Abstract
Background
Clear-cell renal-cell carcinoma (ccRCC) is one of the leading causes of tumour-related death worldwide. Methyltransferase-like 14 (METTL14) is reported to regulate m6A modification in cancers. The aim of this study is to investigate the biological function and molecular mechanism of METTL14 in the pathogenesis of ccRCC.
Methods
Quantitative real-time PCR (qRT-PCR), western blot and immunohistochemical (IHC) assays were used to detect the expression of METTL14 and Pten. METTL14 overexpression or knockdown was used in the in vitro and in vivo studies to investigate the biological functions of METTL14. m6A-RNA immunoprecipitation and RNA immunoprecipitation were used to investigate the m6A modification mediated by METTL14.
Results
METTL14 expression was significantly down-regulated in ccRCC tissues. Functionally, upregulation of METTL14 inhibited ccRCC cells proliferation and migration in vitro. METTL14 overexpression significantly inhibited the activation of the phosphoinositide 3 kinase (PI3K)/AKT signalling pathway. Furthermore, phosphate and tension homology deleted on chromosome ten (Pten) is a target of METTL14. Overexpression of METTL14 increased the m6A enrichment of Pten, and promoted Pten expression. METTL14-enhanced Pten mRNA stability was dependent on YTHDF1.
Conclusions
METTL14-mediated m6A modification of Pten mRNA inhibited tumour progression, suggesting that METTL14 might be a potential prognostic biomarker and effective therapeutic target for ccRCC.
Subject terms: Cancer, Cell biology
Introduction
Kidney cancer is one of the most common cancers in the world [1]. Clear-cell renal-cell carcinoma (ccRCC) is the main subtype of kidney cancer, accounting for more than 80% of all renal cancer, and is the most aggressive subtype of RCC [2]. ccRCC often showed traditional resistance to chemotherapy and radiotherapy, making the treatment for ccRCC is very limited [3], and the prognosis of ccRCC patients is unoptimistic [4]. The underlying molecular mechanisms of ccRCC are largely unclear [4], restraining the development of ccRCC therapy. So, it’s of vital importance to clarify the mechanism of ccRCC pathogenesis and find new treatment targets.
N6-methyladenosine (m6A) RNA methylation is one of the most abundant RNA modifications [5, 6], which plays a critical role in tumour progression [7]. m6A modification is mediated by m6A “writers”, “erasers” and “readers”, which functions in adding, removing, and recognising m6A modifications, respectively [8]. m6A “writer” was composed of methyltransferase-like 3 (METTL3), methyltransferase-like 14 (METTL14), and Wilms’ tumour 1-associating protein (WTAP) [9]. METTL3 functions mainly in catalysing the conversion of adenosine (A) to m6A, while METTL14 functions as an essential component to support METTL3 in recognising RNA substrates [10]. m6A modification plays its role through recruiting m6A “readers”, including YTH domain family (YTHDF), HNRNPA2B1, and insulin-like growth factor 2 mRNA binding protein 2 (IGF2BP) [11, 12], leading to the change of mRNA stability, splicing, translation and microRNA maturation [13, 14]. M6A “writers”, “erasers” and “readers” have been found to play important regulatory roles in various cancer pathogenesis, such as bladder cancer [15], gastric cancer [16] and colorectal cancer [17]. In ccRCC, some m6A methylation regulators were reported to exert potential prognostic value [18, 19]. However, whether m6A methylation regulators take part in the pathogenesis of ccRCC and the underlying mechanism remains unknown.
In this study, we found that METTL14 was downregulated in ccRCC, leading to the ccRCC tumour progression. The mechanism was related to the METTL14-mediated m6A modification of Pten mRNA. METTL14 could promote Pten expression through a YTHDF1-dependent mechanism. Downregulation of METTL14 restrained Pten expression in ccRCC, leading to the tumour progression through activation of the PI3K/AKT signalling pathway.
Methods
Tissues samples collection
A total of 28 pairs of RCC tissues and the corresponding adjacent non-tumour tissues were acquired from The Affiliated Hospital of Zunyi Medical University from 2015 to 2017. All participants were naive to prior chemotherapy or radiotherapy and the diagnosis was confirmed by two independent pathologists. The consent agreement was signed and approved by the Ethics Committee of The Affiliated Hospital of Zunyi Medical University. All patients involved in the present study did not receive chemotherapy or radiotherapy before the surgery. 16 tissues were stored at −80 °C immediately after the surgery, and the other 12 tissues were embedded in paraffin for pathological analysis.
Cell culture and transfection
HK2 cells and human ccRCC cell lines Caki-1 and Caki-2 cells were purchased from ATCC (American Type Culture Collection). Cells were cultured in Dulbecco’s Modification of Eagle’s Medium (DMEM; Gibco, MA, USA) containing 10% foetal bovine serum (FBS; Gibco, MA, USA) at a humid condition of 37 °C, 5% CO2. In all, 0.5 μM BEZ235 (MedChem Express, Dactolisib, USA), a dual PI3K/mTOR inhibitor, was dissolved in dimethyl sulfoxide (DMSO) and then added to Caki-1 cells for24 h.
For overexpression of METTL14, Pten or YTHDF1 in cells, the coding domain sequences of human METTL14, Pten or YTHDF1 mRNA were amplified and inserted into pcDNA3.1 vector to construct the overexpression plasmid. For knockdown of METTL14, Pten or YTHDF1 in cells, the small hairpin RNA (shRNA) targeting METTL14, Pten or YTHDF1 was designed and synthesised by the Genscript (Nanjing, China). The sequences used were provided as follows: sh-METTL14: 5′-ACCTCGCCGTGTTAAATAGCAAAGATTCAAGAGATCTTTGCTATTTAACACGGCTT-3′; sh-Pten: 5′-ACCTCGATCTTGACCAATGGCTAAGTTCAAGAGACTTAGCCATTGGTCAAGATCTT-3′; sh-YTHDF1: 5′-ACCTCGACAGTCAAATCAGAGTAACATCAAGAGTGTTACTCTGATTTGACTGTCTT-3′; sh-NC: 5′- ACCTCACTCAAAAGGAAGTGACAAGATCAAGAGTCTTGTCACTTCCTTTTGAGTTT-3′. The overexpression plasmid or the shRNA and the indicated control were transfected to the cells by using Lipofectamine 3000 (Invitrogen, USA).
m6A RNA methylation quantification
The total RNA was extracted using TRIzol reagent (Invitrogen, USA). Then m6A RNA Methylation Assay Kit (Abcam, MA, USA) was used to detect the overall methylation m6A content according to the manufacturer’s instruction. Briefly, 200 ng total RNA was inputted in per reaction following the diluted capture antibody, detection antibody solution and the enhancer solution adding into per reaction, respectively. The m6A level was quantified by colorimetry, and the absorbance of each reaction was measured at 450 nm.
Quantitative reverse-transcriptase PCR (qRT-PCR)
Total RNA was extracted using TRIzol reagent (Invitrogen). Then the total RNA was used to generate cDNA by HiScript® III 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China) with an oligo-dT primer. Real-time reverse-transcriptase PCR was performed using the SYBR qRT-PCR Master Mix kit (Vazyme, Nanjing, China) as recommended by the manufacturer. The product was analysed on the RT-PCR Detection System (Analytic, Jena, Germany). The quantitative measures were obtained using the ΔΔCT method and GAPDH was served as the internal control. The primers for the target genes were: METTL3, forward: 5′-TTG TCT CCA ACC TTC CGT AGT-3′, reverse: 5′-CCA GAT CAG AGA GGT GGT GTA G-3′; METTL14, forward: 5′-GTT GGA ACA TGG ATA GCC GC-3′, reverse: 5′-CAA TGC TGT CGG CAC TTT CA-3′; Pten, forward: 5′‐GTTCATAACGATGGCTGTGG‐3′, reverse: 5′‐TGCATGCATGACACATTAACA‐3′; GAPDH, forward: 5’-CGC CTT ATT CGA GAG GTG TCA-3′, reverse: 5′-TGG GCT GTC ACT ACG GAA GG-3′.
