Abstract
We investigated the bacterial community structure in an aerated plug-flow lagoon treating pulp and paper mill effluent. For this investigation, we developed a composite method based on analyses of PCR amplicons containing the ribosomal intergenic spacer (RIS) and its flanking partial 16S rRNA gene. Community percent similarity was determined on the basis of RIS length polymorphism. A community succession was evident in the lagoon, indicated by a progressive community transition through seven sample locations. The most abrupt changes in community structure were associated with a temperature change from 39 to 35°C and with increases in dissolved oxygen. The temporal differences in community structure, based on summer and winter samplings, were greater than the spatial differences during either season. Clone libraries of rDNA-RIS amplicons were constructed from each of three summer samples. Among 90 clones analyzed (30 clones from each sample), 56 phylotypes were distinguished by restriction fragment length polymorphism. Indices of phylotype richness, evenness, and diversity all increased in clone libraries from the beginning to the end of the lagoon. A representative clone of each phylotype was phylogenetically analyzed on the basis of its partial 16S rRNA gene sequence (ca. 450 bp). Phylogenetic analysis confirmed the increase in diversity and further indicated increasing richness of bacterial divisions. Pioneers in the community spatial succession appeared to include thermotolerant, microaerophilic methanol-oxidizing bacteria related to the genus Methylobacillus, as well as thermotolerant, microaerophilic nitrogen-fixing bacteria related to the genus Azospirillum.
Aerated lagoons are commonly used for biotreatment of municipal and industrial wastewaters such as pulp and paper mill effluents. Such lagoons harbor complex microbial communities, which are selected by the physicochemical properties of the wastewater, the design and operation of the lagoon, and the ambient environmental conditions. These microbial communities are responsible for pollutant degradation and transformation. Functional stability of these microbial communities is essential for consistent and reliable pollutant degradation. These microbial communities, however, are subject to various perturbations, such as variations of influent pH, temperature, organic loading rates, and toxicant levels, as well as seasonal climate changes (30), and such changes do affect microbial community composition and system performance (45). Thus, temporal changes in microbial communities are expected in aerated lagoons. Further, in lagoons with a plug-flow regime, spatial differences between communities are likely to occur over the course of the system. Despite this realization, current design plans and models assume that microbial communities in biotreatment systems are homogeneous and temporally stable. There are very few reports of investigations of microbial community structure and dynamics in aerated lagoons. Ecological studies of these microbial communities will advance our fundamental understanding of microbial ecology and will provide useful information for improving the design and performance of aerated lagoons.
Bacterial communities have been compared on the basis of rRNA genes (rDNA) to investigate the interactions within ecosystems. Quantitative determinations of microbial community similarities have been made based on denaturing gradient gel electrophoresis (DGGE) banding profiles of PCR-amplified rDNA fragments. In most studies, only the absence or presence of common bands was incorporated in the calculation of similarity coefficients (28, 31, 32). Although these similarity coefficients improved upon qualitative comparison of banding patterns, they likely overestimate community similarity, since they do not take into account band density, a function of the relative abundance of a phylotype. Thus, these coefficients do not reflect evenness and related aspects of community structure (i.e., the phylotypes present are equally weighted whether predominant or barely detectable). Because of various potential PCR biases and variability of rDNA copy numbers among organisms, it would not be appropriate to consider band intensity to directly represent the relative abundance of the population represented by a band. However, band intensity will indicate the relative differences in a population and so will provide useful information for comparing community profiles. Recently, researchers have started to consider band intensity in community comparison (46).
Despite their usefulness in characterizing microbial communities, 16S rDNA sequences are sometimes not divergent enough to distinguish species of the same genus (34). Bacterial strains with distinct physiologies have been reported to have identical 16S rRNA genes (35). Unlike the 16S rDNA, the 16S-23S rDNA ribosomal intergenic spacer (RIS) has a highly variable length (19). Fisher and Triplett (14) examined the 307 RIS sequences then available in the GenBank and found 200 length classes. Also, RIS sequences are much more variable than 16S rDNA sequences. The RIS has been used as a marker to distinguish species and strains of a species (23, 43). Thus, restriction fragment length polymorphism (RFLP) analysis of the RIS may resolve bacteria in environmental samples at the species and, perhaps, subspecies levels, where important functional distinctions may occur. The RIS appears to be genetically stable over many generations of bacteria investigated (17, 18). Although, a few bacteria have unlinked 16S and 23S genes (20, 27, 38), the vast majority of bacteria examined have RISs within rRNA operons (15).
