Abstract
We examined the pathway by which the fungicide biphenyl is metabolized in the imperfect fungus Paecilomyces lilacinus. The initial oxidation yielded the three monohydroxylated biphenyls. Further hydroxylation occurred on the first and the second aromatic ring systems, resulting in the formation of five di- and trihydroxylated metabolites. The fungus could cleave the aromatic structures, resulting in the transformation of biphenyl via ortho-substituted dihydroxybiphenyl to six-ring fission products. All compounds were characterized by gas chromatography-mass spectroscopy and proton nuclear magnetic resonance spectroscopy. These compounds include 2-hydroxy-4-phenylmuconic acid and 2-hydroxy-4-(4′-hydroxyphenyl)-muconic acid, which were produced from 3,4-dihydroxybiphenyl and further transformed to the corresponding lactones 4-phenyl-2-pyrone-6-carboxylic acid and 4-(4′-hydroxyphenyl)-2-pyrone-6-carboxylic acid, which accumulated in large amounts. Two additional ring cleavage products were identified as (5-oxo-3-phenyl-2,5-dihydrofuran-2-yl)-acetic acid and [5-oxo-3-(4′-hydroxyphenyl)-2,5-dihydrofuran-2-yl]-acetic acid. We found that P. lilacinus has a high transformation capacity for biphenyl, which could explain this organism's tolerance to this fungicide.
Biphenyl and 2- and 4-hydroxybiphenyl have been used to limit fungal growth on citrus fruit. Toxic effects to humans after prolonged exposure to air containing biphenyl and after cutaneous application are known (14). Some biphenyl derivatives also are suspected carcinogens and have estrogenic activity (21, 33, 39).
Most research on the degradation of biphenyls has been conducted with bacteria (3, 11, 24, 36). Fungal transformation of biphenyl has been studied to determine how this antifungal substance may be inactivated but, more importantly, as a model of mammalian biphenyl metabolism. Primary oxidation of biphenyl by yeasts of the genera Candida and Debaryomyces (5, 22, 34, 44) and by filamentous fungi of the genera Absidia, Aspergillus, Cunninghamella, Gliocladium, and Helicostylum (7, 9, 12, 37), yields monohydroxylated biphenyls, which can undergo a second hydroxylation step or sugar conjugate formation (5, 7, 9, 12, 34, 37, 44). Usually, the initial hydroxylation favors the 4-position of biphenyl (34, 37). Induction of biphenyl hydroxylase activity in Aspergillus by several biphenyl substrates and also by 4,4′-dihydroxybiphenyl has been reported (28).
Although fungi constitute the majority of microbial biomass in soil, reports on fungal metabolism of biphenyl are limited to a few yeasts and filamentous fungi and usually restricted to hydroxylation processes (9, 34, 37). Since 1993 there have been two reports suggesting that these organisms may transform hydroxylated biphenyls and cleave the aromatic rings (22, 27). Each strain appears to be able to form only a single ring cleavage product, however, and the overall capacity for ring cleavage seemed low.
Our objective in this study was to determine how the filamentous fungus Paecilomyces lilacinus transforms the fungicide biphenyl. The ability of this fungus to form a variety of oxidation products including ring cleavage products not previously described in fungi may be one reason that this fungus appears resistant to high levels of biphenyl.
MATERIALS AND METHODS
Chemicals.
Biphenyl and 2-hydroxybiphenyl were purchased from Merck (Darmstadt, Germany), and 3-hydroxy-, 4-hydroxy-, and 2,5-dihydroxybiphenyl as well as 4,4′-dihydroxybiphenyl were purchased from Aldrich (Steinheim, Germany). 3,4-Dihydroxybiphenyl was obtained from Promochem (Wesel, Germany), and 2,3-dihydroxybiphenyl was obtained from Wako Pure Chemical Industries (Neuss, Germany). All chemicals and solvents were of the highest purity available.
The compounds 2,3,4-trihydroxybiphenyl, 3,4,5-trihydroxybiphenyl, 3,4,4′-trihydroxybiphenyl, 4-phenyl-2-pyrone-6-carboxylic acid, and 2-hydroxy-4-phenylmuconic acid were not commercially available and were synthesized as follows.
2,3,4-Trihydroxybiphenyl was synthesized by bromination of 1,2,3-trimethoxybenzene (17). The Grignard reagent formed from the resulting 1-bromo-2,3,4-trimethoxy benzene was coupled with cyclohexanone. The reaction products were dehydrated with oxalic acid and treated with p-chloranil to produce 2,3,4-trimethoxybiphenyl. Demethylation with AlCl3 yielded the corresponding 2,3,4-trihydroxybiphenyl (2, 23).
3,4,5-Trihydroxybiphenyl was synthesized by Ullmann coupling of iodobenzene with 5-iodo-1,2,3-trimethoxybenzene (10). Separation of the 3,4,5-trimethoxybiphenyl from the mixture of biphenyls followed by demethylation with AlI3 yielded the target product 3,4,5-trihydroxybiphenyl (1).
3,4,4′-trihydroxybiphenyl was synthesized by coupling of veratrole with N′-(4-nitrophenyl)-diazenium tetrafluoroborate and transformation of the nitro group of the resulting compound into the corresponding amino derivative by hydrogenation with a PtO2 catalyst. The p-substituted aniline was transformed to the corresponding phenol via the diazenium salt following conventional methods (29). The 4′-hydroxy-3,4-dimethoxybiphenyl obtained was demethylated with pyridine hydrochloride to trihydroxylated biphenyl (16, 29).
4-Phenyl-2-pyrone-6-carboxylic acid was synthesized starting from trans-β-methyl cinnamic acid chloride as outlined by Rey et al. (32).
2-Hydroxy-4-phenylmuconic acid was produced photochemically from 4-phenyl-2-pyrone-6-carboxylic acid. Photochemical treatment of 2-pyrones in methanol results in the formation of bicyclic lactones and/or ketenes, which add methanol under the reaction conditions (19). Thus, treatment of 4-phenyl-2-pyrone-6-carboxylic acid at 300 nm and 15°C yielded 2-hydroxy-4-phenylmuconic acid in small amounts.