Western blot
Total protein from tumour tissues or cells was extracted by using RIPA lysis buffer plus PMSF on ice, and BCA assay kit (Santa Cruz, California, USA) was used to measure the total protein concentration. A total of 40 μg protein samples were separated by using SDS-PAGE electrophoresis, and then transferred onto PVDF membranes. After blocking with 5% skim milk, the membrane was incubated with prepared primary antibodies at 4 °C overnight. The other day, the membrane was washed and incubated with the secondary antibodies for 1 h at room temperature. Finally, enhanced chemiluminescence (ECL, ThermoFisher, MA, USA) was used to visualise this membrane. The protein band was analysed by using the ImageJ software. Antibodies were listed as follows: anti-METTL3 (#86132, 1:1000, CST, Beverly, Massachusetts, USA); anti-GAPDH (#5174, 1:5000, CST, Beverly, Massachusetts, USA); anti-PI3K (#4249S, 1:1000, CST, Beverly, Massachusetts, USA); anti-phospho-PI3K p85 (Tyr458)/p55 (Tyr199) (p-PI3K, #4228S, 1:1000, CST, Beverly, Massachusetts, USA); anti-Akt (ab18785, 1:1000, Abcam, Cambridge, MA, USA); anti-p-Akt (Ser473) (ab81283, 1:1000, Abcam, Cambridge, MA, USA); anti-mTOR (ab32028, 1:1000, Abcam, Cambridge, MA, USA); anti-p-mTOR (Ser2448) (ab109268, 1:1000, Abcam, Cambridge, MA, USA); anti-S6K (#9202, CTS, Beverly, Massachusetts, USA); anti-p-S6K (Thr389) (#9205, CTS, Beverly, Massachusetts, USA); anti-4EBP1 (#9452, CTS, Beverly, Massachusetts, USA); anti-p-4EBP1 (Thr37/46) (#9459, CTS, Beverly, Massachusetts, USA); anti-METTL14 (ab220031, 1:1000, Abcam, Cambridge, MA, USA); anti-Pten (ab267787, 1:1000, Abcam, Cambridge, MA, USA); anti-YTHDF1 (ab252346, 1:1000, Abcam, Cambridge, MA, USA).
Immunochemical (IHC) staining
The paraffin-embedded tumour sections were dewaxed in xylene and rehydrated in alcohol. 3% H2O2 was used to block the endogenous peroxidase, followed by microwave heating treatment to retrieve the antigen. After blocking with 5% BSA at 37 °C for 1 h, the tumour sections were incubated with primary antibodies against METTL14 or Pten (1:100 dilution, Abcam, USA) at 4 °C overnight. The other day, the sections were washed away with the primary antibody and then subjected to incubation with corresponding secondary antibodies at 37 °C for 1 h. One hour later, the secondary antibodies were washed away, and the tumour sections were stained with diaminobenzidine (DAB) solution and counterstained with hematoxylin. Then slices were visualised and the representative images were taken using a light microscope (Olympus, Tokyo, Japan).
Immunofluorescent (IF) staining
Cells were fixed with 4% paraformaldehyde and blocked with 1% BSA and 0.1% Triton-X at room temperature for 2 h. Then cells were incubated with primary antibodies against Pten (1:100) overnight at 4 °C. The next day, appropriate secondary antibodies (Abcam) were added and incubated with cells for 1 h at room temperature. The nuclei were stained with Hoechst 33342 for 10 min at room temperature. Immunofluorescence was examined under a fluorescence microscope (Nikon, Japan).
CCK-8 assay
Cell Count Kit-8 (CCK-8) (Beyotime, Shanghai, China) was used to examine cell proliferation according to the manufacturer’s instructions. Briefly, Caki-1 and Caki-2 cells which were stably overexpressed or knocked down of METTL14 were cultured in 96-well plates at the density of 1 × 103/well. 24 h later, 10 μL of CCK-8 solution was added to each well of the 96-well plate to incubate the cells for 4 h. The detection was performed at 24, 48, 72 and 96 h, followed by measurement of the 450-nm OD using a microplate reader.
Clone-formation assay
The cells were cultured in a six-well plate (9.6 cm2) at a density of 1 × 103/well. Two weeks later, the clone was macroscopic, and the cells were stained with 0.5% crystal violet for 15 min. After washing off the staining solution, the colonies were observed under a light microscope (Olympus, Tokyo, Japan), and the number of the colonies was calculated in a six-well plate (9.6 cm2).
5-ethynyl-2′-deoxyuridine (EdU) assay
EdU assay was performed by using the Cell Light EdU DNA Imaging Kit (RiboBio, China) according to the manufacturer’s instructions. Briefly, the cells were seeded into a 24-well plate overnight, and 50 µM EdU was added the next day for an additional 2 h. Then the cells were fixed with 4% paraformaldehyde at room temperature for 30 min, followed with a 5-min incubation with the glycine solution and 10-min incubation with 0.5% Trion X-100. After washing twice using PBS, the cells were stained with the fluorescent dye and Hoeschst 33342. The images were captured using fluorescent microscopy (Nikon, Japan), and the percentage of EdU-positive cells was detected from five random fields in three wells.
Cell migration assay
Cell migration was determined by transwell assay. Caki-1 and Caki-2 cells were seeded to the upper transwell chamber at a density of 5 × 104 cells/well into an 8-mm pore transwell plate (Costar, USA). Serum-free medium was added onto the upper chamber, and the culture medium containing 20% FBS was added into the lower chamber. Following 24 h of incubation, the unmigrated cells were cleaned using a cotton swab and then fixed in 4% paraformaldehyde for 20 min and stained with haematoxylin. Images were taken with a light microscope (Olympus, Tokyo, Japan). The number of migrated cells was counted in five random fields (200×) and represented as the average of three independent replicates.
MeRIP and RIP-qRT-PCR
MeRIP was carried out by using Magna MeRIP m6A Kit (Merck Millipore, Darmstadt, Germany) according to the manufacturer’s instruction. Total RNAs were extracted from the cells and subjected to fragment and then incubated with protein A beads pre-incubated with anti-m6A polyclonal antibody (Abcam, MA, USA), YTHDF1 polyclonal antibody (Abcam, MA, USA) or IgG (isotype control) in RIP buffer at 4 °C for 3 h. Then, the samples were washed, followed by purification with Qiagen and subjected to qRT-PCR for the detection of Pten.
RNA stability
To detect the RNA stability in cells, 5 mg/mL actinomycin D (Sigma, MA, USA) was treated to cells overexpressed with YTHDF1 or not. RNA was isolated using TRIzol reagent (Invitrogen, NY, USA) at the time point of 0, 1, 2, 3, 4, 5 and 6 h, and then analysed by real-time PCR, normalised to GAPDH. t1/2 of mRNA was calculated.
Tumour xenograft model
For stable cell line construction, METTL14 shRNA lentivirus was constructed using pLKD-CMV-EGFP vectors, and infected the cells which were selected by treatment with puromycin (1.5 μg/mL) for 14 days. BALB/c nu/nu male mice (4–6 weeks) were subcutaneously injected with the Caki-1 cells (1 × 107 cells/200 μl) with or without METTL14 knockdown into each flank. Overexpression of Pten was performed by using Pten overexpression lentivirus (GenePharma, Shanghai, China) by intratumoral injection of 50 μL virus (4 × 107 IU/mL) after the tumour cells injection. BALB/c nu/nu male mice were randomly divided into three groups: negative control (NC) group (n = 5), METTL14 knockdown (METTL14 KD) group (n = 5), METTL14 knockdown+Pten overexpressio (METTL14 KD + Pten OE) group (n = 5). The mice were sacrificed after 28 days, and the tumours were isolated. The tumour was subjected to haematoxylin and eosin (H&E) staining for tissue histopathological studies. IHC staining and western blot analysis of the tumour were performed to analyse the METTL14, Pten expression and the Akt signalling activation. The animal experiments were approved by the Affiliated Hospital of Zunyi Medical University Animal Care and Use Committee.
Statistical analysis
Data were analysed using the Prism Graphpad 8.0 software and presented as mean ± SEM. Individual experiments were repeated three times. Two-tailed Student’s t test made comparisons between two groups. One-way ANOVA made comparisons between multiple groups. P < 0.05 was considered statistically significant.
