Abstract
A total of 50 isolates of Shiga toxin-producing Escherichia coli (STEC), including 29 O157:H7 and 21 non-O157 STEC strains, were analyzed for antimicrobial susceptibilities and the presence of class 1 integrons. Seventy-eight (n = 39) percent of the isolates exhibited resistance to two or more antimicrobial classes. Multiple resistance to streptomycin, sulfamethoxazole, and tetracycline was most often observed. Class 1 integrons were identified among nine STEC isolates, including serotypes O157:H7, O111:H11, O111:H8, O111:NM, O103:H2, O45:H2, O26:H11, and O5:NM. The majority of the amplified integron fragments were 1 kb in size with the exception of one E. coli O111:H8 isolate which possessed a 2-kb amplicon. DNA sequence analysis revealed that the integrons identified within the O111:H11, O111:NM, O45:H2, and O26:H11 isolates contained the aadA gene encoding resistance to streptomycin and spectinomycin. Integrons identified among the O157:H7 and O103:H2 isolates also possessed a similar aadA gene. However, DNA sequencing revealed only 86 and 88% homology, respectively. The 2-kb integron of the E. coli O111:H8 isolate contained three genes, dfrXII, aadA2, and a gene of unknown function, orfF, which were 86, 100, and 100% homologous, respectively, to previously reported gene cassettes identified in integrons found in Citrobacter freundii and Klebsiella pneumoniae. Furthermore, integrons identified among the O157:H7 and O111:NM strains were transferable via conjugation to another strain of E. coli O157:H7 and to several strains of Hafnia alvei. To our knowledge, this is the first report of integrons and antibiotic resistance gene cassettes in STEC, in particular E. coli O157:H7.
Shiga toxin-producing Escherichia coli (STEC) have been an important cause of foodborne illness worldwide. Human infection with STEC is potentially fatal and may be associated with serious complications such as hemolytic -uremic syndrome (HUS) and hemorrhagic colitis (7). Among STEC strains, O157:H7 is the classical serotype that was first associated with enterohemorrhagic diseases in the early 1980s as a cause of serious foodborne disease outbreaks. Since then, over 100 STEC serotypes, other than O157:H7, have been associated with human illness (17). In the United States approximately 73,000 cases of infections annually have been attributed to O157:H7 strains and more than 36,000 to non-O157:H7 STEC strains (16). There are more than 100 deaths each year due to STEC infections. Several studies in Japan have suggested that initiation of antibiotic therapy early in the stages of STEC infection was able to prevent the disease progression to HUS (5, 11, 30). However, antimicrobial treatment for STEC infections is still regarded as controversial (10, 23, 35). Currently, no specific therapy for HUS is available, and most patients require prolonged clinical and outpatient treatment (31).
Initially, E. coli O157:H7 was found to be susceptible to many antibiotics (3, 24). However, several recent studies have documented antibiotic resistance among E. coli O157:H7 isolates (4, 13, 19, 29). Non-O157 STEC strains isolated from humans and animals also have developed antibiotic resistance, and many are resistant to multiple antimicrobials commonly used in human and veterinary medicine (4, 6, 29). There is currently a great deal of speculation regarding the role that the therapeutic and subtherapeutic use of antimicrobials in animals has played in accelerating the development and dissemination of antimicrobial-resistant bacterial pathogens (1, 33, 34). Research is urgently needed to determine the potential effect of antimicrobials used in animal production environments on the emergence and spread of bacterial antibiotic resistance in both veterinary and human medicine.
A novel system has been identified in multiple resistant bacteria and is postulated to play an important role in the acquisition and dissemination of antibiotic resistance genes (8). This system has been referred to as bacterial integrons. Integrons are mobile DNA elements with a specific structure consisting of two conserved segments flanking a central region containing “cassettes” that usually code for resistance to specific antimicrobials (9). Four classes of integrons have been identified to date (15). The majority of integrons identified among clinical isolates belong to class 1 type (12, 27). In class 1 integrons, the 5′ conserved region encodes a site-specific recombinase (integrase) and a strong promoter or promoters that ensure expression of the integrated cassettes. The 3′ conserved segment carries qacΔE, which specifies resistance to antiseptics and disinfectants; the sul-1 gene, which confers sulfonamide resistance; and an open reading frame of unknown function (9). Multiple cassette insertions and more than 40 distinct cassettes have been identified among integrons (25). Currently, there is a paucity of data regarding the mechanisms of acquisition and dissemination of antibiotic resistance in E. coli O157:H7 and other STEC. This study was initiated to characterize antimicrobial susceptibility patterns among STEC strains, including E. coli O157:H7, isolated from cattle, ground beef, and humans and to determine if resistance phenotypes observed could be attributed to the acquisition of integron-mediated resistance gene cassettes.
MATERIALS AND METHODS
Bacterial strains.
A total of 50 STEC isolates including 29 O157:H7 strains and 21 non-O157 STEC strains from humans, animals, and foods were used in the study (Table 1). The STEC isolates originated from humans (n = 8), cattle (n = 32), and ground beef (n = 10). The 29 E. coli O157:H7 strains were selected among 118 isolates from a previous study (19) and were resistant to at least one antibiotic based on disk diffusion in vitro susceptibility testing (Difco, Detroit, Mich). Salmonella enterica serovar Typhimurium DT104 CVM786 was used as a positive control for integron PCR reactions. Three Hafnia alvei strains isolated from ground beef and one strain of E. coli O157:H7 originating from human stools were used as recipient strains in conjugation experiments. Bacteria were grown on MacConkey agar (Difco) and stored in Trypticase soy broth (Difco) containing 50% glycerol at −80°C until use.
TABLE 1.