Recently, RIS analysis (RISA) has been exploited in investigating bacterial communities (1, 8, 14, 15, 37), mostly by qualitative comparison of the banding patterns resulting from size separation of RIS PCR amplicons from communities. In one case, communities have been compared using Sorensen's index calculated on the basis of RIS amplicon banding patterns (15). As suggested by Fisher and Triplett (14), quantitative analysis of the relative abundance of RIS amplicons has the potential to yield important additional information about community structure. Finally, analyzing the 16S rDNA sequences linked to RISs has the potential to provide additional information about the identity of community members represented by RISs. Since some organisms have multiple, distinct RISs, community RIS profiles may contain multiple bands from some organisms. This fact may bias evaluation of community structure on the basis of RIS profiles, but this would not be expected to cause errors greater than those typical of other culture-based or molecular methods for analysis of microbial communities. Importantly, all potential biases inherent to RISA are not a major impediment to comparing microbial communities analyzed with a consistent method.
In this study we examined the spatial differences in bacterial community structure in a plug-flow aerated lagoon treating pulp mill effluents. We related those differences to physicochemical gradients in the lagoon. We also compared the spatial differences to temporal differences over a 5-month period. We analyzed the community using a new composite RISA method. This method includes analyses of (i) RIS length polymorphism (RIS-LP) to quantify community similarity, (ii) RFLP to identify and enumerate phylotypes in rDNA-RIS clone libraries, and (iii) sequences of the partial 16S rDNA linked to the RIS to identify the phylotypes.
MATERIALS AND METHODS
Sampling and chemical analyses.
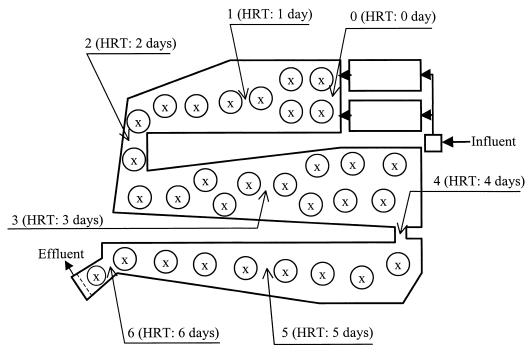
Mixed liquor samples were collected in September 1998 and February 1999 from the aerated lagoon of the Weyerhaeuser Canada pulp and paper mill in Kamloops, British Columbia (119.6°W, 50.7°N). The lagoon is S-shaped, and the influent passes through the lagoon in about 6 days, which is the hydraulic retention time (HRT) (Fig. 1). Sample identifications refer to the month (SEP or FEB) and the location of sampling (locations 1 to 6 indicated in Fig. 1). From a small boat, 1-liter grab samples of mixed liquor were removed from the surface of the lagoon near its center. The samples were shipped to our laboratory on ice, and the biomass was harvested within 24 h of sampling by centrifugation at 4°C for 10 min at 17,000 × g and stored at −70°C until use for DNA extraction. For SEP samples biomass was harvested from 250 ml of mixed liquor, and for FEB samples biomass was harvested from 450 ml. At the sampling locations in the lagoon, the temperature, pH, and dissolved oxygen (DO) were measured in situ. Total organic carbon (TOC) and biomass (as volatile suspended solids [VSS]) were determined according to standard procedures (2). The resin acid concentrations were measured using gas chromatography as reported previously (47).
FIG. 1.
A schematic representation of the plug-flow aerated lagoon investigated in this study. The HRT of the lagoon (not including in the settling pond before the lagoon) was about 6 days. The number of each sampling location (from 0 to 6) corresponds approximately to the HRT (in days) at each location. Circles represent surface aerators.
DNA extraction and PCR.