Results
METTL14 is low expressed in the ccRCC and correlates with poor prognosis
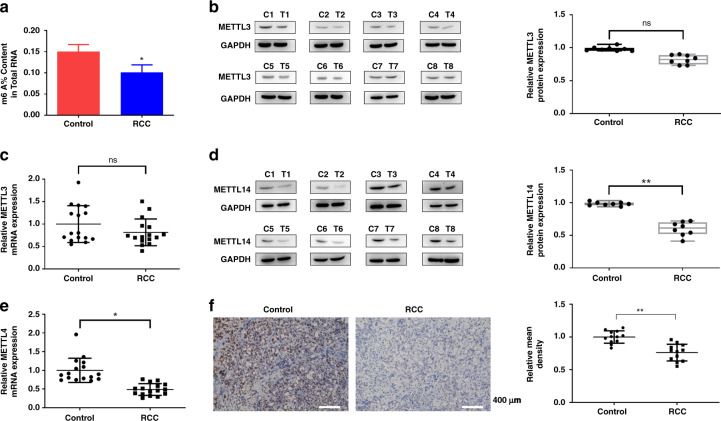
Firstly, we detected the total m6A level in the ccRCC tissues. As shown in Fig. 1a, m6A level showed a significant decrease in the ccRCC tissues. We then detected the m6A methyltransferases METTL3 and METTL14 expression in the ccRCC tissues. Western blot and qRT-PCR showed that expression of METTL3 showed no difference between the normal tissues and the tumour tissues (Fig. 1b, c), while METTL14 expression was significantly downregulated in the tumour tissues (Fig. 1d, e), which were further verified by the IHC staining (Fig. 1f). TCGA database also showed that METTL3 showed no difference between the 72 normal tissues and 533 ccRCC tumour tissues (Supplementary Fig. 1A), while METTL14 was decreased in the 533 ccRCC tumour tissues compared to the 72 normal tissues (Supplementary Fig. 1B). Moreover, TCGA database revealed METTL3 expression had no correlation with the OS of ccRCC patients (Supplementary Fig. 1C), while low expression of METTL14 was associated with poor prognosis of ccRCC patients (Supplementary Fig. 1D). The datasets also showed that METTL3 level was downregulated in the kidney chromophobe (KICH), tissues and increased in the liver hepatocellular carcinoma (LIHC), and lung squamous cell carcinoma (LUSC) tissues (Supplementary Fig. 2A). METTL14 level was decreased in the KICH, kidney renal papillary cell carcinoma (KIRP) and LUSC tissues (Supplementary Fig. 2B). These results suggested that METTL14 might be involved in ccRCC progression.
Fig. 1. METTL14 is low expressed in the ccRCC and correlates with poor prognosis.
a Total m6A content in the ccRCC tissues. n = 16. b Protein levels of METTL3 in the ccRCC tissues (C1-C8) and the adjacent normal tissues (T1-T8) were detected by western blot. Protein expression levels were analysed by ImageJ densitometric analysis. n = 16. c mRNA levels of METTL3 in the ccRCC tissues and the adjacent normal tissues were detected by qRT-PCR. n = 16. d Protein levels of METTL14 in the ccRCC tissues (C1-C8) and the adjacent normal tissues (T1-T8) were detected by western blot. Protein expression levels were analysed by ImageJ densitometric analysis. n = 16. e mRNA levels of METTL14 in the ccRCC tissues and the adjacent normal tissues were detected by qRT-PCR. n = 16. f Immunochemical staining showed METTL14 expression in tumour tissues. Bar = 400 μm. n = 12. Data are presented as mean ± SEM from three independent experiments. *P < 0.05, **P < 0.01,***P < 0.001, compared to the control group.
METTL14 inhibits ccRCC cell proliferation and migration in vitro
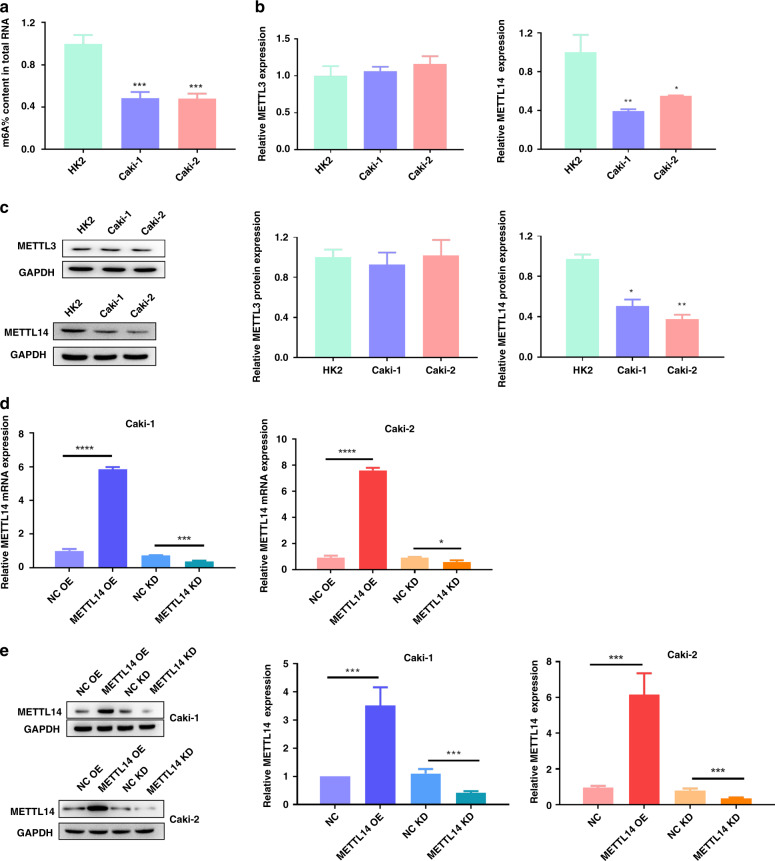
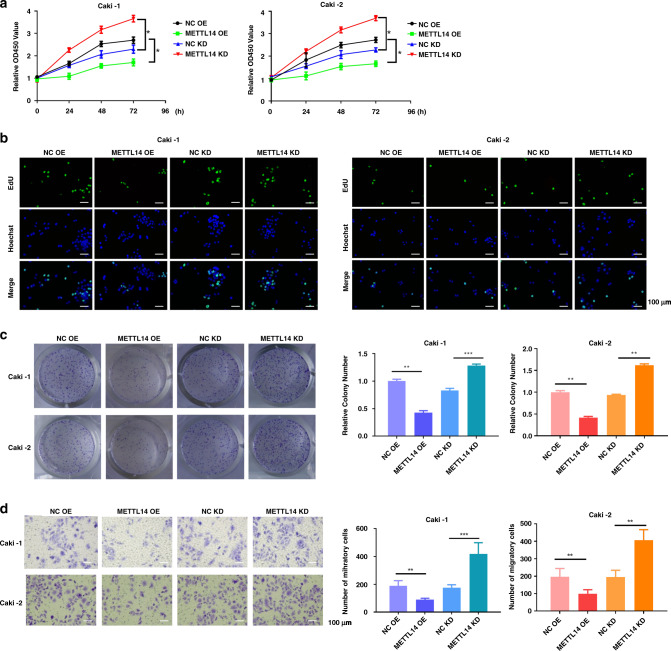
We also detected the total m6A level in the ccRCC cells Caki-1 and Caki-2. Compared to the HK2 cells, the level of m6A methylation in the Caki-1 and Caki-2 was downregulated (Fig. 2a). Then, we examined the expression of METTL3 and METTL14 in the ccRCC cells Caki-1 and Caki-2. qRT-PCR and western blot showed that METTL3 had no differences between normal cell line HK2 and ccRCC cell lines (Caki-1 and Caki-2) (Fig. 2b, c). Compared to the HK2 cells, METTL14 was significantly downregulated in the Caki-1 and Caki-2 cells (Fig. 2b, c). To investigate the regulatory role of METTL14 in ccRCC, we explored the effect of METTL14 on ccRCC cell proliferation and migration in vitro. Then, we knocked down or overexpressed METTL14 in Caki-1 and Caki-2 cells. The knockdown sequence used in the present study was selected after verifying the knockdown efficacy from three shRNA in Caki-1 cells by using qRT-PCR (Supplementary Fig. 2). After verifying the knockdown or overexpression efficiency by using qRT-PCR and western blot (Fig. 2d, e), cell proliferation and migration were measured. As shown by CCK-8 assay in Fig. 3a and EdU assay in Fig. 3b, overexpression of METTL14 inhibited the cell proliferation, while knockdown of METTL14 promoted proliferation of Caki-1 and Caki-2 cells. Consistent with the results of cell proliferation, clone-formation assay showed that overexpression of METTL14 inhibited the clone formation, while knockdown of METTL14 increased the clone-formation rate of Caki-1 and Caki-2 cells (Fig. 3c). Transwell migration assay revealed that upregulation of METTL14 impeded the migratory ability of Caki-1 and Caki-2 cells, downregulation of METTL14 expression significantly elevated the migration ability of Caki-1 and Caki-2 cells (Fig. 3d).