Characteristics of O157:H7 and non-O157 STEC isolates used in the study
| Isolate | Serotype | stx genea | Presence (+) or absence (−) of:
|
Source | Yr | |
|---|---|---|---|---|---|---|
| eae gene | hlyA gene | |||||
| CVM65 | O157:H7 | 1, 2 | + | + | Cattle | 1991 |
| CVM68 | O157:H7 | 1, 2 | + | + | Cattle | 1991 |
| CVM990 | O157:H7 | 1, 2 | + | + | Cattle | 1991 |
| CVM996 | O157:H7 | 1, 2 | + | + | Cattle | 1991 |
| CVM1001 | O157:H7 | 1, 2 | + | + | Cattle | 1991 |
| CVM69 | O157:H7 | 1, 2 | + | + | Cattle | 1992 |
| CVM73 | O157:H7 | 1, 2 | + | + | Cattle | 1992 |
| CVM998 | O157:H7 | 1, 2 | + | + | Cattle | 1992 |
| CVM1002 | O157:H7 | 1, 2 | + | + | Cattle | 1992 |
| CVM1000 | O157:H7 | 1, 2 | + | + | Cattle | 1992 |
| CVM1010 | O157:H7 | 1, 2 | + | + | Cattle | 1992 |
| CVM35 | O157:H7 | 1, 2 | + | + | Cattle | 1993 |
| CVM37 | O157:H7 | 2 | + | + | Cattle | 1993 |
| CVM44 | O157:H7 | 1, 2 | + | + | Cattle | 1993 |
| CVM978 | O157:H7 | 2 | + | + | Cattle | 1993 |
| CVM981 | O157:H7 | 1, 2 | + | + | Cattle | 1993 |
| CVM988 | O157:H7 | 1, 2 | + | + | Cattle | 1993 |
| CVM1006 | O157:H7 | 2 | + | + | Cattle | 1993 |
| CVM1007 | O157:H7 | 1, 2 | + | + | Cattle | 1993 |
| CVM79 | O157:H7 | 1, 2 | + | + | Ground beef | 1988 |
| CVM33 | O157:H7 | 1, 2 | + | + | Ground beef | 1993 |
| CVM1004 | O157:H7 | 1, 2 | + | + | Ground beef | 1993 |
| CVM46 | O157:H7 | 1, 2 | + | + | Ground beef | 1994 |
| CVM47 | O157:H7 | 1, 2 | + | + | Ground beef | 1994 |
| CVM1009 | O157:H7 | 1, 2 | + | + | Ground beef | 1994 |
| CVM48 | O157:H7 | 1, 2 | + | + | Ground beef | 1995 |
| CVM90 | O157:H7 | 1, 2 | + | + | Human | 1983 |
| CVM51 | O157:H7 | 1, 2 | + | + | Human | 1992 |
| CVM1008 | O157:H7 | 1, 2 | + | + | Human | 1992 |
| CVM1871 | O26:H11 | 1 | + | + | Cattle | 1991 |
| CVM1888 | O26:H11 | 1 | + | + | Cattle | 1993 |
| CVM1875 | O103:H2 | 1, 2 | + | + | Cattle | 1993 |
| CVM1876 | O103:H2 | 1 | + | + | Cattle | 1993 |
| CVM1877 | O111:H8 | 1 | + | + | Cattle | 1993 |
| CVM1884 | O111:NM | 1 | + | + | Cattle | 1993 |
| CVM1878 | O111:H11 | 1, 2 | + | + | Cattle | 1994 |
| CVM1879 | O111:H11 | 1 | + | + | Cattle | 1994 |
| CVM1880 | O111:NM | 1, 2 | + | + | Cattle | 1994 |
| CVM1881 | O118:H16 | 1 | + | + | Cattle | 1993 |
| CVM1887 | O126:H8 | 1 | + | − | Cattle | 1991 |
| CVM1874 | O91:H21 | 1, 2 | − | + | Cattle | 1993 |
| CVM1889 | O45:H2 | 1 | + | + | Cattle | 1992 |
| CVM1873 | O88:H49 | 1, | + | + | Ground beef | 1994 |
| CVM1872 | O46:H38 | 1, 2 | − | + | Ground beef | 1991 |
| CVM1870 | O22:H8 | 2 | − | + | Ground beef | 1993 |
| CVM1882 | O125:NM | 1 | + | + | Human | 1990 |
| CVM1883 | O113:K75:H21 | 2 | + | + | Human | 1988 |
| CVM1885 | O103:H2 | 1, 2 | + | + | Human | 1987 |
| CVM1890 | O5:NM | 1 | + | + | Human | 1996 |
| CVM1891 | Orough:H9 | 2 | + | + | Human | 1996 |
1, stx-1 positive; 2, stx-2 positive.
Quantitative antimicrobial susceptibility determination.
Antimicrobial MICs of E. coli isolates were determined via the Sensititre automated antimicrobial susceptibility system (Trek Diagnostic Systems, Westlake, Ohio) and interpreted according to the National Committee for Clinical Laboratory Standards (NCCLS) guidelines for broth microdilution methods (20, 21). Sensititre susceptibility testing was performed according to the manufacturer's instructions. The following antimicrobials were tested: amikacin, amoxicillin-clavulanic acid, ampicillin, apramycin, ceftiofur, ceftriaxone, cephalothin, chloramphenicol, ciprofloxacin, florfenicol, gentamicin, kanamycin, nalidixic acid, streptomycin, sulfamethoxazole, tetracycline, and trimethoprim-sulfamethoxazole. E. coli ATCC 25922 and 35218, Enterococcus faecalis ATCC 29212, Staphyloccoccus aureus ATCC 29213, and Pseudomonas aeruginosa ATCC 27853 were used as controls in antimicrobial MIC determinations.
Bacterial DNA preparation, PCR, and DNA sequencing.