Total community DNA was extracted from the biomass using the method of Yu and Mohn (48). The resultant DNA samples were diluted 10-fold and used in PCR amplification (ca. 200 ng of DNA per PCR reaction) with primers S926f (5′-CTYAAAKGAATTGACGG-3′) and L189r (5′-TACTGAGATGYTTMARTTC-3′), which anneal to positions 910 to 926 of the 16S rRNA gene and positions 189 to 207 of the 23S rRNA gene (Escherichia coli numbering), respectively. The resultant PCR products contain the complete RIS and parts of the flanking rDNAs (ca. 600 bp of 16S rDNA and 190 bp of 23S rDNA) and are referred to as rDNA-RIS. PCR amplification was conducted in a total volume of 50 μl containing 25 pmol of each primer, 200 μM concentrations of each deoxynucleoside triphosphate, 1.5 mM 1× PCR buffer (20 mM Tris-HCl, pH 8.4; 50 mM KCl), 670 μg of bovine serum albumin per ml, and 1.25 U of Taq DNA polymerase (Life Technologies, Burlington, Ontario, Canada). Following a simplified hot start, the DNA templates were first subjected to an initial denaturation at 95°C for 2 min. The subsequent cycles consisted of a 0.5-min denaturation step at 94°C, a 0.5-min annealing step at 47°C, and a 2-min extension step at 72°C. After 30 cycles, there was a final 5-min extension at 72°C. Negative controls containing no DNA template were included in parallel.
The PCR method was optimized to yield the maximum number of rDNA-RIS bands from lagoon samples. A second reverse primer, L23r, targeting the 23S rDNA was tested and rejected. Inclusion in the PCR of betaine (0.5 to 2.5 M) with or without dimethyl sulfoxide (5 to 10%) did not improve the reaction. Concentrations of 1.5 to 2.5 mM MgCl2 were tested. Annealing temperatures of 45 to 60°C were tested.
Community percent similarity based on RIS-LP.
PCR-amplified rDNA-RIS fragments were separated on 3.5% polyacrylamide (38:1) gels, which were then stained with GelStar (MFC BioProducts, Rockland, Maine), which is more sensitive than ethidium bromide. The RIS-LP banding patterns were documented using an AlphaImager 1200 (Alpha Innotech, San Leandro, Calif.). Individual bands were detected by the AlphaEase program (version 4) of the AlphaImager. The relative mobility of each band was calculated using the 800-bp band of the 100-bp molecular size marker as the reference. The density and percent relative abundance of each band was calculated using the 1D-MULTI program of AlphaEase. For band matching, a maximum tolerance of 1% was chosen. Community percent similarity was calculated from pairwise comparisons of the RIS-LP banding patterns. Community percent similarity was calculated as the sum of shared relative abundances of all matching bands. The shared relative abundance of each matching band was defined as the lower of the two percent relative abundances. Subtraction of the community percent similarity from 100% yielded the community dissimilarities, which were then used to create dissimilarity matrices. Dendrograms (UPGMA) were generated from the above dissimilarity matrices using the neighbor-joining program in the Phylip package (13).
Clone library construction, RIS-RFLP analysis, and 16S rDNA sequencing.
Clone libraries of the PCR-amplified rDNA-RIS fragments from SEP-0, SEP-3, and SEP-6 were constructed using the TOPO cloning kit (Invitrogen, Carlsbad, Calif.). From each clone library, 30 white colonies were randomly picked. Clone identifications have two numbers, the first of which indicates the sample of origin (e.g., Kmpls3-5 is the fifth clone picked from sample SEP-3). Clones were screened with PCR for the presence of the rDNA-RIS inserts using primer pairs M13f(−20)-S926f and M13f(−20)-L189r in separate PCR reactions. The length of each rDNA-RIS insert was determined by electrophoresis. The PCR screening also determined the orientation of the rDNA-RIS inserts, which facilitated sequencing of the 16S rDNA region of the rDNA-RIS inserts. The PCR products of the positive clones from the above screening assays were purified using the QIAquick PCR purification kit (Qiagen, Mississauga, Ontario, Canada), and two aliquots were digested separately with MspI and MboI (Life Technologies) for 2 h at 37°C. The above digests were resolved on 2.5% agarose gels, and the RFLP patterns within each clone library were compared and grouped. The RFLP patterns were also compared between the different clone libraries.