Fig. 2. METTL14 level is upregulated in ccRCC.
a Total m6A content in the ccRCC cell lines. b qRT-PCR showed the mRNA expression of METTL3 and METTL14 in ccRCC cell lines (Caki-1 and Caki-2) compared with HK2 cells. c Western blot verified the protein expression of METTL3 and METTL14 in Caki-1 and Caki-2 compared with HK2 cells. METTL14 was overexpressed or knocked down in Caki-1 and Caki-2 cells. d, e The transfection efficacy was verified by qRT-PCR and western blot. *P < 0.05, **P < 0.01, ***P < 0.001, compared to the indicated group. NC OE overexpression plasmid of negative control pcDNA3.1, METTL14 OE overexpression plasmid of METTL14, NC KD shRNA targeting negative control, METTL14 KD shRNA targeting METTL14.
Fig. 3. METTL14 inhibits ccRCC cell proliferation and migration in vitro.
a CCK-8 assay was performed to measure the proliferation of Caki-1 and Caki-2 cells overexpressed or knockdown of METTL14. b EdU staining revealed the proliferation of Caki-1 and Caki-2 cells overexpressed or knockdown of METTL14. Bar = 100 μm. c Representative images and quantification of colony-formation assay of ccRCC cells overexpressed or knockdown of METTL14 in a 6-well plate (9.6 cm2). Magnification, ×200. d Transwell invasion assay was performed to determine the effects of METTL14 on the migration ability of ccRCC cells. Magnification, ×200. Bar = 100 μm. n = 6. Data are presented as mean ± SEM from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, compared to the indicated group. NC OE overexpression plasmid of negative control pcDNA3.1, METTL14 OE overexpression plasmid of METTL14, NC KD shRNA targeting negative control, METTL14 KD shRNA targeting METTL14.
METTL14 inhibits the AKT signalling activation
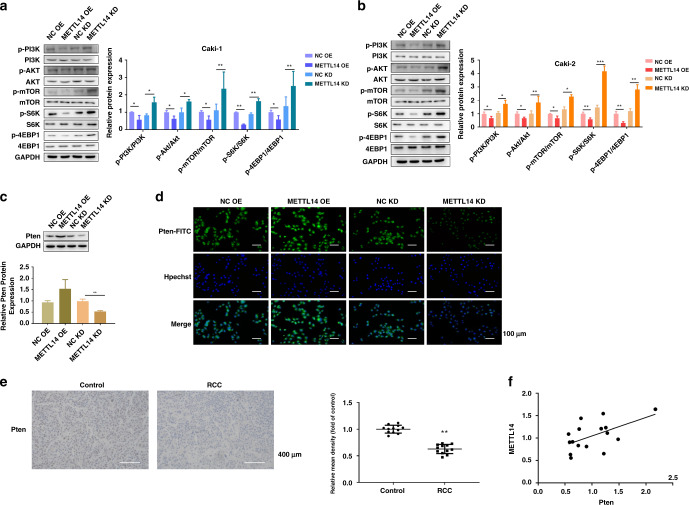
We further explored the mechanism of the effect of METTL14 on cell proliferation and migration. The PI3K/AKT signalling pathway has been shown to regulate cell proliferation, apoptosis, and tumour metastasis in various tumour progression, including RCC [20, 21]. In this study, we found that overexpression of METTL14 decreased the phosphorylation level of PI3K, AKT and mTOR in Caki-1 and Caki-2 cells, indicating the inhibition of the PI3K/AKT signalling pathway (Fig. 4a, b). On the contrary, knockdown of METTL14 enhanced the PI3K/AKT signalling transduction, as indicated by the increasing phosphorylation level of PI3K, AKT and mTOR in Caki-1 and Caki-2 cells (Fig. 4a, b). To ascertain the actual activation of the cascade, the phosphorylation of mTOR targets (S6K and 4EBP1) levels were also detected by western blot assay. The result showed that overexpression of METTL14 decreased the phosphorylation levels of S6K and 4EBP1, while knockdown of METTL14 increased the phosphorylation levels of S6K and 4EBP1 in Caki-1 and Caki-2 cells (Fig. 4a, b).
Fig. 4. Downregulation of METTL14 promotes the AKT signalling activation.
a, b PI3K, AKT, mTOR, S6K, 4EBP1 and the related phosphorylation protein level in Caki-1 and Caki-2 cells were detected by using western blot. c, d Pten protein expression in Caki-1 cells overexpressed or knocked down of METTL14 was detected by using western blot and immunofluorescence. Bar = 100 μm. e Immunochemical staining showed Pten expression in tumour tissues. Bar = 400 μm. n = 12. f Positive correlation of METTL14 and Pten expression in tumour tissues is shown. n = 16. Data are presented as mean ± SEM from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, compared to the indicated group. NC OE overexpression plasmid of negative control pcDNA3.1, METTL14 OE overexpression plasmid of METTL14, NC KD shRNA targeting negative control, METTL14 KD shRNA targeting METTL14.
Pten is a tumour-suppressive protein, which modulates PI3K/Akt signalling pathway negatively [22, 23]. We then detected the effect of METTL14 on Pten expression. Western blot analysis showed that METTL14 overexpression upregulated Pten expression, and knockdown of METTL14 inhibited Pten expression in Caki-1 cells (Fig. 4c), the same as the results from IF staining of Pten in Caki-1 cells (Fig. 4d). Moreover, IHC staining showed that Pten was down-regulated in the ccRCC tissues (Fig. 4e). The expression of METTL14 was significantly correlated with Pten expression (Fig. 4f). All these data suggested that METTL14 positively regulates the Pten expression.
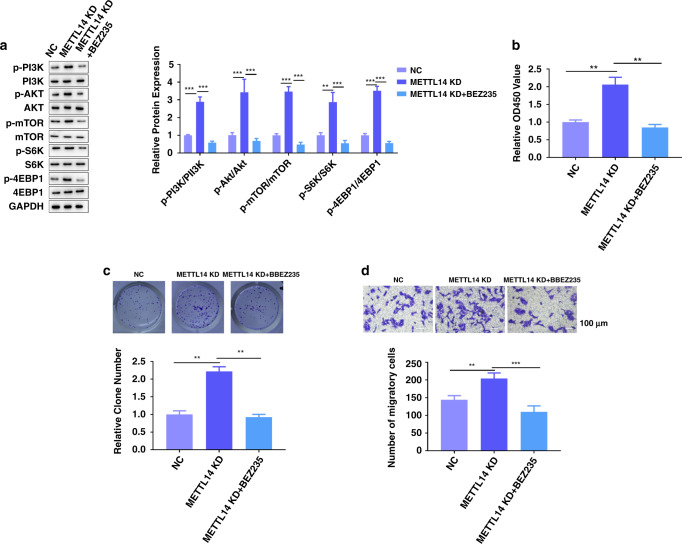
BEZ235 abrogates the effect of METTL14 knockdown in vitro
To further explore whether the effects of METTL14 were mediated through the PI3K/AKT/mTOR pathway, BEZ235, in combination with a dual PI3K/mTOR inhibitor, was added to Caki-1 cells with METTL14 knockdown. Western blot assay showed that knockdown of METTL14 increased the phosphorylation levels of S6K and 4EBP1, which were abolished by BEZ235 (Fig. 5a). Moreover, BEZ235 significantly compensated for the promoting effect of knockdown of METTL14 on Caki-1 cell proliferation, clone formation and migration (Fig. 5b–d). All these data suggested that blocking PI3K/AKT/mTOR pathway abrogates the effect of manipulating METTL14.
Fig. 5. BEZ235 abrogates the effect of METTL14 knockdown in vitro.
BEZ235, in combination with a dual PI3K/mTOR inhibitor, was added to Caki-1 cells with METTL14 knockdown. a PI3K, AKT, mTOR, S6K, 4EBP1 and the related phosphorylation protein level in Caki-1 cells were detected by using western blot. b CCK-8 assay was performed to measure the proliferation of Caki-1 cells of the indicated group. c Representative images and quantification of colony-formation assay of Caki-1 cells of the indicated group. Magnification, ×200. d Transwell invasion assay was performed to determine the effects of BEZ235 on METTL14’s regulation on migration ability of Caki-1 cells. Magnification, ×200. Bar = 100 μm. n = 6. Data are presented as mean ± SEM from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, compared to the indicated group. NC shRNA targeting METTL14 + DMSO, METTL14 KD shRNA targeting METTL14 + DMSO, METTL14 KD + BEZ235 shRNA targeting METTL14 + BEZ235.