Growth and incubation conditions.
P. lilacinus SBUG-M 1093 was isolated from wood chip piles (31) and has been deposited in the strain collection of the Department Biology of Greifswald University (SBUG) (DSMZ accession no. 14052). The strain was maintained on malt agar slants (3% malt; Serva). For incubation experiments, mycelium from an agar plate (1 cm2) was transferred to a 500-ml Erlenmeyer flask with 100 ml of Sabouraud glucose broth containing 40 g of d-glucose and 10 g of peptone of casein (Merck) per liter. After growth for 72 h at 30°C and 180 rpm on a rotary shaker, 5 ml of homogenized mycelium (homogenized three times for 2 s each time at 3,000 U/min in an IKA Ultra-Turrax homogenizer [Janke & Kunkel, Stauffen, Germany]) was used to inoculate 500-ml shake flasks containing 100 ml of a mineral salts medium (MM) (20) and 1 g of d-glucose as a carbon source. The slurry was cultivated as before for another 72 h. Glucose-grown cells were harvested by centrifugation (at 6000 × g for 5 min) and washed twice with sterile MM. The pellet was resuspended in 100 ml of sterilized MM. As substrates, biphenyl, 2-, 3-, or 4-hydroxybiphenyl, 3,4- or 4,4′-dihydroxybiphenyl, 3,4,4′- or 3,4,5-trihydroxybiphenyl, and 4-phenyl-2-pyrone-6-carboxylic acid were added at concentrations ranging from 0.05 or 0.1 to 1 mg/ml. Cells in MM without substrates and biphenyl derivatives in MM were used as controls. All cultures were tested for microbiological purity.
Chemical analysis and identification of intermediates.
For the detection and quantification of metabolites in the aqueous culture supernatant, high-performance liquid chromatography (HPLC) was used. RP (reversed-phase) analytical HPLC was performed on a Hewlett-Packard (Bad Homburg, Germany) 1050 M HPLC apparatus equipped with a quaternary pump system, a 1040 M series 1 diode array detector, and a Hewlett-Packard Chemstation. Gradient elution mode was used for separation in a mobile phase consisting of methanol and 0.1% (wt/vol) (10.2 mM) phosphoric acid starting from an initial ratio of 20% methanol and reaching 100% methanol in 14 min. The flow rate was 1 ml/min; detection wavelengths were 220 nm (UV) and 254 and 280 nm.
Data are reported as means for two separate experiments with replicated batch cultures. Standard deviation was no more than 7%.
Purification of intermediates was carried out on a Merck-Hitachi HPLC (Merck) system equipped with a model L 6200 A Intelligent Pump, a Rheodyne 7161 injection valve with a 100-μl loop, and a model L-4250 absorbance detector operating at 254 nm. Gradient elution mode was used for separation in a mobile phase consisting of methanol and 0.1% (wt/vol) (16.7 mM) acetic acid starting from an initial ratio of 20% methanol and reaching 60% methanol in 18.5 min, at a flow rate of 1 ml/min. For both HPLC systems a LiChroCart 125-4 RP-18 end-capped (5-μm) column (Merck) was used.
For gas chromatographic analysis, whole cultures were lyophilized using an Alpha 1–4 apparatus (Christ, Osterode, Germany). The dried samples were resolved in 10 ml of methanol, centrifuged (at 5,000 × g for 5 min), and evaporated under a vacuum at 40°C. The methanol extracts were examined by gas chromatography-mass spectroscopy (GC-MS). The GC-MS system consisted of a GC 8000 gas chromatograph (Fisons, Mainz, Germany) and a Fisons MD 800 mass spectrometer operating at 70 eV. Helium was used as the carrier gas at a flow rate of 1.9 ml/min. Separation was carried out on a 30-m BPX 5 column (0.25-mm by, 0.33-μm film; SGE, Weiterstadt, Germany) at a temperature program from 80 to 300°C (10°C/min). High-resolution mass spectra were recorded on a Vacuum Generators (Manchester, United Kingdom) analytical instrument, model 70-250 SE.
Derivatization was achieved by methylation with diazomethane as described by De Boer and Backer (8) in a microapparatus (Z 10,100-1; Aldrich-Chemie).
Proton nuclear magnetic resonance (1H NMR) spectroscopy was carried out on a Bruker (Karlsruhe, Germany) ARX 300-MHz spectrometer or on a Bruker DRX500 instrument at 500 MHz. Deuterated methanol (99.99%) was used as a solvent and tetramethyl silane was used as a reference.
RESULTS
We identified 14 metabolites formed by P. lilacinus SBUG-M 1093 from biphenyl and selected biphenyl derivatives in aqueous supernatants or in methanol extracts obtained from lyophilized whole cultures (Table 1). After extraction of supernatants with ethyl acetate, the same pattern of metabolites was found. Seven mono- and dihydroxylated biphenyls were identified by comparison of UV and mass spectra with those of authentic standards. For some additional metabolites (products A through G), standards were not available. These products were characterized by GC-MS, by 1H NMR, and (partially) by comparison with literature data (products B and G).
TABLE 1.