One clone from each RFLP group (phylotype) within each clone library was chosen for sequencing. The recombinant plasmid DNA from the selected clones was isolated using the QIAprep miniprep kit (Qiagen). The 16S rDNA region of those recombinant plasmids was sequenced as described previously (47) using the M13 primer flanking the 16S rDNA region.
Calculation of diversity indices.
On the basis of RFLP phylotypes, indices of phylotype richness, evenness and diversity were calculated according to the method of Atlas and Bartha (5). The diversity of the phylotypes was further examined using rarefaction analysis (22, 42). Rarefaction calculations were performed using the program aRarefactWin, version 1.2 (available at http://www.uga.edu/∼Strata/AnRareReadme.html).
Phylogenetic analysis of rDNA sequences.
The determined partial 16S rDNA sequences (ca. 450 bp, nucleotide positions 910 to 1360, E. coli numbering) were evaluated using the program Chimera Check implemented in the Ribosomol Database Project (RDP [29]) to detect potential chimeric artifacts, and none were detected. The partial 16S rDNA sequences of the representative clones were aligned against the most similar sequence in the RDP using the program Sequence Aligner implemented in the RDP (29). Then, the above alignments were manually edited using GeneDoc (33). Evolutionary distances were calculated according to the model of Jukes and Cantor (24), and neighbor-joining trees (39) were constructed using the Phylip package (13). Only unambiguous positions were used in the phylogenetic analyses. Bootstrap analysis was performed using SEQBOOT with 100 replicates. Sequence identity (%) was calculated from aligned sequences using the program BioEdit (available at http://www.mbio.ncsu.edu/RNaseP/info/programs/BIOEDIT/bioedit.html).
Nucleotide sequence accession numbers.
The partial rDNA sequences determined for the phylotypes have been deposited in the GenBank under the following accession numbers: Kmlps0-1, AF289867; Kmlps0-2, AF289868; Kmlps0-4, AF289869; Kmlps0-12, AF289870; Kmlps0-14, AF289871; Kmlps0-16, AF289872; Kmlps0-17, AF289873; Kmlps0-22 to Kmlps0-24, AF289874 to AF289876; Kmlps0-27, AF289877; Kmlps0-28, AF289878; Kmlps3-1, AF289879; Kmlps3-2, AF289880; Kmlps3-5 to Kmlps3-13, AF289881 to AF289889; Kmlps3-15, AF289890; Kmlps3-19, AF289891; Kmlps3-20, AF289892; Kmlps3-23, AF289893; Kmlps3-24, AF289894; Kmlps3-26, AF289895; Kmlps3-29 to Kmlps3-31, AF289896 to AF289898; Kmlps6-1 to Kmlps6-8, AF289899 to AF289906; Kmlps6-10, AF289907; Kmlps6-12, AF289908; Kmlps6-13, AF289920; Kmlps6-14 to Klps6-18, AF289909 to AF289913; Kmlps6-20, AF289914; Kmlps6-21, AF289915; Kmlps6-23, AF289916; Kmlps6-25, AF289917; Kmlps6-26, AF289921; Kmlps6-29, AF289992; Kmlps6-30, AF289918; and Kmlps6-32, AF289919.
RESULTS
Physicochemical conditions in the lagoon.
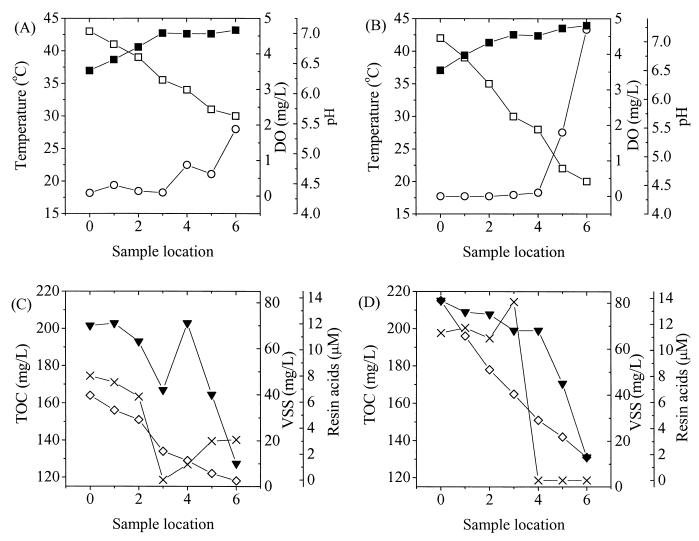
On both sampling dates, from the beginning to the end of the lagoon, there were decreases in temperature, TOC, VSS, and total resin acids, as well as increases in DO and pH (Fig. 2). These spatial gradients were due to the plug-flow regime of the lagoon and were generally gradual and constant. The most abrupt changes occurred in DO, which rose abruptly, presumably when biological oxygen demand was depleted. At each sampling location, the physicochemical conditions differed on the two sampling dates. In general, the gradients were steeper in February 1999 than in September 1998, and the end of the lagoon was 10°C colder on the former date.