Bacterial DNA used for PCR was prepared by boiling a bacterial culture in 500 μl of distilled water for 10 min, followed by centrifugation. Isolates were initially characterized for STEC-associated virulence genes, stx-1 and stx-2 coding for Shiga toxins, eae for intimin that is associated with attaching and effacing lesion, and hlyA for hemolysin (18). The presence of class 1 integrons and the associated sulfonamide resistance gene (sul-1) and integrase gene (int) were detected using the PCR method described by Levesque et al. (14) and Sandvang et al. (28). Class 1 integrons were amplified using the following PCR primers: 5′-CS (5′-GGCATCCAAGCACAAGC-3′) and 3′-CS (5′-AAGCAGACTTGACTGAT-3′). The class 1 integrase gene and sulfonamide resistance gene were also amplified as described above, but the following primers were used: int-F (5′-CCTCCCGCACGATGATC-3′), int-R (5′-TCCACGCATCGTCAGGC-3′), sul1-F (5′-CTTCGATGAGAGCCGGCGGC-3′), and sul1-R (5′-GCAAGGCGGAAACCCGCGCC-3′). All primers were synthesized by Life Technologies (Gaithersburg, Md.). Amplification reactions were carried out with 10 μl of boiled bacterial suspensions, 250 μM deoxynucleoside triphosphate, 2.5 mM MgCl2, 50 pmol of primers, and 1 U of Gold Taq polymerase (Perkin-Elmer, Foster City, Calif.). Distilled water was added to bring the final volume to 50 μl. The PCR cycle for class 1 integron and integrase gene included an initial denaturation at 94°C for 10 min, followed by 30 cycles of denaturation for 1 min at 94°C, primer annealing for 1 min at 54°C, and extension for 2 min at 72°C, and a final extension at 72°C for 10 min. The PCR cycle for amplification of the sul-1 gene was identical to that described above, except for the annealing temperature, which was 59°C. The reaction products were then analyzed by electrophoresis in 1.0% agarose gels stained with ethidium bromide, visualized under UV light, and recorded by using a gel documentation system (Bio-Rad, Hercules, Calif.). For each set of PCR reactions, serovar Typhimurium DT104 CVM786 was included as a positive control. The PCR-generated DNA fragments were then purified using a PCR purification kit (Boehringer Mannheim, Indianapolis, Ind.) and sequenced in an ABI automatic DNA sequencer (Model 377; Perkin-Elmer) at the University of Maryland Biotechnology Institute by using the above-described forward and reverse primers. DNA sequences were analyzed by searching the GenBank database of the National Center for Biotechnology Information via the BLAST network service. The E. coli O157:H7 integron sequence has been assigned the GenBank accession number AF234167.
Southern blot hybridization.
Southern blot hybridizations were performed to determine the locations of identified integrons. Genomic and plasmid DNA were isolated from integron-positive STEC isolates using the High Pure Plasmid Isolation Kit (Boehringer Mannheim). Genomic and plasmid DNA were both digested with BamHI. The class 1 integrase gene was used as a DNA probe and was labeled with digoxigenin using the PCR DIG Probe Synthesis Kit (Boehringer Mannheim). Digested DNA was run in 1% agarose gels and transferred to nylon membranes. Hybridization procedures and conditions were performed as specified with a nonradioactive labeling and detection kit (Boehringer Mannheim).
Conjugation experiments.
E. coli O157:H7 strain CVM990 and E. coli O111:NM strain CVM1884 that possessed integrons were used as donor strains. E. coli O157:H7 strain JM263 and H. alvei strains CVM1202, CVM1203, and CVM1208 were used as recipient strains (see Table 4). Streptomycin and tetracycline resistance were used as selective markers for E. coli O157:H7 strains CVM990 and CVM1884, respectively, based on their antibiotic resistance phenotypes (see Table 4). Nalidixic acid and ampicillin resistance were used as selective markers for E. coli O157:H7 JM263 and H. alvei strains CVM1202, CVM1203, and CVM1208, respectively. Conjugation was performed by mating a donor strain with a recipient strain on Trypticase soy agar (TSA; Difco) plates (36). After incubation at 37°C overnight, the mating mixture was streaked onto a TSA plate containing a combination of selective antibiotics dependent upon the donor and recipient strains used (100 μg of streptomycin and 100 μg of nalidixic acid per ml, 100 μg of streptomycin and 100 μg of ampicillin per ml, 30 μg of tetracycline and 100 μg of nalidixic acid per ml, or 30 μg of tetracycline and 100 μg of ampicillin per ml). Transconjugants were examined for antibiotic susceptibility profiles, and integron transfer was confirmed via PCR assays and/or DNA sequencing.
TABLE 4.
Antimicrobial resistance profiles and MICs for donor, recipient, and transconjugant isolates
| Bacterial strain(s) | MICa (μg/ml) of:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| AMC | AMP | CEF | CHL | KAN | NAL | STR | SUL | TET | |
| Donors | |||||||||
| CVM990 (E. coli O157:H7) | 4 | 4 | 4 | ≤4 | >64 | ≤4 | >256 | >512 | 32 |
| CVM1884 (E. coli O111:NM) | 4 | 4 | 8 | ≤4 | ≤16 | ≤4 | <32 | >512 | >32 |
| Recipients | |||||||||
| JM263 (E. coli O157:H7) | 4 | 4 | 32 | 32 | ≤16 | >256 | ≤32 | ≤128 | 8 |
| CVM1202 (H. alvei) | >32 | >32 | >32 | ≤4 | ≤16 | ≤4 | ≤32 | ≤128 | ≤4 |
| CVM1203 (H. alvei) | >32 | >32 | >32 | ≤4 | ≤16 | ≤4 | ≤32 | ≤128 | ≤4 |
| CVM1208 (H. alvei) | >32 | >32 | >32 | ≤4 | ≤16 | ≤4 | ≤32 | ≤128 | ≤4 |
| Transconjugants | |||||||||
| CVM990-JM263 | 4 | 8 | 32 | 32 | ≤16 | >256 | >256 | >512 | >32 |
| CVM990-CVM1202 | >32 | >32 | >32 | ≤4 | ≤16 | ≤4 | 128 | >512 | >32 |
| CVM990-CVM1203 | >32 | >32 | >32 | ≤4 | ≤16 | ≤4 | 128 | >512 | >32 |
| CVM990-CVM1208 | >32 | >32 | >32 | ≤4 | ≤16 | ≤4 | 128 | >512 | >32 |
| CVM1884-JM263 | 4 | 8 | 32 | 32 | ≤16 | >256 | ≤32 | >512 | >32 |
| CVM1884-CVM1202 | >32 | >32 | >32 | ≤4 | ≤16 | ≤4 | 128 | >512 | >32 |
| CVM1884-CVM1203 | >32 | >32 | >32 | ≤4 | ≤16 | ≤4 | 128 | >512 | >32 |
| CVM1884-CVM1208 | >32 | >32 | >32 | ≤4 | ≤16 | ≤4 | 128 | >512 | >32 |
MICs were determined via microdilution methods according to NCCLS standards (20, 21). Abbreviations: AMC, amoxicillin-clavulanic acid; AMP, ampicillin; CEF, cephalothin; CHL, chloramphenicol; KAN, kanamycin; STR, streptomycin; SUL, sulfamethoxazole; TET, tetracycline. Numbers in boldface represent resistant phenotypes.