Formation of hydroxylated biphenyls and ring cleavage products after incubation of P. lilacinus SBUG-M 1093 with biphenyl, several hydroxylated biphenyls, and a synthesized ring cleavage product
| Producta | Substrate
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| BP | 2-OHBP | 3-OHBP | 4-OHBP | 4,4′-DiOHBP | 3,4-DiOHBP | 3,4,5-TriOHBP | 3,4,4′-TriOHBP | PPCA | |
| Hydroxylated biphenyls | |||||||||
| 2-OHBP | + | ||||||||
| 3-OHBP | + | ||||||||
| 4-OHBP | + | ||||||||
| 4,4′-DiOHBP | + | + | |||||||
| 3,4-DiOHBP | + | + | |||||||
| 2,3-DiOHBP | + | ||||||||
| 2,5-DiOHBP | + | ||||||||
| 3,4,4′-TriOHBP | + | + | + | + | + | ||||
| Ring cleaved products | |||||||||
| PPCA | + | + | + | + | + | ||||
| C | + | + | + | + | + | + | + | + | |
| D | + | ||||||||
| E | + | ||||||||
| F | + | + | + | ||||||
| G | + | ||||||||
BP, biphenyl; OHBP, hydroxybiphenyl; DiOHBP, dihydroxybiphenyl; TriOHBP, trihydroxybiphenyl; PPCA, 4-phenyl-2-pyrone-6-carboxylic acid; C, 4-(4′-hydroxyphenyl)-2-pyrone-6-carboxylic acid; D, 2-hydroxy-4-phenylmuconic acid; E, 2-hydroxy-4-(4′-hydroxyphenyl)-muconic acid; F, [5-oxo-3-(4′-hydroxyphenyl)-2,5-dihydrofuran-2-yl]-acetic acid; G, (5-oxo-3-phenyl-2,5-dihydrofuran-2-yl)-acetic acid.
Formation of hydroxylated biphenyls.
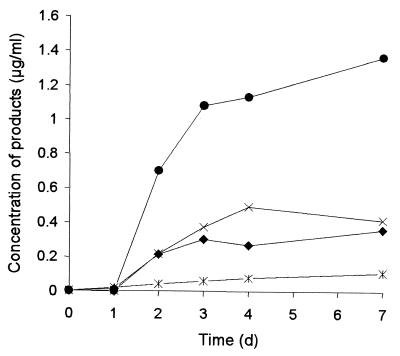
P. lilacinus strain SBUG-M 1093 can tolerate biphenyl at concentrations up to 1 mg/ml, accumulating 4-hydroxybiphenyl and 4,4′-dihydroxybiphenyl. During incubations with a lower concentration of biphenyl (0.1 mg/ml), 4,4′-dihydroxybiphenyl was formed as the major metabolite (Fig. 1). 2- and 4-Hydroxybiphenyl and an additional product (product A) were detected in smaller amounts by HPLC. After 24 to 96 h of incubation, concentrations of 4-hydroxybiphenyl and 4,4′-dihydroxybiphenyl increased. In contrast, the concentration of 2-hydroxybiphenyl increased slowly and at a constant rate (Fig. 1).
FIG. 1.
Time-dependent formation of hydroxylated products from biphenyl by glucose-grown cells of P. lilacinus SBUG-M 1093 during incubation with biphenyl (0.1 mg/ml). Symbols: ✻, 2-hydroxybiphenyl; ⧫, 4-hydroxybiphenyl; ●,4,4′-dihydroxybiphenyl; ×, product A.
Initial oxidation of biphenyl leading to 4-hydroxybiphenyl, 2-hydroxybiphenyl, and traces of 3-hydroxybiphenyl was monitored by HPLC (for 4-hydroxybiphenyl, λmax = 200, 262 nm; for 2-hydroxybiphenyl; λmax = 248, 276 nm; for 3-hydroxybiphenyl, λmax = 206, 252, 290 nm) and GC-MS analysis [m/z 170 (C12H10O+, M+), 141 (C11H9+, M+-CHO), 139 (C11H7+, M+-CH3O), 115 (C9H7+)] in comparison with synthetic standards. Mass spectral data [m/z 186 (C12H10O2+, M+) 157 (C11H9O+, M+-CHO), 128 (C10H8+, M+-2CHO), 115 (C9H7+), 77 (C6H5+)] of the main product after incubation with biphenyl (0.1 mg/ml [Fig. 1]) were consistent with those for an authentic standard of 4,4′-dihydroxybiphenyl.
When P. lilacinus SBUG-M 1093 was incubated with 2-hydroxybiphenyl, 3-hydroxybiphenyl, and 4-hydroxybiphenyl, the cultures accumulated dihydroxylated derivatives of biphenyl, recognized by the fragmentation pattern in their mass spectra after GC separation [m/z 186 (C12H10O2+, M+), 157 (C11H9O+, M+-CHO), 128 (C10H8+, M+-C2H2O2), 115 (C9H7+, M+-C3H3O2), 77 (C6H5+, M+-C6H5O2)]. Incubation of 2-hydroxybiphenyl led to the formation of 2,3- and 2,5-dihydroxybiphenyl; 3-hydroxybiphenyl was transformed to 3,4-dihydroxybiphenyl; and 4-hydroxybiphenyl was transformed to 4,4′-dihydroxybiphenyl. Structural determinations were made following comparison of Rf-values (HPLC), UV spectra, and GC-MS retention times with the data of authentic standards.
GC-MS analyses of a methylated methanol extract of the lyophilized culture supernatant suggest the presence of an additional hydroxylated product (named product A) in cultures incubated with biphenyl (Fig. 1), 3,4-dihydroxybiphenyl (Fig. 2), and 4,4′-dihydroxybiphenyl. The molecular ion peak of the methyl derivative of this compound at m/z 244 and those of the main fragments at m/z 229 (C14H13O3+, M+-CH3), 201 (C13H13O2+, M+-COCH3), and 115 (C9H7+) were consistent with data for trihydroxylated biphenyl. In accordance with this assumption, high-resolution mass spectrometry showed the molecular formula to be C15H16O3 (mass found, 244.109138; mass calculated, 244.109945). By comparing the GC-MS data with those of the synthesized 2,3,4-, 3,4,5-, and 3,4,4′-trihydroxybiphenyls and by coinjection of the methylated standards, product A was identified as 3,4,4′-trihydroxybiphenyl.
FIG. 2.