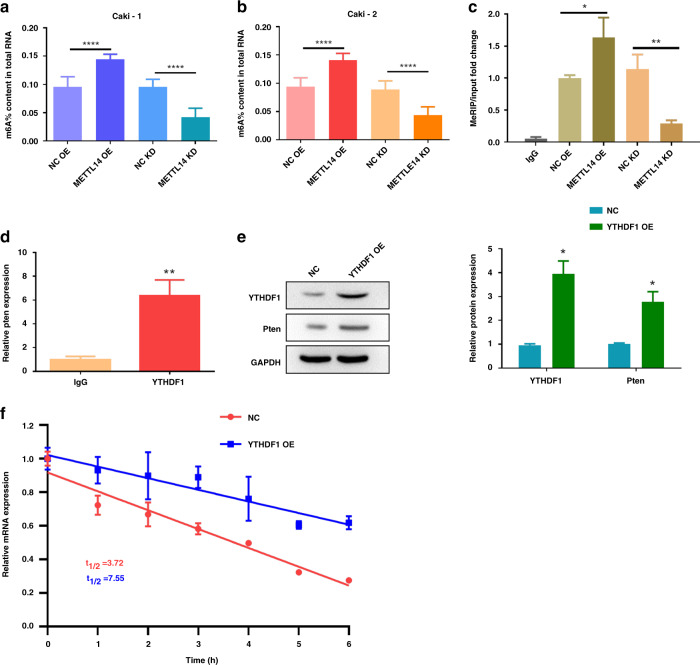
METTL14 overexpression enhances Pten mRNA stability via an m6A-YTHDF1-dependent manner
m6A modification could promote protein expression through enhancing mRNA stability through YTHDF1 recognition [24]. Therefore, we wonder whether METTL14 upregulated Pten expression through increasing its mRNA stability. We found that overexpression of METTL14 increased the total m6A modification level in Caki-1 and Caki-2 cells, while knockdown of METTL14 decreased the m6A modification in Caki-1 and Caki-2 cells (Fig. 6a, b). MeRIP was performed to investigate the interaction of METTL14 and Pten. As shown in Fig. 4c, METTL14 overexpression enhanced the m6A enrichment of Pten mRNA, while knockdown of METTL14 decreased the Pten mRNA m6A modification (Fig. 6c).
Fig. 6. METTL14 overexpression enhances Pten mRNA stability via an m6A-YTHDF1-dependent manner.
a, b m6A level of Caki-1 and Caki-2 cells overexpressed or knocked down of METTL14 was detected. c Effect of METTL14 on Pten mRNA m6A level in Caki-1 cells was measured by MeRIP. d RIP assay followed by qRT-PCR to assay Pten mRNA endogenously associated with YTHDF1 in Caki-1 cells. e YTHDF1 and Pten protein expression in Caki-1 cells overexpressed with YTHDF1 were detected by western blot. f After actinomycin D (5 mg/mL) treatment, RNA stability assay showed the mRNA half-life (t1/2) of Caki-1 cells overexpressed with YTHDF1. n = 6. Data are presented as mean ± SEM from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, compared to the indicated group. NC OE overexpression plasmid of negative control pcDNA3.1, METTL14 OE overexpression plasmid of METTL14, NC KD shRNA targeting negative control, METTL14 KD shRNA targeting METTL14, YTHDF1 OE overexpression plasmid of YTHDF1.
YTHDF1 is the reader of mRNA m6A modification, and facilitates the initiation of mRNA translation [25]. RIP-PCR illustrated that YTHDF1 could directly interact with Pten mRNA, indicating the potential binding of YTHDF1 on Pten mRNA (Fig. 6d). Western blot further showed that YTHDF1 overexpression could increase Pten protein expression (Fig. 6e). The RNA stability assay showed that the mRNA t1/2 was prolonged by YTHDF1 overexpression (Fig. 6f). All these data indicated that METTL14 overexpression enhances Pten mRNA stability via an m6A-YTHDF1-dependent manner.
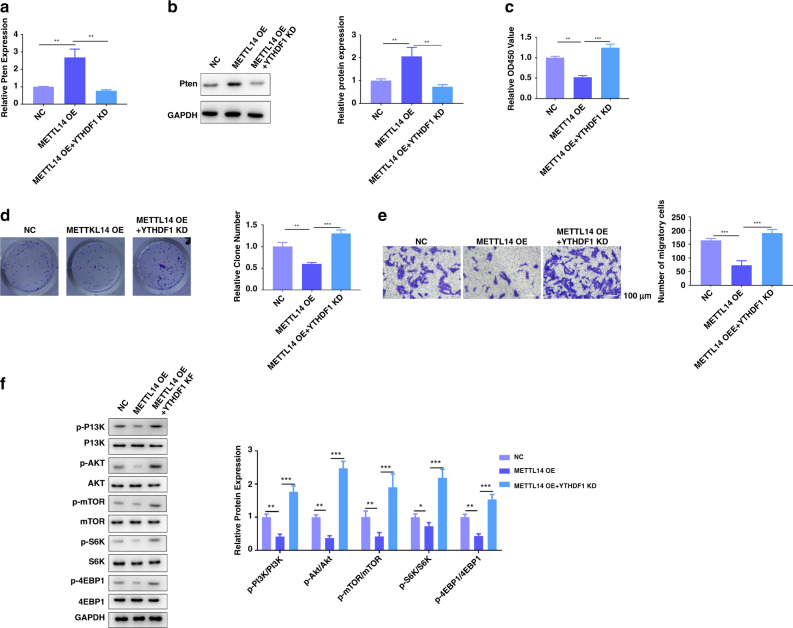
METTL14 inhibits ccRCC cell proliferation, migration and the AKT signalling activation via an m6A-YTHDF1-dependent manner
Then, we assessed whether the silencing of YTHDF1 affected PTEN levels and proliferation in the models of overexpression of METTL14 in Caki-1 cells. The mRNA and protein levels of PTEN were increased by overexpression of METTL14, which were recovered by silencing of YTHDF1 (Fig. 7a, b). Functionally, silencing of YTHDF1 significantly compensated for the inhibitory effect of overexpression of METTL14 on Caki-1 cell proliferation, clone formation and migration (Fig. 7c–e). Furthermore, overexpression of METTL14 decreased phosphorylation of PI3K, AKT, mTOR, S6K and 4EBP1, which were increased by knockdown of YTHDF1 (Fig. 7f). All these data suggested that METTL14 regulates PTEN expression and participates in ccRCC progression via an m6A-YTHDF1-dependent manner.
Fig. 7. METTL14 inhibits ccRCC cell proliferation, migration and the AKT signalling activation via an m6A-YTHDF1-dependent manner.
METTL14 was overexpressed in Caki-1 cells with or without YTHDF1 knockdown. a qRT-PCR assay showed the mRNA expression of Pten in Caki-1 cells of the indicated group. b Western blot showed the expression of Pten in Caki-1 cells of the indicated group. c CCK-8 assay was performed to measure the proliferation of Caki-1 cells of the indicated group. d Representative images and quantification of colony-formation assay of Caki-1 cells of the indicated group. Magnification, ×200. e Transwell invasion assay was performed to determine the effects of YTHDF1 on METTL14’s regulation on migration ability of Caki-1 cells. Magnification, ×200. Bar = 100 μm. f PI3K, AKT, mTOR, S6K, 4EBP1 and the related phosphorylation protein level in Caki-1 cells were detected by western blot. n = 6. Data are presented as mean ± SEM from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, compared to the indicated group. NC overexpression plasmid of negative control pcDNA3.1+shRNA targeting negative control, METTL14 OE overexpression plasmid of METTL14 + shRNA targeting negative control, METTL14 OE + YTHDF1 KD overexpression plasmid of METTL14 + shRNA targeting YTHDF1.
