC5 (22.22–22.36 Mb) were also under selection in XP. SDR9C7 is a well‐recognized gene for skin barrier formation due to its role in dehydrogenation of acylceramides49. RDH16, participating in energy and metabolism processes in adipose tissues in pigs and rats, has been shown to be one of the functional genes involved in the body size of Guizhou small goats50. Thus, it was reasonable to predict that RDH16 may be a promising candidate gene associated with the body size of the XP. TAC3 encoding neurokinin B (NKB) plays an important role in regulating the human reproductive axis. A previous study has demonstrated that TAC3 is a key regulator of kisspeptin expression and GnRH pulse secretion51. A distinctive finding indicate that the variation of A63P in TAC3 may increase the risk of earlier puberty onset in Chinese girls52. The results indicated that TAC3 variants in XP, especially one nonsynonymous variants and one functional-altering variant, may contribute to sexual maturity of XP by regulating kisspeptin expression and GnRH secretion. MYO1A (myosin IA) is a candidate gene that lightens the coat color of cattle, which contributes to heat stress resistance in cattle53. In humans, variants in the MYO1A gene are reported in non-syndromic sensorineural hearing loss54. In the present study, we detected a novel missense variant/splice variant (p.V299I) in MYO1A. The splice variant may affect MYO1A expression. This led us to hypothesize that the p.V299IA splice variant may have a role in local adaptation by regulating coat color and/or hearing in Xiang pigs. LYZ is an antibacterial enzyme which damages bacterial cell walls by catalyzing the hydrolysis of the b-(1,4)-glycosidic linkage between N-acetylglucosamine and the muramic acid of the peptidoglycan layer. LYZ displays antimicrobial, antitumoral and immunomodulatory properties, which have been extensively studied55,56. PDE1A are reported to be one of the hypoxia-related genes in Tibetan pig6.
To accurately detect the positively selected genes under selection in Xiang pig, we further explored the overlapped selected genes between two or three comparisons datasets. We identified seven genes implicated in growth and development processes (IGF1R, PROP1, TBX19, STAC3, RLF, SELENOM, MSTN). In TBX19, we simultaneously observed strong selective sweep signals (Fig. 9a) and 364 high-ΔAF SNPs in the DU vs XP and LW vs XP comparisons (Table S7). Among the list of these high-ΔAF SNPs, three missense mutations (N105T, M158V, and T189A) were within the evolutionary conserved T-box domain and were predicted as function-altering variants (Fig. 9b). This result was well consistent with that found in Chinese domestic pigs23. TBX19 (TPIT) functions by activating pituitary cell differentiation, and is also a positive regulator for pro-opiomelanocortin (POMC) expression, which contributes to the production of the adrenocorticotropic hormone (ACTH) in the hypothalamic–pituitary–adrenal (HPA) axis57. Mutations in this gene may lead to the isolated deficiency of pituitary POMC-derived ACTH57,58. Patients with ACTH deficiency are clinically characterized by severe obesity, weight loss, lack of appetite, hypoglycaemia and low blood pressure58. Previous studies have reported that this gene is related to development and growth in Chinese native pigs, such as Enshi black pig, Bama Xiang59,60. Together with these findings, TBX19 may act as a candidate gene associated with slow development and growth trait in XP. Studies have revealed that IGF1R is associated with growth animal growth and development, such as Hulun Buir Sheep14 and European pigs4. PROP1 is a homeodomain transcription factor that controls the development of the anterior pituitary gland. Mutations within this gene caused dwarfism in mice and humans due to deficiency of pituitary hormone61. Numerous reports have demonstrated association of PROP1 with growth traits in livestock, such as cattle20, sheep62, and horse63. STAC3 is specifically expressed in tissues of skeletal muscle as a nutrient regulated gene and plays a role in vertebrate skeletal muscle development and function25. RLF has been reported to markedly enhance DNA methylation of factors related to transcriptional regulation, which plays an essential role in embryonic muscle development29. SELENOM has been identified as a candidate gene for growth traits in the Anqing six-end-white pig (ASP)36. MSTN (GDF-8) plays a negative role in regulating the formation and differentiation of muscle cells and subsequently inhibits the growth of skeletal muscles40. Mutations or deletions in MSTN can cause the proliferation and hypertrophy of muscle cells and muscle fibers. Interestingly, Bama mini-pigs have a higher level of MSTN mRNA than Landrace pigs64. Similarly, XP is also a mini-type breed. However, further studies are needed to confirm the expression level of MSTN in XP. Therefore, these genes detailed above may be the most promising genomic evidence that explain the small body size of Xiang pigs. Xiang pigs inhabit a