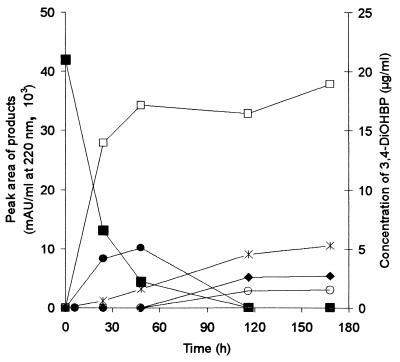
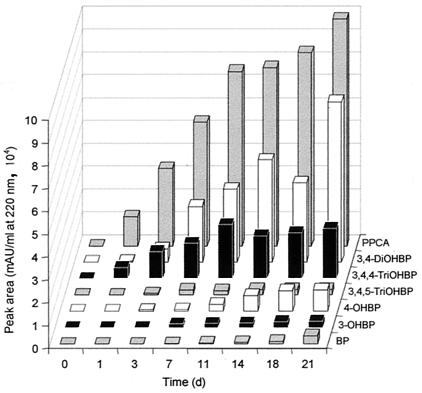
Formation of products by glucose-grown cells of P. lilacinus SBUG-M 1093 during incubation with 3,4-dihydroxybiphenyl (0.05 mg/ml). Symbols: •, product A; ✻, product B; □, product C; ⧫, product F; ○, product G; ■,3,4-dihydroxybiphenyl (3,4-DiOHBP).
Formation of ring cleavage products.
In addition to the hydroxylated biphenyl derivatives, further products (products B and C) were enriched in the supernatant after incubation with biphenyl (0.1 mg/ml). When fungal cells were incubated with hydroxylated intermediates (Table 1), it became clear that products B and C were formed from 3- and 4-hydroxybiphenyl as well as from 3,4-dihydroxybiphenyl. When 3,4-dihydroxybiphenyl was used as the substrate, additional metabolites (products D through G) were detected by HPLC (Fig. 3) or GC-MS analyses. Metabolites B, C, F, and G have HPLC retention values that suggested these compounds were acids. Consequently, methylation of the extracted compounds was necessary for GC-MS analysis.
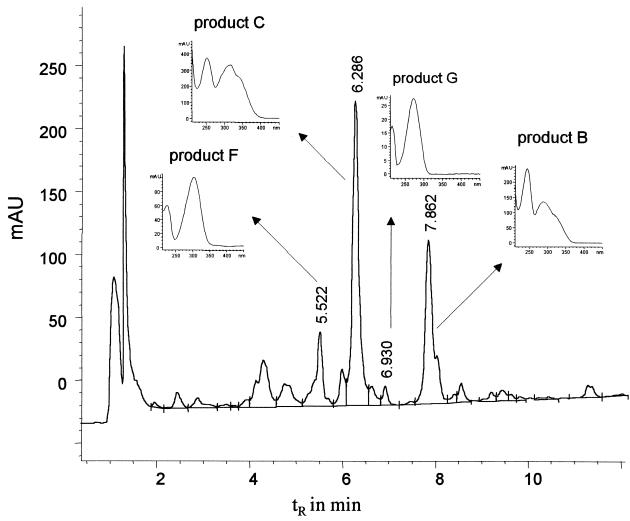
FIG. 3.
HPLC elution profile of the aqueous supernatant after incubation of P. lilacinus SBUG-M 1093 with 3,4-dihydroxybiphenyl (tR = 8.57 min; 0.1 mg/ml; 7 days) and UV absorption spectra of products B (tR = 7.86 min), C (tR = 6.28 min), F (tR = 5.52 min), and G (tR = 6.93 min).
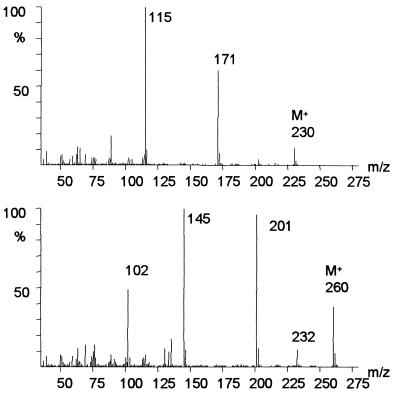
Metabolite B was identified as 4-phenyl-2-pyrone-6-carboxylic acid by GC-MS and 1H NMR analysis. The methyl derivative of this metabolite showed a molecular ion peak at m/z 230 (C13H10O4) and main fragment ions at m/z 202 (C12H10O3+, M+-CO), 171 (C11H7O2+, M+-COOCH3), and 115 (C9H7+, M+-COOCH3-2CO) in GC-MS. The mass spectrum (Fig. 4) and 1H NMR data [6.83 (d, J3,5 = 1.7 Hz, 1H, 5-H), 7.54 (m, J2′4′,4′6′ = 2.1, J3′4′,4′5′ = 8.6 Hz, 1H, 4′-H) 7.55 (m, J3′4′,4′5′ = 8.6 Hz, 2H, 3′, 5′-H), 7.57 (d, J3,5 = 1.7 Hz, 1H, 3-H), 7.78 (dd, J2′4′,4′6′ = 2.1, 2H, 2′, 6′-H) ppm] were consistent with those of a sample of 4-phenyl-2-pyrone-6-carboxylic acid, synthesized for reference.
FIG. 4.
Mass spectra of methylated products B (4-phenyl-2-pyrone-6-carboxylic acid) (top) and C [4-(4′-hydroxyphenyl)-2-pyrone-6-carboxylic acid] (bottom).