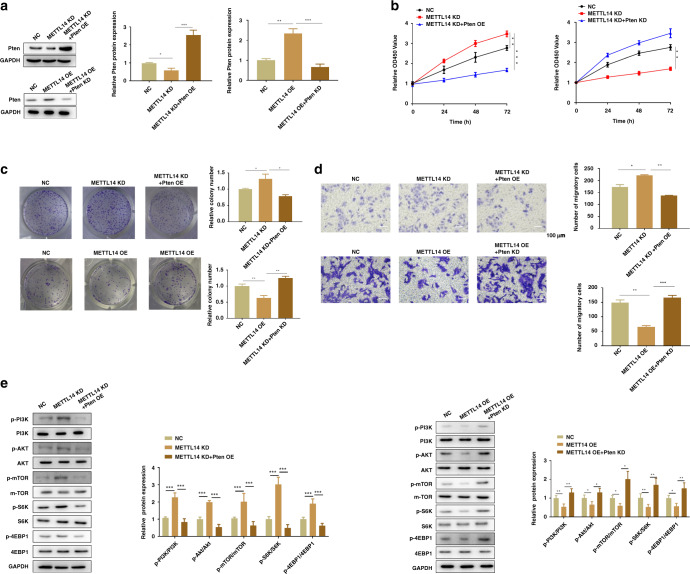
METTL14 inhibits ccRCC cell proliferation, migration and the AKT signalling activation through upregulating Pten in vitro
As we have proved that METTL14 suppressed cell proliferation and migration in Caki-1 and Caki-2 cells, we then investigated whether the effect of METTL14 was dependent on its regulatory effect on Pten expression. We overexpression of METTL14 in Caki-1 cells with or without Pten knockdown. Moreover, we knocked down METTL14 in Caki-1 cells with or without Pten overexpression. Western blot analysis revealed that overexpression of METTL14 increased Pten expression, while knockdown of METTL14 induced downregulation of Pten expression (Fig. 8a). Overexpression of METTL14 suppressed cell proliferation, while knockdown of Pten reversed the role of METTL14 (Fig. 8b). Downregulated METTL14 promoted cell proliferation, which was inhibited by overexpression of Pten (Fig. 8b). Clone-formation assay also showed that overexpression of METTL14 suppressed clone formation, while knockdown of Pten further promoted clone formation (Fig. 8c). Meanwhile, Pten overexpression reversed the promoting effect of METTL14 on the clone formation (Fig. 8c). Transwell assay showed that overexpression of METTL14 inhibited Caki-1 cell migration, which was recovered by knockdown of Pten (Fig. 8d). Knockdown of METT14 enhanced the migration ability of Caki-1 cells, and overexpression of Pten abolished the effect of METTL14 (Fig. 8d). Then, western blot assay suggested that overexpression of METTL14 decreased phosphorylation of PI3K, AKT, mTOR, S6K and 4EBP1, which were increased by knockdown of Pten (Fig. 8e). On the contrary, METTL14 knockdown increased phosphorylation of PI3K, AKT, mTOR, S6K and 4EBP1, while Pten overexpression abrogated the effect of METTL14 on the PI3K/AKT/mTOR signalling pathway (Fig. 8e). All these data suggested that the effect of METTL14 was dependent on the regulation of Pten expression.
Fig. 8. METTL14 inhibits ccRCC cell proliferation, migration and the AKT signalling activation through upregulating Pten in vitro.
METTL14 was overexpressed in Caki-1 cells with or without Pten knockdown. METTL14 was knocked down in Caki-1 cells with or without Pten expression. a Western blot showed the expression of Pten in Caki-1 cells of the indicated group. b CCK-8 assay was performed to measure the proliferation of Caki-1 cells of the indicated group. c Representative images and quantification of colony-formation assay of Caki-1 cells of the indicated group. Magnification, ×200. d Transwell invasion assay was performed to determine the effects of Pten on METTL14’s regulation on migration ability of Caki-1 cells. Magnification, ×200. Bar = 100 μm. e PI3K, AKT, mTOR, S6K, 4EBP1 and the related phosphorylation protein level in Caki-1 cells were detected by using western blot. n = 6. Data are presented as mean ± SEM from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, compared to the indicated group. NC overexpression plasmid of negative control pcDNA3.1+shRNA targeting negative control, METTL14 OE overexpression plasmid of METTL14 + shRNA targeting negative control, METTL14 OE + Pten KD overexpression plasmid of METTL14 + shRNA targeting Pten, METTL14 KD shRNA targeting METTL14 + overexpression plasmid of negative control pcDNA3.1, METTL14 KD + Pten OE shRNA targeting METTL14 + overexpression plasmid of Pten.
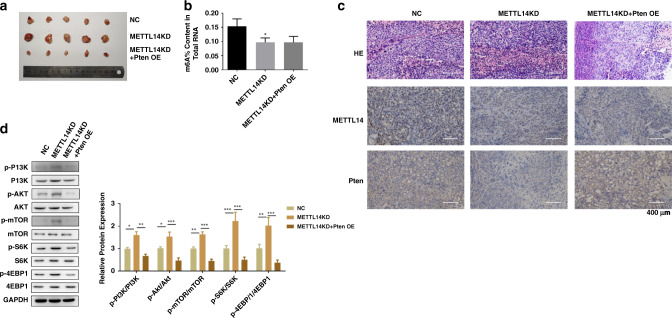
Pten reverses the effect of METTL14 knockdown in vivo
We then verified the effect and mechanism of METTL14/Pten on ccRCC progression in vivo. The mice were divided into three groups, with ten mice in each group. Caki-1 cells with or without METTL14 knockdown were subcutaneously injected into each flank of the mice to contrast the ccRCC xenograft model. Pten overexpression lentivirus was administrated by intratumoral injection after the tumour cells injection to overexpression Pten in the tumour tissues. As shown in Fig. 9a, mice in METTL14 knockdown group showed an obvious larger tumour size than the control group, while overexpression of Pten inhibited the METTL14 knockdown-induced tumour progression. Knockdown of METTL14 induced decreasing level of m6A modification in the tumour tissues (Fig. 9b). IHC staining showed that knockdown of METTL14 decreased the expression of Pten, which was recovered by overexpression of Pten (Fig. 9c). Western blot analysis further showed that knockdown of METTL14 increased the phosphorylation level of PI3K, AKT, mTOR, S6K and 4EBP1, which were inhibited by Pten overexpression, suggesting that Pten overexpression abrogated the activation of PI3K/AKT signalling induced by METTL14 knockdown (Fig. 9d).
Fig. 9. Pten reverses the effect of METTL14 knockdown in vivo.
Caki-1 cells with or without METTL14 knockdown were subcutaneously injected into each flank of the mice to establish the ccRCC xenograft model. Pten overexpression lentivirus was administrated by intratumoral injection after the tumour cells injection to overexpression Pten in the tumour tissues. a Pictures of the isolated tumours of the indicated group. b Total m6A level of tumours in the indicated group. c H&E and IHC staining of METTL14 and Pten of the tumour tissues. Bar = 400 μm. d PI3K, AKT, mTOR, S6K, 4EBP1 and the related phosphorylation protein level in tumour tissues were detected by using western blot. Data are presented as mean ± SEM from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, compared to the indicated group. NC overexpression plasmid of negative control pcDNA3.1+shRNA targeting negative control, METTL14 KD shRNA targeting METTL14 + overexpression plasmid of negative control pcDNA3.1, METTL14 KD + Pten OE shRNA targeting METTL14 + overexpression plasmid of Pten.
Discussion
ccRCC is a type of highly aggressive renal malignant tumour [26]. Traditional radiotherapy and chemotherapy often exhibit low benefits for treating ccRCC, making it important to find the new mechanism and new targets for ccRCC [27]. Recently, some m6A methylation regulators have been reported to exert potential prognostic value [18, 19]. Studies have indicated that overexpression of METTL3, METTL14, METTL16, WTAP or other components, can regulate several cancers development [28–31]. For instance, Chen et al. suggested overexpression of METTL14 inhibited colorectal cancer cells migration, invasion and metastasis in vitro [32]. However, whether m6A methylation regulators take part in the pathogenesis of ccRCC and the underlying mechanism remains unknown. In this study, we found that downregulation of METTL14 was involved in the pathogenesis of ccRCC. Downregulation of METTL14 restrained Pten expression in ccRCC by regulating m6A modification of Pten mRNA, leading to the tumour progression through activation of the PI3K/AKT signalling pathway.