remote mountainous area in Guizhou Province, where is known for its high elevation and high humidity. During long-term adaptation to the harsh environments, Xiang pigs have evolved their unique adaptive characteristics. By combining the results from Fst and θπ statistical analyses, we identified seven genes that participate in the immune response (SERPINB2, ADGRE5, STAT6, IL2, CD80, RHBDD3, PIK3IP1)17,19,24,31,33,34,38, which may be involved in shaping the particular disease-resistance characteristics of Xiang pigs. Additionally, we also uncovered genes, which may play different roles in adaptation processes of Xiang pigs. These genes include NR2E1 for behavioral defense response12, SERPINB8 for maintaining the mechanical stability of skin15, SERPINB10 involved in the UV-induced cellular response16, UVSSA for response to ultra-violet (UV) radiation30, VEGFC responsible for regulating oxygen homeostasis6. The positive selection signatures identified here provided new insights into the potential adaptive mechanisms in XP. Apart from these findings associated with growth, immunity, and environmental adaptation, some important functional genes associated with reproduction (CCNB2, TRPM6, EYA3, CYP7B1, LIMK2, RSPO1, ADAM32, SPAG16)13,18,22,26,26–28,39,41 and meat quality (DECR1, EWSR1)21,35 were also identified under selection pressure in Xiang pigs (Table 2). Of these, EYA3, LIMK2 and ADAM32 showed particularly strong signs of selective sweeps presumably associated with the domestication of Xiang pig. EYA3 (eyes absent 3) is an ancient retinal-determining gene, and its expression is induced by long-day stimulation in mammals. It is clear that EYA3 is an essential factor for modulating the expression of the TSHβ, which regulates changes in seasonal reproductive biology26. It has been reported that EYA3 is an important candidate gene in male and female seasonal estrus27. LIMK2, especially the testis-specific isoform tLIMK2, is essential for the proper progression of spermatogenesis. Additionally, LIMK1/2 has shown to participate in mouse embryo early cleavage and blastocyst formation by regulating the phosphorylation level of cofilin in mouse embryos65. In humans, three deleterious nonsynonymous substitutions in LIMK2 are involved in male infertility37. When comparing with the European commercial breeds (DU and LW), LIMK2 showed strong signs of selective sweeps in XP. Notably, the derived alleles of 115 SNPs in LIMK2 exhibited high AF (AF = 1) in XPs, whereas these derived alleles displayed low AF (AF < 0.35) in European breeds (DU and LW) (Table S10). ADAM32 has been identified as contributing to fertilization in mouse39. Compared with the MS, DU and LW breeds, XP exhibited lower prolificacy. Therefore, EYA3, LIMK2 and ADAM32 genes might be promising candidate genes involved in the reproduction traits of Xiang pig.
Based on the genome-wide SNP allele frequency (AF) analysis, we identified a suite of promising genes. We further found that some of these genes from ΔAF test colocalized with the selected regions detected in the Fst-θπ analysis (Table S11). These overlapped candidate genes may have undergone strong selection, and potentially affected phenotypic traits of Xiang pig, such as ZZHC2, LIMK2 and TBX19 mentioned above. Apart from the overlapped genes, we further focused specifically on the candidate genes detected only in the ΔAF analysis that contained missense variants. Among these candidate genes, two well-known genes (NR6A1, LTBP2) were of particular significance, as they are both involved in affecting the numbers of vertebrae in swine. The NR6A1 (nuclear receptor subfamily 6, group A, member 1) gene is also known as the germ cell nucleus factor (GCNF). Thus far, a large number of studies have confirmed the association between NR6A1 gene polymorphisms and lumbar vertebrae numbers in pigs and other species42,66. In the present study, one nonsynonymous mutation (c.T485C, rs326780270) was identified in the coding region of exon-4 of NR6A1 gene. The derived C allele frequency at this SNP locus was 1.0 in XP, whereas it was 0.0 in the European breeds (DU and LW). LTBP2 is expressed prominently in the outer lamellar layers of the annulus fibrosus of the fetal intervertebral disk in humans67. LTBP2 regulates the activity of growth differentiation factor Gdf11 by inhibiting the extracellular processing of proGdf11. Gdf11 knock-out mice exhibit an increase in the numbers of ribs from 13 (wild type) to 1868. SNP (c.4481A > C) in LTBP2 is associated with thoracic vertebrae numbers in an F2 intercross population between Landrace and Korean native pigs43. In our study, we observed one nonsynonymous mutation (c.T2732C, rs328662847) in the coding region of exon-17 of LTBP2 gene. The derived C allele frequency at this SNP locus was 1.0 in XP, whereas it was 0.0 in the European breeds (DU and LW). Given the importance of NR6A1 and LTBP2 to vertebrae numbers, the two SNPs: rs326780270: T > C in NR6A1 and rs328662847: T > C, may affect vertebrae development in XP.