Metabolite C, the main product resulting from transformation of 3,4-dihydroxybiphenyl (Fig. 2), also was accumulated during incubation of P. lilacinus with 4,4′-dihydroxybiphenyl, 3,4,5-trihydroxybiphenyl, 3,4,4′-trihydroxybiphenyl, and 4-phenyl-2-pyrone-6-carboxylic acid (Fig. 5). On the basis of the MS and 1H NMR data, the formation of this compound from different p,p′-hydroxylated biphenyls, and its direct formation by hydroxylation of 4-phenyl-2-pyrone-6-carboxylic acid (Table 1), the structure of metabolite C was determined to be 4-(4′-hydroxyphenyl)-2-pyrone-6-carboxylic acid. The mass spectral data of the methylated compound (Fig. 4) showed a molecular ion at m/z 260 and fragment ions at m/z 232 (C13H12O4, M+-CO), 201 (C12H9O3, M+-COOCH3), and 145 (C10H9O, M+-COOCH3-2CO). High-resolution mass spectrometry indicated that the molecular formula of product C was C14H12O5 after methylation. The 1H NMR data (500 MHz) obtained after separation of the lyophilized extract by preparative HPLC were as follows: δ 6.70 (s, 1H, 5-H), 6.95 (d, 2H, 3′,5′-H), 7.57 (s, 1H, 3-H), 7.70 (d, 2H, 2′,6′-H) ppm. The 1H NMR spectrum showed two signals with single intensity and two signals with double intensity, pointing to six protons. The doublet signals at δ = 6.95 and 7.70 ppm with a coupling constant of J2′3′;5′6′ = 8.7 Hz indicate a phenyl ring with a hydroxyl group in the para position due to two pairs of protons in ortho positions of the aromatic ring.
FIG. 5.
Formation of product C [4-(4′-hydroxyphenyl)-2-pyrone-6-carboxylic acid] after incubation of glucose-grown cells of P. lilacinus SBUG-M 1093 with different biphenyl derivatives (BP, biphenyl; OHBP, hydroxybiphenyl; DiOHBP, dihydroxybiphenyl; TriOHBP, trihydroxybiphenyl; PPCA, 4-phenyl-2-pyrone-6-carboxylic acid).
Metabolites D and E, formed from 3,4-dihydroxybiphenyl, were detected only in the methanol extract by GC-MS. Product D was identified as 2-hydroxy-4-phenylmuconic acid as follows. According to high-resolution mass spectrometry analysis, the molecular ion peak m/z 276 of the methylated intermediate D had the molecular formula C15H16O5 (mass found, 276.101303; mass calculated, 276.099774). The base peak at m/z 217, assigned to C13H13O3, resulted in the loss of COOCH3 from the molecular ion. Further fragment ions at m/z 261 (C14H13O5+, M+-CH3), 245 (C14H13O4+, M+-OCH3), 202 (C12H10O3+, M+-COOCH3-CH3), 185 (C12H9O2+, M+-COOCH3-CH4O), and 59 (COOCH3+) correspond with a structure containing one or two methyl esters and one methoxy function, indicating that intermediate D is a hydroxylated (di)carboxylic acid. This compound was identical to a sample of 2-hydroxy-4-phenylmuconic acid, derived from the photochemical treatment of 4-phenyl-2-pyrone-6-carboxylic acid, by GC-MS analysis. This structural assignment was further corroborated by the fact that products B and D can be chemically interconverted.
Product E, formed from 3,4-dihydroxybiphenyl and 4-phenyl-2-pyrone-6-carboxylic acid (Table 1), was identified as 2-hydroxy-4-(4′-hydroxyphenyl)-muconic acid. After methylation, the mass spectrum of this compound showed a molecular ion peak at m/z 306 and the main fragment ions at m/z 275 (C15H15O5+, M+-OCH3), 247 (C14H15O4+, M+-COOH), 232 (C13H12O4+, M+-COOCH3-CH3), 145 (C10H9O+), and 59 (COOCH3+). The similarities in the fragmentation pattern to that of metabolite D and the difference of m/z 30 between the distinctive fragments of both metabolites suggests, a doubly hydroxylated muconic acid with one hydroxy group at the para position of the aromatic ring system.
Metabolite F, formed after incubation with 3,4-dihydroxybiphenyl (Fig. 2 and 3), 4,4′-dihydroxybiphenyl, and 3,4,4′-trihydroxybiphenyl, was separated by HPLC and identified by GC-MS as well as by 1H NMR as [5-oxo-3-(4′-hydroxyphenyl)-2,5-dihydrofuran-2-yl]-acetic acid. The molecular ion peak of the methylated compound and the fragment ions were detected at m/z 262 (C14H14O5, M+), 234 (C13H14O4+, M+-CO), 189 (C11H9O3+, M+-CH2COOCH3), 161 (C10H9O2+, M+-CH2COOCH3-CO), 133 (C9H9O+, M+-CH2COOCH3-2CO), and 132 (C9H8O+). The 1H NMR data were as follows: δ 2.38 (dd, J1,2 = 16.4 Hz, J1, 3 = 9.5 Hz, 1H, 1-H), 2.96 (dd, J1,2 = 16.4 Hz, J2,3 = 2.7 Hz, 1H, 2-H), 5.95 (ddd, J1,3 = 9.3 Hz, J2,3 = 2.7 Hz, J3,4 = 1.4 Hz, 1H, 3-H), 6.27 (d, J3,4 = 1.4 Hz, 1H, 4-H), 6.92 (m, 2H, 2′,6-H), 7.52 (m, 2H, 3′5′-H) ppm.
Metabolite G resulted from the degradation of 3,4-dihydroxybiphenyl (Fig. 2 and 3) to (3-phenyl-5-oxo-2,5-dihydrofuran-2-yl)-acetic acid. The mass spectrum of the methylated compound showed the molecular ion peak at m/z 232 (C13H12O4) and the main fragment ions at m/z 204 (C12H12O3+, M+-CO), 159 (C10H7O2+, M+-CH2COOCH3), 131 (C9H7O+, M+-CH2COOCH3-CO), 103 (C8H7+, M+-CH2COOCH3-2CO), and 102 (C8H6+). Four signals in the 1H NMR spectrum [δ 2.38 (dd, J1,2 = 16.2 Hz, J1,3 = 9.3 Hz, 1H, 1-H), 2.92 (dd, J1,2 = 16.2 Hz, J2,3 = 2.8 Hz, 1H, 2-H), 6.04 (ddd, J1,3 = 9.3 Hz, J2,3 = 2.8 Hz, J3,4 = 1.5 Hz, 1H, 3-H), 6.45 (d, J3,4 = 1,5 Hz, 1H, 4-H) ppm] showed a system of nonaromatic protons similar to that concluded from the 1H NMR spectrum of compound F. In contrast, the aromatic proton signals at δ 7.49 (d, 1H, 4′-H), 7.51 (m, 2H, 2′,6-H), and 7.65 (m, 2H, 3′5′-H) ppm indicated a monosubstituted and hence unhydroxylated aromatic ring system. HPLC, GC-MS, and 1H NMR spectra, corresponded to a metabolite isolated and characterized after incubation of Trichosporon mucoides SBUG-M 801, a yeast that accumulates large amounts of intermediate G after incubation with 3,4-dihydroxybiphenyl (R. Sietmann et al., unpublished data).