PFGE.
Pulsed-field gel electrophoresis (PFGE) was performed to determine DNA fingerprinting profiles of donors, recipients, and transconjugates and to verify the gene transfer from donor to recipient strains. The PFGE procedure of the Centers for Disease Control and Prevention was used. Briefly, bacteria were grown on TSA plates supplemented with 5% defibrinated sheep blood (Becton Dickinson Microbiology Systems, Cockeysville, Md.) at 37°C for 18 h. Bacterial colonies were suspended in cell suspension buffer (100 mM Tris HCl, 100 mM EDTA; pH 8.0) and adjusted to an optical density of 0.50 to 0.54 using Dade MicroScan Turbidity Meter (Dade Behring, Inc., West Sacramento, Calif.). The cell suspension (200 μl) was mixed with 10 μl of proteinase K (10 mg/ml) and an equal volume of melted 1% SeaKem Gold agarose (FMC BioProducts, Rockville, Maine) containing 1% sodium dodecyl sulfate. The mixture was carefully dispensed into a sample mold (Bio-Rad). After solidification, the plugs were transferred to a tube containing 5 ml of lysis buffer (50 mM Tris HCl; 50 mM EDTA, pH 8.0; 1% sarcosyl) and 0.1 mg of proteinase K per ml. Cells were lysed overnight in a water bath at 54°C with agitation at 180 rpm. After lysis, the plugs were washed twice with distilled water and four times with TE buffer (10 mM Tris HCl, 1 mM EDTA; pH 8.0) for 15 min per wash at 50°C with agitation at 180 rpm. Agarose-embedded DNA was digested with 50 U of XbaI (Boehringer Mannheim) overnight in a water bath at 37°C. The plugs were placed in a 1% SeaKem Gold agarose gel. Restriction fragments were separated by electrophoresis in 0.5× TBE (Tris-borate-EDTA) buffer at 14°C for 18 h using a Chef Mapper (Bio-Rad) with pulse times of 2.16 to 54.17 s. The gel was stained with ethidium bromide, and DNA bands were visualized with a UV transilluminator.
RESULTS
Antimicrobial susceptibility profiles.
The antimicrobial susceptibilities of 21 non-O157 and 29 O157:H7 STEC isolates were determined using microbroth dilution (Table 2). All E. coli O157:H7 isolates tested were resistant to at least one antibiotic, which agreed with results from our previous study using disk diffusion in vitro susceptibility testing (19). The majority of the E. coli O157:H7 isolates were resistant to several antimicrobials tested, particularly sulfamethoxazole (93%), tetracycline (93%), and streptomycin (76%). E. coli O157:H7 isolates were also resistant to kanamycin (21%) and ampicillin (14%). Several isolates were resistant to multiple antimicrobial agents, and the most common pattern was resistance to streptomycin, sulfamethoxazole, and tetracycline. Four E. coli O157:H7 isolates were multiple resistant to five antibiotics: ampicillin, kanamycin, streptomycin, sulfamethoxazole, and tetracycline. Two of these isolates (CVM73 and CVM1001) were isolated from cattle in 1991 and 1992, respectively, one (CVM90) was from a human sample, and the fourth (CVM79) was obtained from ground beef in 1988.
TABLE 2.
Antimicrobial resistance of E. coli O157:H7 and non-O157 STEC isolates
| Class and/or antimicrobial | % Resistant isolatesa
|
|
|---|---|---|
| O157:H7 | Other STEC | |
| β-Lactams | ||
| Ampicillin | 14 | 33 |
| Amoxicillin-clavulanic acid | 0 | 5 |
| Cephalosporins | ||
| Cephalothin | 0 | 19 |
| Ceftiofur | 0 | 0 |
| Ceftriaxone | 0 | 0 |
| Phenicols | ||
| Chloramphenicol | 0 | 29 |
| Florfenicolb | 0 | 10 |
| Tetracycline | 93 | 43 |
| Aminoglycosides | ||
| Amikacin | 0 | 0 |
| Apramycin | 0 | 0 |
| Kanamycin | 21 | 19 |
| Gentamicin | 0 | 0 |
| Streptomycin | 76 | 43 |
| Sulfonamides and/or trimethoprim | ||
| Sulfamethoxazole | 93 | 48 |
| Trimethoprim-sulfamethoxazole | 0 | 19 |
| Quinolones (including fluoroquinolones) | ||
| Nalidixic acid | 0 | 0 |
| Ciprofloxacin | 0 | 0 |
The majority of the non-O157:H7 isolates displayed resistance to multiple antimicrobials compared to the E. coli O157:H7 isolates (Table 2). Resistance to chloramphenicol (29%), trimethoprim-sulfamethoxazole (19%), cephalothin (19%), florfenicol (10%), and amoxicillin-clavulanic acid (5%) was only observed among non-O157:H7 isolates (Table 2). CVM1877 (O111:H8) and CVM1889 (O45:H2) were resistant to seven antibiotics: ampicillin, chloramphenicol, kanamycin, streptomycin, sulfamethoxazole, tetracycline, and either cephalothin or trimethoprim-sulfamethoxazole. CVM1879 (O111:H11) was multiply resistant to nine antibiotics, including ampicillin, cephalothin, chloramphenicol, florfenicol, kanamycin, streptomycin, sulfamethoxazole, tetracycline, and trimethoprim-sulfamethoxazole. Four non-O157:H7 isolates (CVM1877, CVN1878, CVM1879, and CVM1889) were resistant to kanamycin and chloramphenicol.