For each comparison, we subsequently investigated the functions associated with the annotated genes from both Fst and θπ statistical approaches by analyzing over-represented GO term and KEGG pathway analysis. In the TT vs XP comparison, the most significant pathway was “Cellular senescence” (Fig. 10a), which is often regarded as an anti-cancer mechanism69. In the MS vs XP comparison, the KEGG analysis revealed that “pathways in cancer” represented the greatest enrichment. We also detected calcium signaling pathway, which plays a crucial role in muscle function and plasticity and is involved in many processes during animal embryonic development. Of note, 15 significant GO terms related to the immune system in XP were identified, such as the adaptive immune response, response to virus, toll-like receptor 2 signaling pathway, and interleukin-2 receptor binding (Table S13-2). Other GO terms of interest were related to adaptability, including aggresomal, locomotor behavior, cellular response to heat, cellular response to UV. The results were consistent with the fact that XP exhibits strong disease resistance and high adaptability. In the DU vs XP comparison, the results of KEGG enrichment analysis (Fig. 10c) revealed that eight pathways involved in the immune system or infectious diseases were significantly enriched, including Th1 and Th2 cell differentiation, graft-versus-host disease, allograft rejection, Th17 cell differentiation, intestinal immune network for IgA production, human immunodeficiency virus 1 infection, autoimmune thyroid disease, and human T-cell leukemia virus 1 infection. This may provide a better mechanism for the disease resistance in XP. Compared with Duroc pigs, Xiang pigs are smaller and not as tall. We identified seven significant pathways that were related to developmental and metabolic processes including Wnt signaling pathway, histidine metabolism, inositol phosphate metabolism, metabolic pathways, beta-alanine metabolism, ether lipid metabolism, and arginine and proline metabolism. Among the significant enriched GO categories (Table S13-3), eight terms (GO:0060612 GO:0048536 GO:0001889 GO:0003143 GO:0061303 GO:0055013 GO:0060430 GO:0060322) were associated with development processes. These results indicated that differences in development and metabolism between XPs and DUs. In the LW vs XP comparison, the most significant pathway was “Wnt signaling pathway” (Fig. 10d). Wnt signaling pathway participates in many developmental events during embryogenesis and is essential for muscle fiber growth and maintenance58,70. Wnt signaling pathway is also involved in satellite cell proliferation and differentiation during adult skeletal muscle regeneration71. We also found significantly over-represented GO terms associated with male reproduction (GO:0007283 GO:0007288 GO:0009566) and growth (GO:0030308 GO:0040008) (Table S13-4). Our findings reflect that the candidate genes are mainly involved in disease resistance, adaptability, and developmental and metabolic processes in XPs. The largest number of terms related to disease resistance and adaptation, which further indicate that Xiang pigs underwent natural selection pressure to adapt to their environments.
Conclusions
We conducted full resequencing of 25 Xiang pigs individually, resulting in a comprehensive whole-genome map and the identification of 21 M heritable SNPs. Moreover, we detected the genomic signatures and found that the genes positively selected in Xiang pigs were involved in crucial biological processes. We highlighted some genes/regions under possible selection relevant to disease resistance (ZCCHC2), early maturation (TAC3), reproduction (LIMK2, EYA3, ADAM32) and growth (TBX19). We mainly exemplified striking evidence of selection at ZCCHC2 and TBX19, which are correlated with diseases, and growth and early development, respectively. Additionally, important missense mutations with high ΔAF were identified within NR6A1 and LTBP2 genes that mainly contribute to vertebrae numbers of Xiang pigs. These findings will facilitate the understanding of the germplasm characteristics and support further investigation of the mechanisms underlying selection in XP pig breed.
Materials and methods
Ethics statement
All animal procedures were approved by Guizhou University Subcommittee of Experimental Animal Ethics (EAE-GIU-2021-P009) and were conducted the rules of animal experimental ethics. The study is also in accordance with ARRIVE guidelines.
Sample collection and sequencing
In this study, we obtained a total of 25 unrelated Xiang pigs (XPs) based on pedigree records from the pig farm in Congjiang County, Guizhou Province. For each pig, the whole blood sample was collected for DNA extraction. The high-quality DNA was used for the whole-genome resequencing. The Illumina DNA libraries (Paired-end, 2 × 150 bp) were constructed and sequenced on the Illumina Hiseq 2500 sequencing platform by BGI-Shenzhen, China. Additionally, we downloaded 100 publicly-available pig genome sequences from NCBI database, including Tibetan (TT, n = 25), Meishan (MS, n = 25), Duroc (DU, n = 25), and Yorkshire (LW, n = 25).
Mapping, SNP calling and annotation
To avoid low-quality reads, the raw paired-end reads were filtered and trimmed using the NGSQC Toolkit with default parameters16. High-quality reads were then aligned to the Sscrofa11.1 reference sequence using the Burrows-Wheeler Aligner (BWA) with the “bwa-mem” algorithms72. SAMtools was used for sorting and indexing the aligned BAM files73. Potential duplicates were removed using Picard tools. Finally, we jointly used “HaplotypeCaller”, “CombineGVCFs” and “GenotypeGVCFs” with default parameters in GATK to call variants, which generated genotype calls in Variant Call Formats (VCF)74.
To exclude possible false positives, we filtered the variants according to the strict filter criteria. We removed those INDELs in the VCF file using the following options: QUAL < 30.0, QD < 5.0, FS > 200, ReadPosRankSum < − 20.075. High-quality SNPs were identified according to the filtering criteria: QUAL > 30, OD > 5.0, FS < 60.0, MQ > 40.0, MQRankSum > − 12.5, ReadPosRankSum > − 8.075. Variants that passed quality control filter were functionally annotated with the ANNOVAR softwares76 based on the corresponding pig genome annotation file from the Ensembl database.
Population structure and linkage disequilibrium
To better understand the genetic relationships among the four pig populations in our study, we performed PCA, NJ tree and ADMIXTURE analysis. Principal component analysis (PCA) was carried out using the GCTA software77, and the first two eigenvectors were plotted in the ggplot2 package under the R platform. For phylogenetic tree analysis, PLINK was used to calculate genetic distance matrix78. Next, the phylogenetic tree was constructed with the neighbor joining (NJ) algorithm in MEGA79 and displayed with FigTree80. Moreover, the population ancestry was estimated using ADMIXTURE software81with K set from 2 to 5. The genome-wide linkage disequilibrium (LD) decay between pairwise SNPs was assessed and visualized using PopLDdecay software82.