DISCUSSION
P. lilacinus SBUG-M 1093 can metabolize biphenyl to monohydroxylated biphenyls (2-, 3-, and 4-hydroxybiphenyl), suggesting the action of monooxygenases during the initial oxidation step. Similar initial oxidation during transformation of biphenyl is known for yeasts of the genera Candida (5) and Debaryomyces (22) and for genera of the filamentous fungi Absidia, Aspergillus, Cunninghamella, Gliocladium, and Helicostylum (9, 12, 37). Secondary hydroxylation occurs on the first hydroxylated aromatic ring to produce 2,3-, 2,5-, and 3,4-dihydroxybiphenyl (4, 22). Furthermore, Mobley et al. (27) have reported the formation of 3,4-catechols of substituted biphenyls. As reported for several fungi, P. lilacinus favors the initial hydroxylation in the 4-position, yielding 4-hydroxybiphenyl, and the second hydroxylation in the 4′-positions, yielding 4,4′-dihydroxybiphenyl. This fungus also can perform a third hydroxylation of biphenyl to produce 3,4,4′-trihydroxybiphenyl. These results suggest that primary oxidation steps leading to monohydroxylated and dihydroxylated biphenyls and to 3,4,4′-trihydroxybiphenyl are similar to the metabolic conversion of biphenyl in mammals (25, 26, 38).
We detected two new ring fission products, 2-hydroxy-4-phenylmuconic acid and 2-hydroxy-4-(4′-hydroxyphenyl)muconic acid, as intermediates during degradation of 3,4-dihydroxybiphenyl by P. lilacinus. Formation of similar intermediates with a 2-hydroxy muconic acid structure was reported for degradation of phloroglucinol by Fusarium solani (43), of gallic acid by Aspergillus flavus (13), of dibenzofuran by T. mucoides (15), and of diphenyl ether by Trametes versicolor (18).
Presumably ring cleavage of the aromatic structure can take place after introduction of a third hydroxyl group into the dihydroxylated aromatic ring (22). After incubation with 3,4,5-trihydroxybiphenyl, only the lactone structure 4-phenyl-2-pyrone-6-carboxylic acid was detected. The formation of 2-hydroxy-4-phenylmuconic acid is possible but not detectable after this incubation.
After incubation with 3,4-dihydroxybiphenyl, lactonic structures were prominent products (Fig. 2). However, 4-(4′-hydroxyphenyl)-2-pyrone-6-carboxylic acid also was formed as a result of further hydroxylation of 4-phenyl-2-pyrone-6-carboxylic acid. The enrichment of further lactonic structures, (5-oxo-3-phenyl-2,5-dihydrofuran-2-yl)acetic acid and [5-oxo-3-(4′-hydroxyphenyl)-2,5-dihydrofuran-2-yl]acetic acid, was demonstrated. The latter product also was found after incubation of Aspergillus parasiticus with biphenyl and 4,4′-dihydroxybiphenyl (27). In this case the authors suggested that each biphenyl ring must be para-hydroxylated as a prerequisite for lactone formation. In contrast, in P. lilacinus a muconolactone structure, (5-oxo-3-phenyl-2,5-dihydrofuran-2-yl)acetic acid, was formed from 3,4-dihydroxybiphenyl after hydroxylation of one of the aromatic ring systems followed by ring cleavage. Lactonization of substituted and unsubstituted muconic acids yielding corresponding muconolactone structures was reported by nonenzymatic isomerization under acidic conditions (30) as well as by the action of muconate cycloisomerases from various bacteria (40, 41, 42).
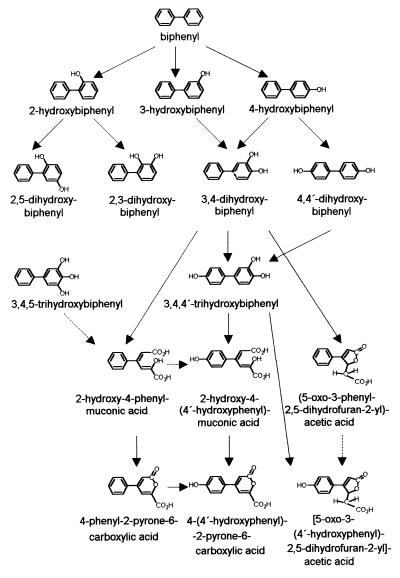
The accumulation of 4-(4′-hydroxyphenyl)-2-pyrone-6-carboxylic acid and [5-oxo-3-(4′-hydroxyphenyl)-2,5-dihydrofuran-2-yl]acetic acid and the formation of 2-hydroxy-4-(4′-hydroxyphenyl)muconic acid suggest that P. lilacinus can further transform ring cleavage products B, D, and G. On the other hand, the trihydroxylated intermediate (3,4,4′-trihydroxybiphenyl) was cleaved by this filamentous fungus, leading directly to the hydroxylated lactone structures (Fig. 6).
FIG. 6.
Proposed pathway for the initial steps of biphenyl degradation in P. lilacinus SBUG-M 1093. →, transformation of P. lilacinus; ––→ proposed transformation step, but intermediate not observed.
The accumulation of hydroxyphenyl lactones suggests that they are deadend products in the pathway of the metabolism of biphenyl and substituted biphenyls. Comparison of the toxicity of hydroxylated biphenyls and the lactonic structures formed, using the method of Singer-Bohne et al. (35), pointed to detoxification of hydroxylated biphenyls by ring cleavage accompanied by lactone formation.