Presence of class one integrons and integron-associated genes.
Class 1 integrons were identified among nine STEC isolates representing multiple serotypes: O157:H7, O111:H11, O111:H8, O111:NM, O103:H2, O45:H2, and O26:H11 (Table 3). The majority of integron amplified fragments were 1 kb in size, except for one E. coli O111:H8 isolate which contained a 2-kb amplicon. DNA sequence analysis revealed that the integrons identified within the O111:H11, O111:H8, O111:NM, O45:H2, and O26:H11 isolates contained the aadA gene encoding resistance to streptomycin-spectinomycin. Integrons identified among the O157:H7 and O103:H2 isolates also possessed a similar aadA gene; however, DNA sequencing revealed only 86 and 88% homology, respectively. The 2-kb integron of the E. coli O111:H8 isolate contained three genes, i.e., dfrXII encoding trimethoprim resistance, aadA2 encoding resistance to streptomycin and spectinomycin, and a gene of unknown function, orfF, which were 86, 100, and 100% homologous to previously reported gene cassettes identified in integrons found in Citrobacter freundii and Klebsiella pneumoniae, respectively. All integron-positive STEC strains were positive for int-1 and sul-1. Figure 1 shows the PCR amplicons of sul-1 (417 bp) and int-1 (280 bp) genes that are present in class 1 integrons. Additionally, Southern blot hybridizations indicated that class 1 integrons were located on plasmids among the STEC isolates (data not shown).
TABLE 3.
Characterization of class 1 integron-associated genes among STEC isolates
| Isolate | Serotype | Size of integron amplicon (kb) | Inserted gene cassettesa | Presence (+) of
|
|
|---|---|---|---|---|---|
| sul gene | int gene | ||||
| CVM990 | O157:H7 | 1 | aadA6 | + | + |
| CVM1878 | O111:11 | 1 | aadA1 | + | + |
| CVM1879 | O111:11 | 1 | aadA1 | + | + |
| CVM1877 | O111:H8 | 2 | aadA2-dfr12 | + | + |
| CVM1880 | O111:NM | 1 | aadA1 | + | + |
| CVM1884 | O111:NM | 1 | aadA1 | + | + |
| CVM1875 | O103:H2 | 1 | aadA1 | + | + |
| CVM1889 | O45:H2 | 1 | aadA1 | + | + |
| CVM1871 | O26:H11 | 1 | aadA1 | + | + |
aadA, aminoglycoside adenyltransferase; dfr, dihydrofolate reductase.
FIG. 1.
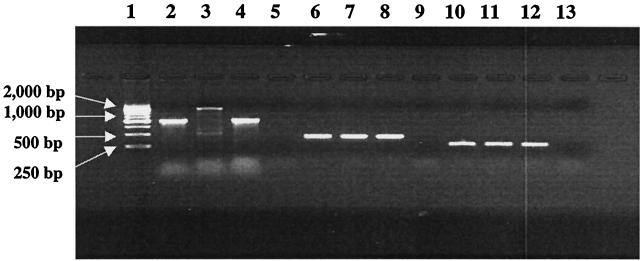
PCR amplicons of STEC integrons. Lanes 2 to 5 were generated with the CS primers specific for class 1 integron. Lanes 6 to 9 were obtained using the sul primers specific for the sul gene that confers sulfonamide resistance. Lanes 10 to 13 were obtained using the primers specific for integrase. Specifically, lanes 2, 6, and 10 show positive controls (serovar Typhimurium DT104); lanes 3, 7, and 11 show E. coli O111:H8 strain CVM1877; lanes 4, 8, and 12 show E. coli O157:H7 strain CVM990; and lanes 5, 9, and 13 show negative controls (E. coli K-12).
Conjugal transfer of plasmids harboring class 1 integrons.
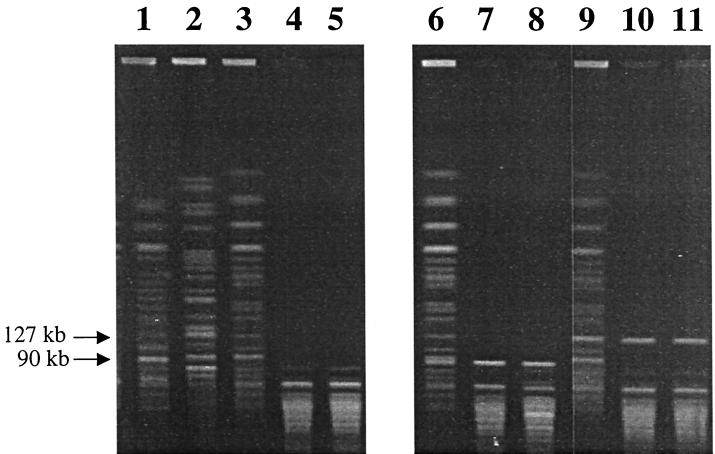
Conjugal transfer of plasmids carrying class 1 integrons occurred from donor strains (O157:H7 CVM990 and O111:NM CVM1884) to recipient strains (O157:H7 JM263 and H. alvei CVM1202, CVM1203, and CVM1208). Class 1 integrons were detected among transconjugants of the recipient strains using integron-specific PCR assays (data not shown). DNA fingerprinting by PFGE revealed that several transconjugants, i.e., CVM990-JM263, CVM990-CVM1202, and CVM990-CVM1208, acquired a 90-kb band when CVM990 was used as the donor strain, whereas transconjugants CVM1884-JM263, CVM1884-CVM1202, and CVM1884-CVM1208 gained a 127-kb band when CVM1884 was used as the donor strain (Fig. 2)
FIG. 2.