Selective sweeps and functional enrichment analyses
Before detecting selection signals, we filtered the SNPs with minor allele frequency (MAF) < 0.05 and call rates < 0.90 and excluded sites with a missing rate > 20% using PLINK. In order to identify the candidate regions under positive selection in XPs, we first calculated the fixation statistics (Fst ) and population nucleotide diversity ratio (θπ) according to the the procedure described by Li et al.6. Both the Fst and θπ statistics require the usage of the standard genotype data. The Fst value was estimated based on the differences in allelic frequencies between populations83. This method is effective for identifying the genomic pattern affected by the divergent selection84. Domestication and selection resulted in decreased nucleotide diversity in populations. Thus, the method based on θπ can serve as efficient statistic to identify signatures of selection in populations6,74. In fact, numerous studies have been performed to detect the selective signal sweep regions in animals by using both Fst and θπ methods6,75,85. The average Fst and θπ were calculated by VCFtools (v0.1.13) in 100-kb sliding windows with a 10-kb step size6,85 between XP and the other four control breeds. To avoid spurious selection signals, windows containing less than 10 SNPs were discarded. Putative selection targets were identified as the candidate regions in fully overlapping windows with high Fst (Fst > 95%) and θπ (θπ > 95%) values85. The selected regions were annotated using Bedtools (v2.17.0).
The highly differentiated SNPs are likely to be directly targeted by selection or to occur near loci under selection86. Therefore, we also estimated allele frequency of single SNP with a genome scan for each pig population and measured the absolute allele frequency difference (ΔAF) to further detect putative selection signatures5. The ΔAF method is best suited for detection of fixed or approximately fixed alleles in Xiang pigs87. The ΔAF value per SNP between the Xiang population and the other four pig populations was calculated using the formulas: ΔAF = abs (AltAFXP − AltAFTT), ΔAF = abs (AltAFXP − AltAFMS), ΔAF = abs (AltAFXP − AltAFDU), and ΔAF = abs (AltAFXP − AltAFLW), respectively. After comparisons of ΔAF value between Xiang and each of the other four groups, we searched for Xiang-specific SNPs across the entire genome with the criteria of AFXiang > 80% and AF non-Xiang < 20%88. By appliying the absolute allele frequency difference (ΔAF) analysis, we could not only furthe evaluate the reliability of the results from Fst and θπ statistics, but also mine the potational selected gene containg interesting candidate mutations.
To investigate the biological enrichment of genes under selective pressure, KEGG pathway and GO classes were analyzed based on the candidated genes from Fst and θπ methods by using KOBAS (v3.0)89. The terms with p-value smaller than 0.05 were considered to be statistically significant.
Supplementary Information
Acknowledgements
The authors would like to acknowledge the funding bodies of this research and TopEdit LLC (https://www.topeditsci.com) for the linguistic editing of this manuscript.
Author contributions
X.R. and J.W. involved in conceptualization and experimental design. X.W., S.H. and X.N. contributed to computational analyses. S.L. and S.H. collected samples and prepared for sequencing. X.W., X.R. and J.W. wrote the manuscript. X.R. involved in funding acquisition and project supervision. All authors read and approved the manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (31672390, 31960641), the National High Technology Research and Development Program of China (863 Program) [2013AA102503], the Guizhou Provincial Science and Technology Projects (QKHPTRC[2019]5615, QKHRC[2016]4012, QKHZC [2017]2585, QKHZC [2017]2587).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xueqin Ran, Email: xqran@gzu.edu.cn.
Jiafu Wang, Email: jfwang@gzu.edu.cn.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-14686-w.