The results show that P. lilacinus oxidizes biphenyl to a broad variety of products that were not previously known from filamentous fungi. Six different ring cleavage products were identified. Their structures indicate a ring fission mechanism going via di- and trihydroxylated intermediates to muconic acid structures which can lactonize. Since the same ring fission mechanism had been described for Aspergillus (27) and Debaryomyces (22)—genera that are not related to Paecilomyces—the strain-specific ability of this feature seems to be widely distributed among different fungal species. The ability to produce di- or trihydroxylated biphenyls which can undergo ring fission to nontoxic products may be the reason for the resistance of several filamentous fungi to the fungicide biphenyl.
ACKNOWLEDGMENTS
This study was supported by the Deutsche Bundesstiftung Umwelt.
We thank M. Kindermann and S. Siegert (Institute of Chemistry and Biochemistry, University of Greifswald) for recording NMR data, S. Franke (Institute of Organic Chemistry, University of Hamburg) for acquiring high-resolution mass spectrometry data, and R. Jack (Institute of Immunology, University of Greifswald) for revising the manuscript.
REFERENCES
- 1.Andersson S. An improvement of the aluminium iodide method for ether cleavage: catalysis by quaternary ammonium iodides. Synthesis. 1985;4:437–439. [Google Scholar]
- 2.Bruce J M, Sutcliffe F K. Synthetic and oxidative studies in the polyhydroxydiphenyl series. Part I. J Chem Soc. 1955;2:4435–4440. [Google Scholar]
- 3.Catelani D, Colombi A, Sorlini C, Treccani V. Metabolism of biphenyl. Biochem J. 1973;134:1063–1066. doi: 10.1042/bj1341063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cerniglia C E. Aromatic hydrocarbons. Metabolism by bacteria, fungi and algae. Rev Biochem Toxicol. 1980;3:321–361. [Google Scholar]
- 5.Cerniglia C E, Crow S A. Metabolism of aromatic hydrocarbons by yeasts. Arch Microbiol. 1981;129:9–13. [Google Scholar]
- 6.Coutrot P, Snoussi M, Savignac P. An improvement in the Wittig-Horner synthesis of 2-alkenoic acids. Synthesis. 1978;1978:33–134. [Google Scholar]
- 7.Cox J C, Golbeck J H. Hydroxylation of biphenyl by Aspergillus parasiticus: approaches to yield improvement in fermentor cultures. Biotechnol Bioeng. 1985;27:1395–1402. doi: 10.1002/bit.260271002. [DOI] [PubMed] [Google Scholar]
- 8.De Boer T D, Backer H J. Diazomethane. Org Synth. 1956;36:14–16. [Google Scholar]
- 9.Dodge R H, Cerniglia C E, Gibson D T. Fungal metabolism of biphenyl. Biochem J. 1979;178:223–230. doi: 10.1042/bj1780223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erdtman H, Eriksson G, Norin T, Forsen S. Aucuparin and methoxyaucuparin, two phenolic biphenyl derivatives from the heartwood of Sorbus aucuparia (L.) Acta Chem Scand. 1963;17:1151–1156. [Google Scholar]
- 11.Gibson D T, Roberts R L, Wells M C, Kobal V M. Oxidation of biphenyl by a Beijerinckia species. Biochem Biophys Res Commun. 1973;50:211–219. doi: 10.1016/0006-291x(73)90828-0. [DOI] [PubMed] [Google Scholar]
- 12.Golbeck J H, Albaugh S A, Radmer R. Metabolism of biphenyl by Aspergillus toxicarius: induction of hydroxylating activity and accumulation of water-soluble conjugates. J Bacteriol. 1983;156:49–57. doi: 10.1128/jb.156.1.49-57.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gurujeyalakshmi G, Mahadevan A. Degradation of gallic acid by Aspergillus flavus. Zentbl Mikrobiol. 1987;142:187–192. [PubMed] [Google Scholar]
- 14.Häkkinen E, Siltanen I, Hernberg S, Seppalainen A M, Karli P, Vikkula E. Diphenyl poisoning in fruit paper production. Arch Environ Health. 1973;26:70–74. doi: 10.1080/00039896.1973.10666226. [DOI] [PubMed] [Google Scholar]
- 15.Hammer E, Krowas D, Schäfer A, Specht M, Francke W, Schauer F. Isolation and characterization of a dibenzofuran-degrading yeast: identification of oxidation and ring cleavage products. Appl Environ Microbiol. 1998;64:2215–2219. doi: 10.1128/aem.64.6.2215-2219.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horner L, Weber K-H. Darstellungen und Eigenschaften weiterer Chinone des Biphenyls. Chem Ber. 1967;100:2842–2853. [Google Scholar]
- 17.Horning E C, Parker J A. Polymethoxybromobenzenes. J Am Chem Soc. 1952;74:2107–2108. [Google Scholar]
- 18.Hundt K, Jonas U, Hammer E, Schauer F. Transformation of diphenyl ethers by Trametes versicolor and characterization of ring cleavage products. Biodegradation. 1999;10:279–286. [Google Scholar]
- 19.Javaheripour H, Neckers D C. Solid phase and solution photochemistry of coumalate esters. J Org Chem. 1977;42:1844–1851. [Google Scholar]
- 20.Kaufman D D, Blake J. Microbial degradation of several acetamide, acylanilide, carbamate, toluidine, and urea pesticides. Soil Biol Biochem. 1973;5:297–308. [Google Scholar]
- 21.Körner W, Hanf V, Schuller W, Bartsch H, Zwirner M, Hagemaier H. Validation and application of a rapid in vitro assay for assessing the estrogenic potency of halogenated phenolic chemicals. Chemosphere. 1998;12:2395–2407. doi: 10.1016/s0045-6535(98)00297-5. [DOI] [PubMed] [Google Scholar]