FIG. 2.
Chemical and physical parameters of the lagoon samples taken in September 1998 (A and C) and February 1999 (B and D). Symbols: □, temperature; ■, pH; ○, DO; ◊, TOC; ▾, VSS; ×, resin acids.
Spatial and temporal community changes based on RIS-LP analysis.
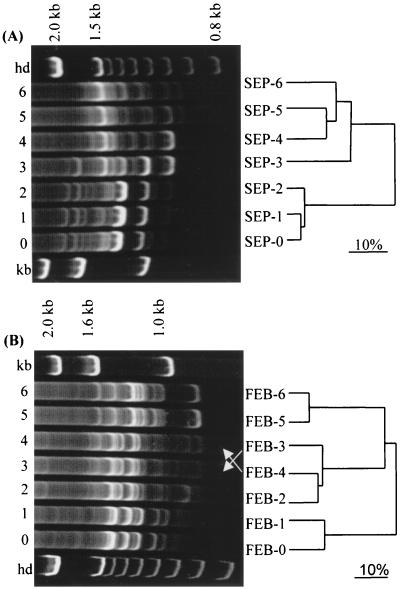
Electrophoretic separation of the total rDNA-RIS amplicons from each sample resulted in distinct RIS-LP banding patterns (Fig. 3). These patterns comprised 14 to 20 bands, including one or more major bands in each sample. Community percent similarities indicated spatial and temporal differences between communities (Fig. 3). On each sampling date, samples from adjacent locations generally had the highest levels of community similarity. Cluster analysis confirmed this trend of a progressive community transition (Fig. 3). While replicate samples were not analyzed, the pattern of transition observed indicates that any random spatial variability of RIS-LP patterns was less than the differences measured between patterns from different sample locations.
FIG. 3.
Comparison by RIS-LP analysis of different communities sampled in September 1998 (A) and February 1999 (B). The gels show the RIS-LP banding patterns; the UPGMA dendrograms show clustering analysis based on percent similarities of the communities. hd, 100-bp DNA molecular marker; kb, 1-kb DNA molecular marker. The scale bar represents 10% community dissimilarity.
Among the September samples, the greatest community discontinuity (56% similarity) occurred between SEP-2 and SEP-3 (Fig. 3). Samples SEP-3, SEP-4, SEP-5, and SEP-6 then evidenced a more gradual community transition. Among the February samples, the greatest community discontinuities occurred between FEB-1 and FEB-2 (34% similarity) and between FEB-4 and FEB-5 (38% similarity). In general, the samples from each date had greater community similarity among themselves (22 to 93% similarity) than with samples from the other date (9 to 51% similarity), indicating greater temporal than spatial community changes in the lagoon ecosystem.
RIS-RFLP analysis of clone libraries.
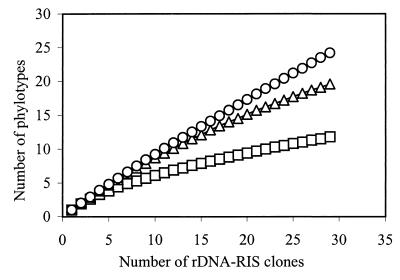
Each RFLP pattern was considered to represent a unique phylotype. Indices of RIS-RFLP phylotype richness, evenness, and diversity increased in libraries originating from the beginning to the end of the lagoon (Table 1). The largest increase in diversity was between SEP-0 and SEP-3 and was due to increases in both richness and evenness. A smaller increase in diversity occurred between SEP-3 and SEP-6 and was mainly due to an increase in richness. Rarefaction analysis confirmed the relative phylotype richness of each sample (Fig. 4). The rarefaction curves did not reach a plateau, indicating that the collections of 30 clones per sample did not represent all of the less-abundant phylotypes in those samples, particularly in samples SEP-3 and SEP-6. Thus, the indices based on these collections should be regarded as conservative estimates of the actual values.