References
- 1.Lunney JK. Advances in swine biomedical model genomics. Int. J. Biol. Sci. 2007;3(3):179–184. doi: 10.7150/ijbs.3.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang H, et al. Molecular cloning, tissue distribution and ontogenetic expression of Xiang pig chemerin and its involvement in regulating energy metabolism through Akt and ERK1/2 signaling pathways. Mol. Biol. Rep. 2012;39(2):1887–1894. doi: 10.1007/s11033-011-0934-8. [DOI] [PubMed] [Google Scholar]
- 3.Larson G, et al. Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science. 2005;307(5715):1618–1621. doi: 10.1126/science.1106927. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Z, Xiao Q, Zhang QQ, Sun H, Chen JC, Li ZC, Xue M, Ma PP, Yang HJ, Xu NY, Wang QS, Pan YC. Genomic analysis reveals genes affecting distinct phenotypes among different Chinese and western pig breeds. Sci. Rep. 2018;8:13352. doi: 10.1038/s41598-018-31802-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao P, et al. Evidence of evolutionary history and selective sweeps in the genome of Meishan pig reveals its genetic and phenotypic characterization. Gigascience. 2018;7(5):giy058. doi: 10.1093/gigascience/giy058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li M, et al. Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat. Genet. 2013;45(12):1431–1438. doi: 10.1038/ng.2811. [DOI] [PubMed] [Google Scholar]
- 7.Yang H. Livestock development in China: Animal production, consumption and genetic resources. J. Anim. Breed. Genet. 2013;130(4):249–251. doi: 10.1111/jbg.12045. [DOI] [PubMed] [Google Scholar]
- 8.Xie J, Li R, Li S, Ran X, Wang J, Jiang J, Zhao P. Identification of copy number variations in Xiang and Kele pigs. PLoS ONE. 2016;11(2):e0148565. doi: 10.1371/journal.pone.0148565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu C, Ran X, Wang J, Li S, Liu J. Detection of genomic structural variations in Guizhou indigenous pigs and the comparison with other breeds. PLoS ONE. 2018;13(3):e0194282. doi: 10.1371/journal.pone.0194282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang LT, Ran XQ, Mao N, Zhang FP, Niu X, Ruan YQ, Yi FL, Li S, Wang JF. Analysis of alternative splicing events by RNA sequencing in the ovaries of Xiang pig at estrous and diestrous. Theriogenology. 2018;119:60–68. doi: 10.1016/j.theriogenology.2018.06.022. [DOI] [PubMed] [Google Scholar]
- 11.Ran X, Hu F, Mao N, Ruan Y, Yi F, Niu X, Huang S, Li S, You L, Zhang F, Tang LT, Wang JF, Liu J. Differences in gene expression and variable splicing events of ovaries between large and small litter size in Chinese Xiang pigs. Porcine Health Manag. 2021;7:52. doi: 10.1186/s40813-021-00226-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ai H, Yang B, Li J, Xie X, Chen H, Ren J. Population history and genomic signatures for high-altitude adaptation in Tibetan pigs. BMC Genomics. 2014;15(1):834. doi: 10.1186/1471-2164-15-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai FN, et al. Whole-genome scanning for the litter size trait associated genes and SNPs under selection in dairy goat (Capra hircus) Sci. Rep. 2016;6:38096. doi: 10.1038/srep38096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding N, Tian D, Li X, Zhang Z, Tian F, Liu S, Han B, Liu D, Zhao K. Genetic polymorphisms of IGF1 and IGF1R genes and their effects on growth traits in hulun buir sheep. Genes. 2022;13(4):666. doi: 10.3390/genes13040666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pigors M, Sarig O, Heinz L, Plagnol V, Fischer J, Mohamad J, Malchin N, Rajpopat S, Kharfi M, Lestringant GG, Sprecher E, Kelsell DP, Blaydon DC. Loss-of-function mutations in SERPINB8 linked to exfoliative ichthyosis with impaired mechanical stability of intercellular adhesions. Am. J. Hum. Genet. 2016;99(2):430–436. doi: 10.1016/j.ajhg.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Majoros H, Borsos BN, Ujfaludi Z, Páhi ZG, Mórocz M, Haracska L, Boros IM, Pankotai T. SerpinB10, a serine protease inhibitor, is implicated in UV-induced cellular response. Int. J. Mol. Sci. 2021;22(16):8500. doi: 10.3390/ijms22168500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schroder WA, Major L, Suhrbier A. The role of SerpinB2 in immunity. Crit. Rev™. Immunol. 2011;31(1):15–30. doi: 10.1615/CritRevImmunol.v31.i1.20. [DOI] [PubMed] [Google Scholar]
- 18.Hernandez-Montiel, W. et al. Genome-wide association study reveals candidate genes for litter size traits in pelibuey sheep. Animals10(3), 434. 10.3390/ani10030434 (2020). [DOI] [PMC free article] [PubMed]
- 19.Spendlove I, Sutavani R. The role of CD97 in regulating adaptive T-cell responses. Adv. Exp. Med. Biol. 2010 doi: 10.1007/978-1-4419-7913-1_12. [DOI] [PubMed] [Google Scholar]
- 20.Pan C, et al. A critical functional missense mutation (H173R) in the bovine PROP1 gene significantly affects growth traits in cattle. Gene. 2013;531(2):398–402. doi: 10.1016/j.gene.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 21.Amills M, Vidal O, Varona L, Tomàs A, Gil M, Sànchez A, Noguera JL. Polymorphism of the pig 2,4-dienoyl CoA reductase 1 gene (DECR1) and its association with carcass and meat quality traits. J. Anim. Sci. 2005;83(3):493–498. doi: 10.1051/gse:2004046. [DOI] [PubMed] [Google Scholar]