- 22.Lange J, Hammer E, Specht M, Francke W, Schauer F. Biodegradation of biphenyl by the ascomycetous yeast Debaryomyces vanrijiae. Appl Microbiol Biotechnol. 1998;50:364–368. doi: 10.1007/s002530051305. [DOI] [PubMed] [Google Scholar]
- 23.Lotspeich F J, Karickhoff S. The synthesis and stereochemistry of 1,2,3,4,4a,11b-hexahydro-9,10,11-trimethoxydibenzo[b,d]thiepin-7(6H)-one. J Org Chem. 1966;31:2183–2187. [Google Scholar]
- 24.Lunt D, Evans W C. The microbial metabolism of biphenyl. Biochem J. 1970;118:54. doi: 10.1042/bj1180054pb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer T, Larsen J C, Hansen E V, Scheline R R. The metabolism of biphenyl. III. Phenolic metabolites in the pig. Acta Pharmacol Toxicol. 1976;39:433–441. doi: 10.1111/j.1600-0773.1976.tb03194.x. [DOI] [PubMed] [Google Scholar]
- 26.Meyer T, Scheline R R. The metabolism of biphenyl. II. Phenolic metabolites in the rat. Acta Pharmacol Toxicol. 1976;39:419–432. doi: 10.1111/j.1600-0773.1976.tb03193.x. [DOI] [PubMed] [Google Scholar]
- 27.Mobley D P, Finkbeiner H L, Lockwood S H, Spivack J. Synthesis of 3-aryl muconolactones using biphenyl metabolism in Aspergillus. Tetrahedron. 1993;49:3273–3280. [Google Scholar]
- 28.Mobley D P. Study of biphenyl hydroxylase activity in Aspergillus parasiticus using a colorimetric assay. Appl Microbiol Biotechnol. 1994;40:622–628. [Google Scholar]
- 29.Musso H, Pietsch H. Zur Struktur von 3,3′-Dihydroxy-diphenochinonen, Chem. Ber. 1967;100:2854–2869. [Google Scholar]
- 30.Pieken W A, Kozarich J W. Lactonization of cis, cis-3-halomuconates: influence of pH and halo substituent on the regiochemistry. J Org Chem. 1990;55:3029–3035. [Google Scholar]
- 31.Reinhard A. Characterization of fungi isolated from woody-chip piles, especially thermophilic and thermotolerant isolates. Microbiol Res. 1994;149:75–83. doi: 10.1016/S0944-5013(11)80142-4. [DOI] [PubMed] [Google Scholar]
- 32.Rey M, Dunkelblum E, Allain R, Dreiding A S. Synthesen von 2-Pyronen aus α,β-ungesättigten Säurechloriden und tertiären Aminen. Helv Chim Acta. 1970;53:2159–2175. [Google Scholar]
- 33.Schultz T W, Kraut D H, Sayler G S, Layton A C. Estrogenicity of selected biphenyls evaluated using a recombinant yeast assay. Environ Toxicol Chem. 1998;17:1727–1729. [Google Scholar]
- 34.Schwartz R D, Williams A L, Hutchinson D B. Microbial production of 4,4′-dihydroxybiphenyl: biphenyl hydroxylation of fungi. Appl Environ Microbiol. 1980;39:702–708. doi: 10.1128/aem.39.4.702-708.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singer-Bohne B, Hofeneder M, Koch H P. Der Hefetest: Eine Ergänzungsmethode zur Bestimmung der akuten Toxizität von Arzneistoffen und Umweltgiften. BIOforum. 1993;16:244–248. [Google Scholar]
- 36.Smith M R, Ratledge C. Catabolism of biphenyl by Pseudomonas sp. NCIB 10643 and Nocardia sp. 10503. Appl Microbiol Biotechnol. 1989;30:395–401. [Google Scholar]
- 37.Smith R V, Davis P J, Clark A M, Glover-Milton S. Hydroxylation of biphenyl by fungi. J Appl Bacteriol. 1980;49:65–73. doi: 10.1111/j.1365-2672.1980.tb01044.x. [DOI] [PubMed] [Google Scholar]
- 38.Smith R V, Rosazza J P. Microbial models of mammalian metabolism. Aromatic hydroxylation. Arch Biochem Biophys. 1974;161:551–558. doi: 10.1016/0003-9861(74)90338-5. [DOI] [PubMed] [Google Scholar]
- 39.Soto A M, Sonnenschein K L, Chung K L, Fernandez N, Olea N, Olea-Serrano M F. The E-Screen assay as a tool to identify estrogens: an update on estrogenic environmental pollutants. Environ Health Perspect. 1995;103:113–122. doi: 10.1289/ehp.95103s7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vollmer M D, Schlömann M. Conversion of 2-chlor-cis, cis-muconate and its metabolites 2-chloro- and 5-chloromuconolactone by chloromuconate cycloisomerase of pJP4 and pAC27. J Bacteriol. 1995;177:2938–2941. doi: 10.1128/jb.177.10.2938-2941.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vollmer M D, Hoier H, Hecht H-J, Schell U, Gröning J, Goldman A, Schlömann M. Substrate specificity of and product formation by muconate cycloisomerase: an analysis of wild-type enzymes and engineering variants. Appl Environ Microbiol. 1998;64:3290–3299. doi: 10.1128/aem.64.9.3290-3299.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vollmer M D, Schell U, Seibert V, Lakner S, Schlömann M. Substrate specificities of chloromuconate cycloisomerase from Pseudomonas sp. B13, Ralstonia eutropha JMP134 and Pseudomonas sp. P51. Appl Microbiol Biotechnol. 1998;51:598–605. doi: 10.1007/s002530051438. [DOI] [PubMed] [Google Scholar]
- 43.Walker J R L, Taylor B G. Metabolism of phloroglucinol by Fusarium solani. Arch Microbiol. 1983;134:123–126. [Google Scholar]
- 44.Wiseman A, Gondal J, Sims P. 4-Hydroxylation of biphenyl by yeasts containing cytochrome P-450. Biochem Soc Trans. 1975;3:278–285. doi: 10.1042/bst0030278. [DOI] [PubMed] [Google Scholar]