PFGE profiles with XbaI digestion of donor, recipient, and transconjugant strains. Lanes 1 to 5 show, respectively, results obtained with donor strains CVM990 (E. coli O157:H7) and CVM1884, (E. coli O111:NM) and recipient strains JM263 (E. coli O157:H7), H. alvei CVM1202, and H. alvei CVM1203 before the conjugations. Lanes 6 to 8 show results with the transconjugants CVM990-JM263, CVM990-CVM1202, and CVM990-CVM1203, respectively, that have gained a 90-kb band. Lanes 9 to 11 show results with the transconjugants CVM1884-JM263, CVM1884-CVM1202, and CVM1884-CVM1203, respectively, that have gained a 127-kb band.
Antimicrobial susceptibility testing of donor, recipient, and transconjugant isolates revealed that all Hafnia transconjugants acquired resistance to streptomycin, sulfamethoxazole, and tetracycline regardless of the donor strain (Table 4). Interestingly, in contrast to CVM1884-JM263, transconjugants CVM1884-CVM1202, CVM1884-CVM1203, and CVM1884-CVM1208 acquired streptomycin resistance even though the donor strain (CVM1884) was susceptible. DNA sequencing of the integrons amplified from both CVM1884 and transconjugant CVM1884-CVM1208 identified the identical aadA1 gene that encodes resistance to streptomycin and spectinomycin. This most likely indicates that although the aadA1 gene was present in the donor strain (CVM1884), it was not being efficiently expressed from the integron promoter. None of the gene cassettes identified within the integrons from the STEC isolates conferred resistance to tetracycline, despite the fact that all of the transconjugants acquired tetracycline resistance. This may be due to the presence of the tetracycline resistance gene on the same plasmid which contained the integrons or on a different plasmid that was cotransferred by conjugation with the integron-containing plasmid into the recipient cells.
DISCUSSION
The emergence and dissemination of antimicrobial resistance among STEC strains may have potential negative clinical implications, although the diarrheal phase of illnesses associated with STEC strains is usually self-limiting and the role of early antimicrobial therapy in the prevention of HUS is still unclear (7). Trimethoprim-sulfamethoxazole and β-lactam antibiotics are commonly used for the treatment of gastroenteritis. In the case of STEC infections, antibiotic treatment is not recommended. However, such antibiotics are often administrated before the disease is diagnosed. For children with acute bloody diarrhea, the most widely accepted recommendation is to obtain a stool culture and initiate empirical antibiotic treatment, because appropriate treatment shortens the duration of the diarrhea, decreases the incidence of complications, and reduces the risk of transmission by shortening the duration of bacterial shedding (22).
Unfortunately, several studies, including the present study, have revealed that many STEC strains have developed resistance to trimethoprim-sulfamethoxazole, β-lactams, and other antibiotics. Additionally, the present study also demonstrated that STEC strains possess integrons which encode antibiotic resistance genes conferring resistance to trimethoprim and sulfamethoxazole. Antibiotic resistance and resistance integrons in STEC would not only complicate future antibiotic therapy but could also potentially stimulate the transfer of resistance genes. Also, antibiotic-resistant STEC strains could possibly possess selective advantages over other bacteria colonizing the gastrointestinal tracts of animals that are treated with antibiotics (therapeutically or subtherapeutically). Resistant STEC strains could then become the predominant E. coli present under antibiotic selective pressures. This could result in STEC population increases and perhaps greater shedding, which could lead to greater contamination of animal food products with STEC.
Integrons and gene cassettes have been found primarily in gram-negative bacterial species belonging to the family Enterobacteriaceae and to the Pseudomonas genus (8, 27). Several studies have reported the presence of class 1 integrons among clinical isolates of K. pneumoniae, K. oxytoca, P. aeruginosa, E. coli, and C. freundii (12). Numerous studies have also revealed that integron-borne gene cassettes are present among S. enterica serotype Typhimurium isolates. A recent study (26) reported that integrons were found in all 45 human and 21 animal isolates of Salmonella serovar Typhimurium DT104 isolated between 1984 and 1997, including 58 isolates from the United Kingdom and 8 isolates from other European countries, the United States, Trinidad, and South Africa. All strains had the pentamer R-type of ampicillin-chloramphenicol-streptomycin-sulfonamides-tetracycline (ACSSuT) and contained two integrons of approximately 1 and 1.2 kb. These isolates also contained the same inserted gene cassettes, irrespective of source and country of origin, suggesting the spread of an epidemic clone. Integrons have also been identified among veterinary E. coli that were isolated from the normal intestinal flora of swine (32) and diseased poultry (2). The ant(3")-Ia (aadA) gene responsible for resistance to streptomycin-spectinomycin was identified in all of the integrons present in the swine E. coli. Other integron-associated genes identified among the swine E. coli isolates included dfr-1 encoding resistance to trimethoprim and the β-lactamase gene oxa-1. Integrons from 8 isolates were determined to be located on plasmids and were transferred to an E. coli DH5 laboratory strain. Additionally, Bass et al. (2) reported that 63% of 100 clinical isolates of avian E. coli exhibited class 1 integrons of approximately 1 kb. These integrons were determined to contain the aadA1 gene cassette conferring resistance to streptomycin-spectinomycin. This wide distribution has presumably been achieved by the transposition of integrons to broad-host-range plasmids. However, the role of integrons in the acquisition and spread of antibiotic resistance has not yet been fully investigated.