- 22.Oyola MG, Zuloaga DG, Carbone D, Malysz AM, Acevedo-Rodriguez A, Handa RJ, Mani SK. CYP7B1 enzyme deletion impairs reproductive behaviors in male mice. Endocrinology. 2015;156(6):2150–2161. doi: 10.1210/en.2014-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu Y, Li W, Yang B, Zhang Z, Ai H, Ren J, Huang L. Signatures of selection and interspecies introgression in the genome of Chinese domestic pigs. Genome Biol. Evol. 2017;9(10):2592–2603. doi: 10.1093/gbe/evx186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for the development of Th2 cells. Immunity. 1996;4(3):313–319. doi: 10.1016/S1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 25.Bower NI, de la Serrana DG, Cole NJ, Hollway GE, Lee HT, Assinder S, Johnston IA. Stac3 is required for myotube formation and myogenic differentiation in vertebrate skeletal muscle. J. Biol. Chem. 2012;287(52):43936–43949. doi: 10.1074/jbc.M112.361311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood S, Loudon A. Clocks for all seasons: Unwinding the roles and mechanisms of circadian and interval timers in the hypothalamus and pituitary. J. Endocrinol. 2014;222(2):R39–59. doi: 10.1530/JOE-14-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia Q, Di R, He XY, Wei CH, Chu MX. Expression analysis of DIO2, EYA3, KISS1 and GPR54 genes in year-round estrous and seasonally estrous rams. Arch. Anim. Breed. 2020;63(2):451–460. doi: 10.5194/aab-63-451-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biason-Lauber A. WNT4, RSPO1, and FOXL2 in sex development. Semin. Reprod. Med. 2012;30(5):387–395. doi: 10.1055/s-0032-1324722. [DOI] [PubMed] [Google Scholar]
- 29.Harten SK, et al. The recently identified modifier of murine metastable epialleles, Rearranged L-Myc Fusion, is involved in maintaining epigenetic marks at CpG island shores and enhancers. BMC Biol. 2015;13:21. doi: 10.1186/s12915-015-0128-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grilz-Seger G, Neuditschko M, Ricard A, Velie B, Lindgren G, Mesarič M, Cotman M, Horna M, Dobretsberger M, Brem G, Druml T. Genome-wide homozygosity patterns and evidence for selection in a set of European and near eastern horse breeds. Genes. 2019;10(7):491. doi: 10.3390/genes10070491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spolski R, Li P, Leonard WJ. Biology and regulation of IL-2: From molecular mechanisms to human therapy. Nat. Rev. Immunol. 2018;18(10):648–659. doi: 10.1038/s41577-018-0046-y. [DOI] [PubMed] [Google Scholar]
- 32.Yudin NS, Larkin DM, Ignatieva EV. A compendium and functional characterization of mammalian genes involved in adaptation to Arctic or Antarctic environments. BMC Genetic. 2017;18:111. doi: 10.1186/s12863-017-0580-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taddio MF, et al. Synthesis and structure-affinity relationship of small molecules for imaging human CD80 by positron emission tomography. J. Med. Chem. 2019;62(17):8090–8100. doi: 10.1021/acs.jmedchem.9b00858. [DOI] [PubMed] [Google Scholar]
- 34.Liu J, et al. Rhbdd3 controls autoimmunity by suppressing the production of IL-6 by dendritic cells via K27-linked ubiquitination of the regulator NEMO. Nat. Immunol. 2014;15(7):612–622. doi: 10.1038/ni.2898. [DOI] [PubMed] [Google Scholar]
- 35.Sosa-Madrid BS, et al. Genomic regions influencing intramuscular fat in divergently selected rabbit lines. Anim. Genet. 2020;51(1):58–69. doi: 10.1111/age.12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang W, et al. Identification of signatures of selection by whole-genome resequencing of a Chinese native pig. Front. Genet. 2020;11:566255. doi: 10.3389/fgene.2020.566255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuzmin A, et al. Identification of potentially damaging amino acid substitutions leading to human male infertility. Biol. Reprod. 2009;81(2):319–326. doi: 10.1095/biolreprod.109.076000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeFrances MC, Debelius DR, Cheng J, Kane LP. Inhibition of T-cell activation by PIK3IP1. Eur. J. Immunol. 2012;42(10):2754–2759. doi: 10.1002/eji.201141653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim T, Oh J, Woo JM, Choi E, Im SH, Yoo YJ, Kim DH, Nishimura H, Cho C. Expression and relationship of male reproductive ADAMs in mouse. Biol. Reprod. 2006;74(4):744–750. doi: 10.1095/biolreprod.105.048892. [DOI] [PubMed] [Google Scholar]
- 40.Stinckens A, et al. Characterization of the complete porcine MSTN gene and expression levels in pig breeds differing in muscularity. Anim. Genet. 2008;39(6):586–596. doi: 10.1111/j.1365-2052.2008.01774.x. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Z, Zariwala MA, Mahadevan MM, Caballero-Campo P, Shen X, Escudier E, Duriez B, Bridoux AM, Leigh M, Gerton GL, Kennedy M, Amselem S, Knowles MR, Strauss JF. A heterozygous mutation disrupting the SPAG16 gene results in biochemical instability of central apparatus components of the human sperm axoneme. Biol. Reprod. 2007;77(5):864–871. doi: 10.1095/biolreprod.107.063206. [DOI] [PubMed] [Google Scholar]
- 42.Burgos C, Latorre P, Altarriba J, Carrodeguas JA, Varona L, López-Buesa P. Allelic frequencies of NR6A1 and VRTN, two genes that affect vertebrae number in diverse pig breeds: a study of the effects of the VRTN insertion on phenotypic traits of a Duroc x Landrace-Large White cross. Meat Sci. 2015;100:150–155. doi: 10.1016/j.meatsci.2014.09.143. [DOI] [PubMed] [Google Scholar]