Dysregulation of m6A modification has been shown to play important roles through modifying many target genes in various cancer pathogenesis [33, 34]. m6A may exhibit oncogenic or suppressive functions depending on the cellular environment [35]. As a core component of m6A “writer”, METTL14 expression levels were reported to correlate with various cancer development. In RCC, METTL14 was reported negatively correlated with RCC pathological and clinical stages, and positively correlated with OS [36]. In the present study, we found that METTL14 was significantly down-regulated in the ccRCC tissues and cells, and correlated to poor prognosis, indicating that METTL14 may possess a regulatory role in ccRCC. While METTL13 had shown no difference between the normal tissues and ccRCC.
METTL14 functioned in various cancer cell proliferation and progression. In breast cancer cells, overexpression of METTL14 promoted cell proliferation, indicating METTL14 served as an oncogene through regulation of m6A methylation [37]. In colorectal cancer, overexpression of METTL14 suppressed tumour metastasis, and METTL14-mediated N6-methyladenosine modification of SOX4 mRNA [38]. So, the effect of METTL14 could be variegated and debatable in different cancer. In the present study, we found that METTL14 overexpression inhibited the Caki-1 and Caki-2 cells proliferation and migration, while METTL14 knockdown promoted the tumour cells malignancy, indicating that METTL14 served as a tumour suppressor in ccRCC independently of METTL3. Xu et al. showed that METTL14 acted as a key gene to modulate tumour immunity, and indicated METTL14 might become a potential therapeutic target for ccRCC [39]. It is worth noting that Caki-1 cells have wild-type von Hippel-Lindau (VHL), while Caki-2 cells express a non-functional mutant VHL [40]. Based on these, METTL14 seems unlikely to be related to VHL tumour suppressor function.
The PI3K/AKT/mTOR pathway plays a significant role in cell proliferation, growth, and survival [41, 42]. A recent bioinformatics study by using TCGA database demonstrated that METTL14 pathway regulates m6A in ccRCC cells via the PI3K/AKT/mTOR and p53 signalling pathways [43]. PI3K is actives via the combination of a variety of ligands and other growth factors to the receptor tyrosine kinases [44]. Activated PI3K phosphorylates phosphatidylinositol 2 phosphoric acid (PIP2) to induce PIP3 [44, 45]. PIP3 recruits Akt and phosphoinositol-dependent protein kinase to activate Akt [44, 45]. Akt activation also depends on phosphorylation by a second kinase called mTOR, on ser473 [44, 45]. The mammalian rapamycin complex 1 (mTORC1) promotes the phosphorylation of 4EBP1 and S6K1, which are downstream targets of signal transduction and stimulate ribosome biogenesis and protein synthesis [44, 46]. Consistently, we found that METTL14 knockdown induced activation of PI3K/AKT signalling pathway, and METTL14 overexpression inhibited the PI3K/AKT signalling activation. Pten is a tumour-suppressive protein that modulates PI3K/Akt signalling pathway negatively [22, 23]. METTL14 has also been reported to associate with Pten expression in the previous study [36]. In this study, we found that METTL14 upregulation enhanced Pten expression, and knockdown of METTL14 reduced Pten expression in ccRCC cells. Moreover, Pten was downregulated in the ccRCC tissues, which showed a positive correlation with METTL14 expression. Further in vitro and in vivo studies showed that overexpression of Pten abolished the effect of METTL14 knockdown on promoting the cell proliferation, migration and tumour progression, indicating that the cell proliferation promoting effect of METTL14 knockdown was dependent on the consequence of downregulation of Pten.
m6A modification influences protein expression through change of mRNA stability, splicing, translation and microRNA maturation [13, 14]. We then performed MeRIP and found that METTL14 overexpression induced significant m6A enrichment of Pten. YTHDF1 is the reader of mRNA m6A modification, and facilitates the initiation of mRNA translation [25]. We found that YTHDF1 could directly interact with Pten mRNA. The RNA stability assay further showed that the mRNA t1/2 was prolonged by YTHDF1 overexpression, suggesting that YTHDF1 mediated Pten stability through an m6A-dependent manner.
To date, there are still some limitations in the study. It is currently unclear what mediates METTL14 downregulation in ccRCC. Besides, the regulating network of METTL14-m6A in ccRCC is complicated. The effect of METTL14 manipulation on total cellular m6A indicates that it must be affecting m6A at the majority of sites. However, the mechanisms of METTL14 whether participates in ccRCC through other genes remain unclear. In the future study, we will compare changes in PTEN-m6A, with changes in m6A on other genes. In addition, we wonder whether other readers of m6A exist in ccRCC to interact with Pten. For instance, IGF2BP2, as m6A readers regulate stability by inhibiting mRNA degradation or enhancing mRNA storage [47]. In the context of ccRCC, IGFBP2 has prognostic associations and would be of particular interest. Both the m6A-demethylases, FTO and ALKBH5, have been described as direct transcriptional targets of hypoxia-inducible factor (HIF), which in turn is directly downstream of the VHL tumour suppressor gene in ccRCC [48]. Therefore, we wonder whether erasers of m6A exist in ccRCC to interact with Pten. Furthermore, we wonder whether m6A on PTEN also affects any other properties of the transcript such as splicing, nuclear export or translation. Moreover, the therapeutic targeting of METTL14 is still in its infancy. Targeting of METTL14 for clinical application in ccRCC has not been reported, to the best of our knowledge. Continuing efforts are still needed to design and optimise strategies for targeting of METTL14 for ccRCC. With the deepening of the understanding of the role and mechanism of METL14 in ccRCC, it is promising to develop METTL14 targeted agents in future.
In conclusion, we found that METTL14 was downregulated in ccRCC, leading to the ccRCC tumour progression. The mechanism was related to the METTL14-mediated m6A modification of Pten mRNA. METTL14 could promote Pten expression through a YTHDF1-dependent mechanism. Downregulation of METTL14 restrained Pten expression in ccRCC, leading to the tumour progression through activation of the PI3K/AKT signalling pathway. Our study provides a new understanding of ccRCC development, and new insights for ccRCC therapy.
Supplementary information
Author contributions
All authors contributed to this review with conception and design, literature review, drafting and critical revision, editing and approval of the final version.
Funding
The study was supported by the National Natural Science Foundation of China (31560327 and 31560325). We would like to thank all the researchers and study participants for their contributions.
Data availability
The data used to support the findings of this study are available from the corresponding author upon request.
Ethics approval and consent to participate
The consent agreement was signed and approved by the Ethics Committee of The Affiliated Hospital of Zunyi Medical University. All patients involved in the present study didn’t receive chemotherapy or radiotherapy before the surgery.
Consent to publish
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-022-01757-y.