The present study reports the presence of class 1 integrons conferring multiple resistance phenotypes among STEC strains isolated from cattle, ground beef, and humans, which is, to our knowledge, the first report on the presence of integrons in O157:H7 and non-O157 STEC strains. More significantly, the antibiotic resistance phenotypes observed in STEC isolates were also transferable, by conjugal plasmids, within species and between STEC and Hafnia sp. (horizontal transfer). The integron-borne aadA gene was silent in E. coli O111:NM, whereas it was fully expressed when it was transferred to H. alvei, suggesting that horizontal transfer may enhance the expression of a resistance gene. As this and other studies have shown, many integron-associated gene cassettes are identical, indicating that they may originate from a common source and are readily disseminated among bacteria. It is also evident that integrons conferring the same resistance phenotype can be genetically different since more than one gene cassette can encode resistance to one particular antibiotic. Some STEC strains in our study contain an aadA cassette that confers resistance to streptomycin and spectinomycin, but the gene cassettes were only 86% identical, suggesting that they may have been evolving independently for some time before their emergence in antibiotic-resistant STEC.
E. coli O157:H7 is considered a newly emerged pathogen, which may explain in part why integrons were only found in one strain among the 29 antibiotic-resistant strains tested. It is likely that integrons will be identified among additional STEC isolates in the future since this and other studies have demonstrated that integrons are readily transferable through conjugation. However, integrons and their associated gene cassettes did not always account for the total phenotypic resistance exhibited by the STEC isolates. Several STEC isolates displayed resistance to multiple antibiotics but did not contain any gene cassettes conferring resistance. Clearly, other mechanisms also contribute to the STEC antibiotic resistance phenotypes.
In summary, STEC strains have developed resistance to multiple classes of antimicrobials. Integrons not only are associated with multiple antibiotic resistance but also may play a significant role in the dissemination of resistance genes. Although the findings of this study strongly support the hypothesis that integrons provide a very efficient strategy for the acquisition and dissemination of new antibiotic resistance genes, a prospective long-term investigation of the natural evolution of gene transfer among bacterial pathogens inhabiting food animals is required.
REFERENCES
- 1.Aarestrup F M, Wegener H C. The effects of antibiotic usage in food animals on the development of antimicrobial resistance of importance for humans in Campylobacter and Escherichia coli. Microb Infect. 1999;1:639–644. doi: 10.1016/s1286-4579(99)80064-1. [DOI] [PubMed] [Google Scholar]
- 2.Bass L, Liebert C A, Lee M D, Summers A O, White D G, Thayer S G, Maurer J J. Incidence and characterization of integrons, genetic elements mediating multiple-drug resistance in avian Escherichia coli. Antimicrob Agents Chemother. 1999;43:2925–2929. doi: 10.1128/aac.43.12.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bopp C, Greene K, Downes F, Sowers E, Wells J, Wachsmuth I. Unusual verotoxin-producing Escherichia coli associated with hemorrhagic colitis. J Clin Mcirobiol. 1987;25:1486–1489. doi: 10.1128/jcm.25.8.1486-1489.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farina C, Goglio A, Conedera G, Minelli F, Caprioli A. Antimicrobial susceptibility of Escherichia coli O157 and other enterohaemorrhagic Escherichia coli isolated in Italy. Eur J Clin Microbiol Infect Dis. 1996;15:351–353. doi: 10.1007/BF01695674. [DOI] [PubMed] [Google Scholar]
- 5.Fukushima H, Hashizume T, Morita Y, Tanaka J, Azuma K, Mizumoto Y, Kaneno M, Matsuura M, Konma K, Kitani T. Clinical experiences in Sakai City Hospital during the massive outbreak of enterohemorrhagic Escherichia coli O157 infections in Sakai City, 1996. Pediatr Int. 1999;41:213–217. doi: 10.1046/j.1442-200x.1999.4121041.x. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez E A, Blanco J. Serotypes and antibiotic resistance of verotoxigenic (VTEC) and necrotizing (NTEC) Escherichia coli strains isolated from calves with diarrhoea. FEMS Microbiol Lett. 1989;51:31–36. doi: 10.1111/j.1574-6968.1989.tb03414.x. [DOI] [PubMed] [Google Scholar]
- 7.Griffin P M. Escherichia coli O157:H7 and other enterohemorrhagic Escherichia coli. In: Blaser M J, Smith P D, Ravdin J I, Greenberg H B, Guerrant R L, editors. Infections of gastrointestinal tract. New York, N.Y: Raven Press; 1995. pp. 739–761. [Google Scholar]
- 8.Hall R M. Mobile gene cassettes and integrons: moving antibiotic resistance genes in gram-negative bacteria. Ciba Found Symp. 1997;207:192–202. doi: 10.1002/9780470515358.ch12. [DOI] [PubMed] [Google Scholar]
- 9.Hall R M, Stokes H W. Integrons: novel DNA elements which capture genes by site-specific recombination. Genetica. 1993;90:115–132. doi: 10.1007/BF01435034. [DOI] [PubMed] [Google Scholar]
- 10.Igarashi T, Inatomi J, Wake A, Takamizawa M, Katayama H, Iwata T. Failure of pre-diarrheal antibiotics to prevent hemolytic uremic syndrome in serologically proven Escherichia coli O157:H7 gastrointestinal infection. J Pediatr. 1999;135:768–769. doi: 10.1016/s0022-3476(99)70100-9. [DOI] [PubMed] [Google Scholar]
- 11.Ikeda K, Ida O, Kimoto K, Takatorige T, Nakanishi N, Tatara K. Effect of early fosfomycin treatment on prevention of hemolytic uremic syndrome accompanying Escherichia coli O157:H7 infection. Clin Nephrol. 1999;52:357–362. [PubMed] [Google Scholar]