- 43.Park HB, Han SH, Lee JB, Cho IC. Rapid Communication: High-resolution quantitative trait loci analysis identifies encoding latent transforming growth factor beta binding protein 2 associated with thoracic vertebrae number in a large F intercross between Landrace and Korean native pigs. J. Anim. Sci. 2017;95(5):1957–1962. doi: 10.2527/jas2017.1390. [DOI] [PubMed] [Google Scholar]
- 44.Chen K, Baxter T, Muir WM, Groenen MA, Schook LB. Genetic resources, genome mapping and evolutionary genomics of the pig (Sus scrofa) Int. J. Biol. Sci. 2007;3(3):153–165. doi: 10.7150/ijbs.3.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dai H, Yan M, Li Y. The zinc-finger protein ZCCHC2 suppresses retinoblastoma tumorigenesis by inhibiting HectH9-mediated K63-linked polyubiquitination and activation of c-Myc. Biochem. Bioph. Res. Co. 2020;521(2):533–538. doi: 10.1016/j.bbrc.2019.10.163. [DOI] [PubMed] [Google Scholar]
- 46.Velie BD, Shrestha M, Franҫois L, Schurink A, Tesfayonas YG, Stinckens A, Blott S, Ducro BJ, Mikko S, Thomas R, Swinburne JE, Sundqvist M, Eriksson E, Buys N, Lindgren G. Using an inbred horse breed in a high density genome-wide scan for genetic risk factors of insect bite hypersensitivity (IBH) PLoS ONE. 2016;11(4):e0152966. doi: 10.1371/journal.pone.0152966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang ZW, Jiang YH, Ma C, Silvestri G, Bosinger SE, Li BL, Jong A, Zhou YH, Huang SH. Coexpression network analysis of benign and malignant phenotypes of SIV-infected sooty mangabey and rhesus macaque. PLoS ONE. 2016;11(6):e0156170. doi: 10.1371/journal.pone.0156170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang M, Yang B, Chen H, Zhang H, Wu Z, Ai H, Ren J, Huang L. The fine-scale genetic structure and selection signals of Chinese indigenous pigs. Evol. Appl. 2020;13(2):458–475. doi: 10.1111/eva.12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takeichi T. SDR9C7 plays an essential role in skin barrier function by dehydrogenating acylceramide for covalent attachment to proteins. J. Dermatol. Sci. 2020;98(2):82–87. doi: 10.1016/j.jdermsci.2020.03.005. [DOI] [PubMed] [Google Scholar]
- 50.Wang X, et al. Whole-genome sequencing of eight goat populations for the detection of selection signatures underlying production and adaptive traits. Sci. Rep. 2016;6:38932. doi: 10.1038/srep38932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rance NE. Menopause and the human hypothalamus: evidence for the role of kisspeptin/neurokinin B neurons in the regulation of estrogen negative feedback. Peptides. 2009;30(1):111–122. doi: 10.1016/j.peptides.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xin X, Zhang J, Chang Y, Wu Y. Association study of TAC3 and TACR3 gene polymorphisms with idiopathic precocious puberty in Chinese girls. J. Pediatr. Endocrinol. Metab. 2015;28:65–71. doi: 10.1515/jpem-2013-0460. [DOI] [PubMed] [Google Scholar]
- 53.Jia P, et al. Four novel SNPs of MYO1A gene associated with heat-tolerance in Chinese cattle. Animals. 2019;9(11):964. doi: 10.3390/ani9110964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Talebi F, Mardasi FG, Asl JM, Tizno S, Zadeh MN. Identification of novel PTPRQ and MYO1A mutations in an iranian pedigree with autosomal recessive hearing loss. Cell J. 2018;20(1):127–131. doi: 10.22074/cellj.2018.4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ibrahim HR, Thomas U, Pellegrini A. A helix-loop-helix peptide at the upper lip of the active site cleft of lysozyme confers potent antimicrobial activity with membrane permeabilization action. J. Biol. Chem. 2001;276(47):43767–43774. doi: 10.1074/jbc.M106317200. [DOI] [PubMed] [Google Scholar]
- 56.Gorbenko GP, Ioffe VM, Kinnunen PKJ. Binding of lysozyme to phospholipid bilayers: Evidence for protein aggregation upon membrane association. Biophys. J. 2007;93(1):140–153. doi: 10.1529/biophysj.106.102749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu J, et al. Tbx19, a tissue-selective regulator of POMC gene expression. Proc. Natl. Acad. Sci. 2001;98(15):8674–8679. doi: 10.1073/pnas.141234898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lefebvre H, Thomas M, Duparc C, Bertherat J, Louiset E. Role of ACTH in the interactive/paracrine regulation of adrenal steroid secretion in physiological and pathophysiological conditions. Front. Endocrinol. 2016;7:98. doi: 10.3389/fendo.2016.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fu Y, et al. Genomic analysis reveals selection in Chinese native black pig. Sci. Rep. 2016;6:36354. doi: 10.1038/srep36354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang Y, Adeola AC, Xie HB, Zhang YP. Genomic and transcriptomic analyses reveal selection of genes for puberty in Bama Xiang pigs. Zool. Res. 2018;39(6):424–430. doi: 10.24272/j.issn.2095-8137.2018.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deladoëy J, et al. “Hot spot” in the PROP1 gene responsible for combined pituitary hormone deficiency. J. Clin. Endocrinol. Metab. 1999;84(5):1645–1650. doi: 10.1210/jcem.84.5.5681. [DOI] [PubMed] [Google Scholar]