References
- 1.Akhtar M, Al-Bozom IA, Al Hussain, T. Molecular and metabolic basis of clear cell carcinoma of the kidney. Adv Anat Pathol. 2018;25:189–96. [DOI] [PubMed]
- 2.Cheville JC, Lohse CM, Zincke H, Weaver AL, Blute ML. Comparisons of outcome and prognostic features among histologic subtypes of renal cell carcinoma. Am J surgical Pathol. 2003;27:612–24. doi: 10.1097/00000478-200305000-00005. [DOI] [PubMed] [Google Scholar]
- 3.Lopez JI, Guarch R, Larrinaga G, Corominas-Cishek A, Orozco R. Cell heterogeneity in clear cell renal cell carcinoma. APMIS: Acta Pathologica, Microbiologica, et Immunologica Scandinavica. 2013;121:1187–91. doi: 10.1111/apm.12073. [DOI] [PubMed] [Google Scholar]
- 4.Luo Y, Shen D, Chen L, Wang G, Liu X, Qian K, et al. Identification of 9 key genes and small molecule drugs in clear cell renal cell carcinoma. Aging. 2019;11:6029–52. doi: 10.18632/aging.102161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA. 1974;71:3971–5. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5’ terminus of HeLa cell messenger RNA. Cell. 1975;4:379–86. doi: 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- 7.Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomedicine Pharmacother = Biomedecine Pharmacotherapie. 2019;112:108613. doi: 10.1016/j.biopha.2019.108613. [DOI] [PubMed] [Google Scholar]
- 8.Zhao W, Qi X, Liu L, Ma S, Liu J, Wu J, et al. Epigenetic regulation of m(6)A modifications in human cancer. Mol Ther Nucleic Acids. 2020;19:405–12. doi: 10.1016/j.omtn.2019.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell. 2018;22:191–205.e199. doi: 10.1016/j.stem.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Śledź P, Jinek M. Structural insights into the molecular mechanism of the m(6)A writer complex. eLife. 2016;5:e18434. [DOI] [PMC free article] [PubMed]
- 11.Xu K, Yang Y, Feng GH, Sun BF, Chen JQ, Li YF, et al. Mettl3-mediated m(6)A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 2017;27:1100–14. doi: 10.1038/cr.2017.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhuang M, Li X, Zhu J, Zhang J, Niu F, Liang F, et al. The m6A reader YTHDF1 regulates axon guidance through translational control of Robo3.1 expression. Nucleic Acids Res. 2019;47:4765–77. doi: 10.1093/nar/gkz157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–5. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, Fang X, Zhong P, Song Z, Hu X. N6-methyladenosine modifications: interactions with novel RNA-binding proteins and roles in signal transduction. RNA Biol. 2019;16:991–1000. [DOI] [PMC free article] [PubMed]
- 15.Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18:110. [DOI] [PMC free article] [PubMed]
- 16.Xu D, Shao W, Jiang Y, Wang X, Liu Y, Liu X, et al. FTO expression is associated with the occurrence of gastric cancer and prognosis. Oncol Rep. 2017;38:2285–92. doi: 10.3892/or.2017.5904. [DOI] [PubMed] [Google Scholar]
- 17.Nishizawa Y, Konno M, Asai A, Koseki J, Kawamoto K, Miyoshi N, et al. Oncogene c-Myc promotes epitranscriptome m(6)A reader YTHDF1 expression in colorectal cancer. Oncotarget. 2018;9:7476–86. doi: 10.18632/oncotarget.23554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, Yu K, Zhong G, Shen W. Identification of a m(6)A RNA methylation regulators-based signature for predicting the prognosis of clear cell renal carcinoma. Cancer Cell Int. 2020;20:157. [DOI] [PMC free article] [PubMed]
- 19.Zhang QJ, Luan JC, Song LB, Cong R, Ji CJ, Zhou X, et al. m6A RNA methylation regulators correlate with malignant progression and have potential predictive values in clear cell renal cell carcinoma. Exp Cell Res. 2020;392:112015. doi: 10.1016/j.yexcr.2020.112015. [DOI] [PubMed] [Google Scholar]
- 20.Polivka J, Jr., Janku F. Molecular targets for cancer therapy in the PI3K/AKT/mTOR pathway. Pharmacol Therapeutics. 2014;142:164–75. doi: 10.1016/j.pharmthera.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 21.Guo H, German P, Bai S, Barnes S, Guo W, Qi X, et al. The PI3K/AKT pathway and renal cell carcinoma. J Genet Genomics = Yi chuan xue bao. 2015;42:343–53. doi: 10.1016/j.jgg.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin X, Zhou X, Liu D, Yun L, Zhang L, Chen X, et al. MicroRNA-29 regulates high-glucose-induced apoptosis in human retinal pigment epithelial cells through PTEN. In vitro Cell Dev Biol Animal. 2016;52:419–26. doi: 10.1007/s11626-015-9990-z. [DOI] [PubMed] [Google Scholar]
- 23.Shinde SR, Maddika S. PTEN modulates EGFR late endocytic trafficking and degradation by dephosphorylating Rab7. Nat Commun. 2016;7:10689. doi: 10.1038/ncomms10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao W, Cui Y, Liu L, Ma X, Qi X, Wang Y, et al. METTL3 facilitates oral squamous cell carcinoma tumorigenesis by enhancing c-Myc stability via YTHDF1-mediated m(6)A modification. Mol Ther Nucleic Acids. 2020;20:1–12. doi: 10.1016/j.omtn.2020.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–99. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Wang T, Chen C, Wu Z, Bai P, Li S, et al. Serum exosomal miR-210 as a potential biomarker for clear cell renal cell carcinoma. J Cell Biochem. 2018; 10.1002/jcb.27347. [DOI] [PubMed]
- 27.Ghatalia P, Zibelman MR, Geynisman DM, Plimack ER. Evolving landscape of the treatment of metastatic clear cell renal cell carcinoma. Clin Adv Hematol Oncol. 2018;16:677–86. [PubMed] [Google Scholar]
- 28.Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020;69:1193–205. [DOI] [PubMed]
- 29.Yang X, Zhang S, He C, Xue P, Zhang L, He Z, et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. 2020;19:46. [DOI] [PMC free article] [PubMed]
- 30.Brown JA, Kinzig CG, DeGregorio SJ, Steitz JA. Methyltransferase-like protein 16 binds the 3’-terminal triple helix of MALAT1 long noncoding RNA. Proc Natl Acad Sci USA. 2016;113:14013–8. doi: 10.1073/pnas.1614759113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen S, Li Y, Zhi S, Ding Z, Wang W, Peng Y, et al. WTAP promotes osteosarcoma tumorigenesis by repressing HMBOX1 expression in an m(6)A-dependent manner. Cell Death Dis. 2020;11:659. doi: 10.1038/s41419-020-02847-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen X, Xu M, Xu X, Zeng K, Liu X, Pan B, et al. METTL14-mediated N6-methyladenosine modification of SOX4 mRNA inhibits tumor metastasis in colorectal cancer. Mol Cancer. 2020;19:106. [DOI] [PMC free article] [PubMed]
- 33.Deng X, Su R, Weng H, Huang H, Li Z, Chen J. RNA N6-methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018;28:507–17. doi: 10.1038/s41422-018-0034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang G, Sun Z, Zhang N. Reshaping the role of m6A modification in cancer transcriptome: a review. Cancer Cell Int. 2020;20:353. [DOI] [PMC free article] [PubMed]
- 35.Tao Z, Zhao Y, Chen X. Role of methyltransferase-like enzyme 3 and methyltransferase-like enzyme 14 in urological cancers. PeerJ. 2020;8:e9589. doi: 10.7717/peerj.9589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Q, Zhang H, Chen Q, Wan Z, Gao X. Identification of METTL14 in kidney renal clear cell carcinoma using bioinformatics analysis. Disease Markers. 2019;2019:5648783. [DOI] [PMC free article] [PubMed]
- 37.Sun T, Wu Z, Wang X, Wang Y, Hu X, Qin W, et al. LNC942 promoting METTL14-mediated m(6)A methylation in breast cancer cell proliferation and progression. Oncogene. 2020;39:5358–72. [DOI] [PubMed]
- 38.Chen X, Xu M, Xu X, Zeng K, Liu X, Pan B, et al. METTL14-mediated N6-methyladenosine modification of SOX4 mRNA inhibits tumor metastasis in colorectal cancer. Mol Cancer. 2020;19:106. doi: 10.1186/s12943-020-01220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu T, Gao S, Ruan H, Liu J, Liu Y, Liu D, et al. METTL14 acts as a potential regulator of tumor immune and progression in clear cell renal cell carcinoma. Front Genet. 2021;12:609174. doi: 10.3389/fgene.2021.609174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pasha M, Sivaraman SK, Frantz R, Agouni A, Munusamy S. Metformin induces different responses in clear cell renal cell carcinoma caki cell lines. Biomolecules. 2019;9:113. [DOI] [PMC free article] [PubMed]
- 41.O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–8. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–30. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- 43.Zhou J, Wang J, Hong B, Ma K, Xie H, Li L, et al. Gene signatures and prognostic values of m6A regulators in clear cell renal cell carcinoma - a retrospective study using TCGA database. Aging. 2019;11:1633–47. doi: 10.18632/aging.101856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aoki M, Fujishita T. Oncogenic roles of the PI3K/AKT/mTOR axis. Curr Top Microbiol Immunol. 2017;407:153–89. doi: 10.1007/82_2017_6. [DOI] [PubMed] [Google Scholar]
- 45.Jhanwar-Uniyal M, Wainwright JV, Mohan AL, Tobias ME, Murali R, Gandhi CD, et al. Diverse signaling mechanisms of mTOR complexes: mTORC1 and mTORC2 in forming a formidable relationship. Adv Biol Regul. 2019;72:51–62. doi: 10.1016/j.jbior.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 46.Ma L, Zhang R, Li D, Qiao T, Guo X. Fluoride regulates chondrocyte proliferation and autophagy via PI3K/AKT/mTOR signaling pathway. Chem-Biol Interact. 2021;349:109659. doi: 10.1016/j.cbi.2021.109659. [DOI] [PubMed] [Google Scholar]
- 47.Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–95. [DOI] [PMC free article] [PubMed]
- 48.Xiao Y, Thakkar KN. The m(6)A RNA demethylase FTO is a HIF-independent synthetic lethal partner with the VHL tumor suppressor. Proc Natl Acad Sci USA. 2020;117:21441–9. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.