- 12.Jones M E, Peters E, Weersink A M, Fluit A, Verhoef J. Widespread occurrence of integrons causing multiple antibiotic resistance in bacteria. Lancet. 1997;349:1742–1743. doi: 10.1016/S0140-6736(05)62954-6. [DOI] [PubMed] [Google Scholar]
- 13.Kim H H, Samadpour M, Grimm L, Clausen C R, Besser T E, Baylor M, Kobayashi J M, Neill M A, Schoenknecht F D, Tarr P I. Characteristics of antibiotic-resistant Escherichia coli O157:H7 in Washington State, 1984–1991. J Infect Dis. 1994;170:1606–1609. doi: 10.1093/infdis/170.6.1606. [DOI] [PubMed] [Google Scholar]
- 14.Levesque C, Piche L, Larose C, Roy P H. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995;39:185–191. doi: 10.1128/aac.39.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mazel D, Dychinco B, Webb V A, Davies J. A distinctive class of integron in the Vibrio cholerae genome. Science. 1998;280:605–608. doi: 10.1126/science.280.5363.605. [DOI] [PubMed] [Google Scholar]
- 16.Mead P S, Slutsker L, Dietz V, McCaig L F, Bresee J S, Shapiro C, Griffin P M, Tauxe R V. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng J, Doyle M P. Microbiology of Shiga toxin-producing Escherichia coli in foods. In: Kaper J, O'Brien A, editors. Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. Washington, D.C.: ASM Press; 1998. pp. 92–111. [Google Scholar]
- 18.Meng J, Zhao S, Doyle M P. Virulence genes of Shiga toxin-producing Escherichia coli isolated from food, animals and humans. Int J Food Microbiol. 1998;45:229–235. doi: 10.1016/s0168-1605(98)00163-9. [DOI] [PubMed] [Google Scholar]
- 19.Meng J, Zhao S, Doyle M P, Joseph S W. Antibiotic resistance of Escherichia coli O157:H7 and O157:NM isolated from animals, food, and humans. J Food Prot. 1998;61:1511–1514. doi: 10.4315/0362-028x-61.11.1511. [DOI] [PubMed] [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7–A5. Wayne, Pa: National Committee for Clinical Laboratory Standards; 2000. [Google Scholar]
- 21.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. Approved standard M31-A. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1999. [Google Scholar]
- 22.O'Ryan M, Prado V. Risk of the hemolytic-uremic syndrome after antibiotic treatment of Escherichia coli O157:H7 infections. N Engl J Med. 2000;343:1271. [PubMed] [Google Scholar]
- 23.Proulx F, Turgeon J P, Delage G, Lafleur L, Chicoine L. Randomized, controlled trial of antibiotic therapy for Escherichia coli O157:H7 enteritis. J Pediatr. 1992;121:299–303. doi: 10.1016/s0022-3476(05)81209-0. [DOI] [PubMed] [Google Scholar]
- 24.Ratnam S, March S, Ahmed R, Bezanson G, Kasatiya S. Characterization of Escherichia coli serotype O157:H7. J Clin Microbiol. 1988;26:2006–2012. doi: 10.1128/jcm.26.10.2006-2012.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Recchia G D, Hall R M. Origins of the mobile gene cassettes found in integrons. Trends Microbiol. 1997;5:389–394. doi: 10.1016/S0966-842X(97)01123-2. [DOI] [PubMed] [Google Scholar]
- 26.Ridley A, Threlfall E J. Molecular epidemiology of antibiotic resistance genes in multiresistant epidemic Salmonella Typhimurium DT104. Microb Drug Resist. 1998;4:113–118. doi: 10.1089/mdr.1998.4.113. [DOI] [PubMed] [Google Scholar]
- 27.Sallen B, Rajoharison A, Desvarenne S, Mabilat C. Molecular epidemiology of integron-associated antibiotic resistance genes in clinical isolates of enterobacteriaceae. Microb Drug Resist. 1995;1:195–202. doi: 10.1089/mdr.1995.1.195. [DOI] [PubMed] [Google Scholar]
- 28.Sandvang D, Aarestrup F M, Jensen L B. Characterisation of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol Lett. 1998;160:37–41. doi: 10.1111/j.1574-6968.1998.tb12887.x. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt H, von Maldeghem J, Frosch M, Karch H. Antibiotic susceptibilities of verocytotoxin-producing Escherichia coli O157 and non-O157 strains isolated from patients and healthy subjects in Germany during 1996. J Antimicrob Chemother. 1998;42:548–550. doi: 10.1093/jac/42.4.548. [DOI] [PubMed] [Google Scholar]
- 30.Shiomi M, Togawa M, Fujita K, Murata R. Effect of early oral fluoroquinolones in hemorrhagic colitis due to Escherichia coli O157:H7. Pediatr Int. 1999;41:228–232. doi: 10.1046/j.1442-200x.1999.4121038.x. [DOI] [PubMed] [Google Scholar]
- 31.Slutsker L, Ries A, Greene K, Wells J, Hutwagner L, Griffin P. Escherichia coli O157:H7 diarrhea in the United States: clinical and epidemiologic features. Ann Intern Med. 1997;126:505–513. doi: 10.7326/0003-4819-126-7-199704010-00002. [DOI] [PubMed] [Google Scholar]
- 32.Sunde M, Sorum H. Characterization of integrons in Escherichia coli of the normal intestinal flora of swine. Microb Drug Resist. 1999;5:279–287. doi: 10.1089/mdr.1999.5.279. [DOI] [PubMed] [Google Scholar]
- 33.Tollefson L, Altekruse S F, Potter M E. Therapeutic antibiotics in animal feeds and antibiotic resistance. Rev Sci Tech. 1997;16:709–715. doi: 10.20506/rst.16.2.1054. [DOI] [PubMed] [Google Scholar]
- 34.Witte W. Medical consequences of antibiotic use in agriculture. Science. 1998;279:996–997. doi: 10.1126/science.279.5353.996. [DOI] [PubMed] [Google Scholar]
- 35.Yoh M, Honda T. The stimulating effect of fosfomycin, an antibiotic in common use in Japan, on the production/release of verotoxin-1 from enterohaemorrhagic Escherichia coli O157:H7 in vitro. Epidemiol Infect. 1997;119:101–103. doi: 10.1017/s0950268897007541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao S, Meng J, Doyle M P, Meinersman R, Wang G, Zhao P. A low molecular weight outer-membrane protein of Escherichia coli O157:H7 associated with adherence to INT407 cells and chicken caeca. J Med Microbiol. 1996;45:90–96. doi: 10.1099/00222615-45-2-90. [DOI] [PubMed] [Google Scholar]