- 62.Ekegbu UJ, Burrows L, Amirpour-Najafabadi H, Zhou H, Hickford JG. Gene polymorphisms in PROP1 associated with growth traits in sheep. Gene. 2019;683:41–46. doi: 10.1016/j.gene.2018.10.024. [DOI] [PubMed] [Google Scholar]
- 63.Srikanth K, et al. Comprehensive genome and transcriptome analyses reveal genetic relationship, selection signature, and transcriptome landscape of small-sized Korean native Jeju horse. Sci. Rep. 2019;9(1):16672. doi: 10.1038/s41598-019-53102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Y, et al. Signaling pathways related to protein synthesis and amino acid concentration in pig skeletal muscles depend on the dietary protein level, genotype and developmental stages. PLoS ONE. 2015;10(9):e0138277. doi: 10.1371/journal.pone.0138277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Duan X, Zhang HL, Wu LL, Liu MY, Pan MH, Ou XH, Sun SC. Involvement of LIMK1/2 in actin assembly during mouse embryo development. Cell Cycle. 2018;17(11):1381–1389. doi: 10.1080/15384101.2018.1482138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang X, et al. Association analysis of polymorphism in the NR6A1 gene with the lumbar vertebrae number traits in sheep. Genes Genomics. 2019;41(10):1165–1171. doi: 10.1007/s13258-019-00843-5. [DOI] [PubMed] [Google Scholar]
- 67.Hayes AJ, Smith SM, Gibson MA, Melrose J. Comparative immunolocalization of the elastin fiber-associated proteins fibrillin-1, LTBP-2, and MAGP-1 with components of the collagenous and proteoglycan matrix of the fetal human intervertebral disc. Spine. 2011;36(21):E1365–1372. doi: 10.1097/BRS.0b013e31821fd23e. [DOI] [PubMed] [Google Scholar]
- 68.Lee YJ, McPherron A, Choe S, Sakai Y, Chandraratna RA, Lee SJ, Oh SP. Growth differentiation factor 11 signaling controls retinoic acid activity for axial vertebral development. Dev. Biol. 2010;347(1):195–203. doi: 10.1016/j.ydbio.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kowald A, Passos JF, Kirkwood TB. On the evolution of cellular senescence. Aging Cell. 2020;19(12):e13270. doi: 10.1111/acel.13270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tee JM, Van Rooijen C, Boonen R, Zivkovic D. Regulation of slow and fast muscle myofibrillogenesis by Wnt/beta-catenin and myostatin signaling. PLoS ONE. 2009;4(6):e5880. doi: 10.1371/journal.pone.0005880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Otto A, et al. Canonical Wnt signalling induces satellite-cell proliferation during adult skeletal muscle regeneration. J. Cell Sci. 2008;121(17):2939–2950. doi: 10.1242/jcs.026534. [DOI] [PubMed] [Google Scholar]
- 72.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McKenna A, et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Z, et al. Whole-genome resequencing reveals signatures of selection and timing of duck domestication. Gigascience. 2018;7(4):giy027. doi: 10.1093/gigascience/giy027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang K, Li MY, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang J, Li SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81(3):559–575. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kumar S, Nei M, Dudley J, Tamura K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008;9(4):299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012;29(8):1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Falush, D, et al. Traces of human migrations in helicobacter pylori populations. Science. 2003;299(5612):1582–1585. doi: 10.1093/molbev/mss075. [DOI] [PubMed] [Google Scholar]
- 82.Zhang C, Dong SS, Xu JY, He WM, Yang TL. PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics. 2019;35(10):1786–1788. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- 83.Porto-Neto LR, Lee SH, Lee HK, Gondro C. Detection of signatures of selection using FST. Methods. Mol. Biol. 2013;1019:423–436. doi: 10.1007/978-1-62703-447-019. [DOI] [PubMed] [Google Scholar]
- 84.Zhao F, McParland S, Kearney F, Du L, Berry DP. Detection of selection signatures in dairy and beef cattle using high-density genomic information. Genet. Sel. Evol. 2015;47(1):49. doi: 10.1186/s12711-015-0127-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Qin M, Li C, Li Z, Chen W, Zeng Y. Genetic diversities and differentially selected regions between Shandong indigenous pig breeds and western pig breeds. Front. Genet. 2020;10:1351. doi: 10.3389/fgene.2019.01351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Carneiro M, et al. Rabbit genome analysis reveals a polygenic basis for phenotypic change during domestication. Science. 2014;345(6200):1074–1079. doi: 10.1126/science.1253714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li WT, et al. Whole-genome resequencing reveals candidate mutations for pig prolificacy. Proc. R. Soc. B Biol. Sci. 2017;284(1869):20172437. doi: 10.1098/rspb.2017.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tong X. Whole genome sequence analysis reveals genetic structure and X-chromosome haplotype structure in indigenous Chinese pigs. Sci. Rep. 2020;10:1. doi: 10.1098/rspb.2017.2437(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xie C, et al. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2020;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.