TABLE 1.
Diversity indices determined based on RIS-RFLP for three clone libraries from lagoon samples taken in September 1998
| Sample | No. of unique phylotypes | Phylotype
|
||
|---|---|---|---|---|
| Richnessa | Evennessb | Diversityc | ||
| SEP-0 | 12 | 7.44 | 1.93 | 2.08 |
| SEP-3 | 20 | 12.86 | 2.20 | 2.87 |
| SEP-6 | 24 | 16.25 | 2.24 | 3.10 |
The number of different phylotypes relative to the number of individuals sampled.
The evenness of distribution of individuals among the phylotypes.
Shannon-Wiener index, which incorporates richness and evenness.
FIG. 4.
Rarefaction curves for the different phylotypes of rDNA-RIS clones. The number of different phylotypes in each clone library was determined by RIS-RFLP using MspI and MboI in separate restriction digestions. Rarefaction curves were calculated with the analytical approximation algorithm described by Hurlbert (22). Symbols: □, SEP-0; ▵, SEP-3; ○, SEP-6.
Two predominant RIS-RFLP phylotypes were found in sample SEP-0, while SEP-3 and SEP-6 did not have such predominant phylotypes (Fig. 5 to 7). Several phylotypes occurred in more than one sample (Table 2). Comparison of the RIS-LP bands (Fig. 3) and the clone insert sizes (not shown) indicated that some RIS-LP bands corresponded to more than one RIS-RFLP phylotype. For this reason, the most dense RIS-LP band in a sample did not always correspond to the most abundant RIS-RFLP phylotype. However, all of the major RIS-LP bands in each sample did correspond to an rDNA-RIS clone insert of equal size and the most abundant RIS-LP band from each sample corresponded to the most abundant clone insert from that sample. This observation indicates that any cloning bias was not severe.
FIG. 5.
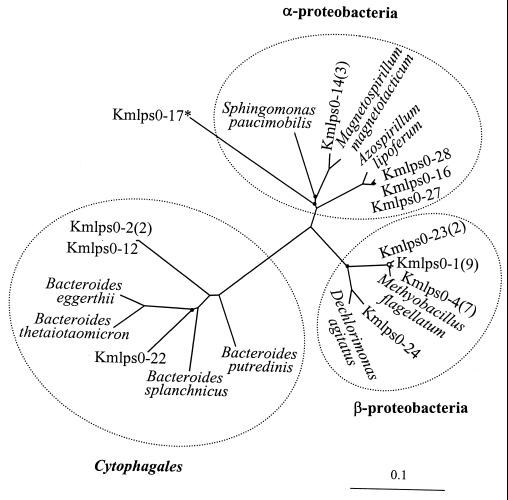
Phylogenetic tree (unrooted) showing the affiliations of the partial 16S rDNA sequences (E. coli positions 910 to 1360) determined from the SEP-0 sample. The most similar RDP sequence is included as a reference for each sequence. The reference strains are S. paucimobilis IFO 13935, M. magnetotacticum DSM 3856, A. lipoferum NCIMB 11861, M. flagellatum KT1, D. agitatus CKB, B. putredinis ATCC 29800, B. splanchnicus NCTC 10825, B. thetaiotaomicron ATCC 29148, and B. eggerthii NCTC 11185. Branch points with open circles have bootstrap values of less than 50%; branch points with filled circles have bootstrap values of 50 to 74%. Other branch points have bootstrap values greater than 75%. Numbers in parentheses following clone identifications indicate the number of members of the RIS-RFLP phylotype represented by that clone. The asterisk indicates the sequence that is most similar to mitochondrial 16S-like rDNA of R. americana. The scale bar corresponds to an estimated 0.1 mutation per nucleotide position.
FIG. 7.
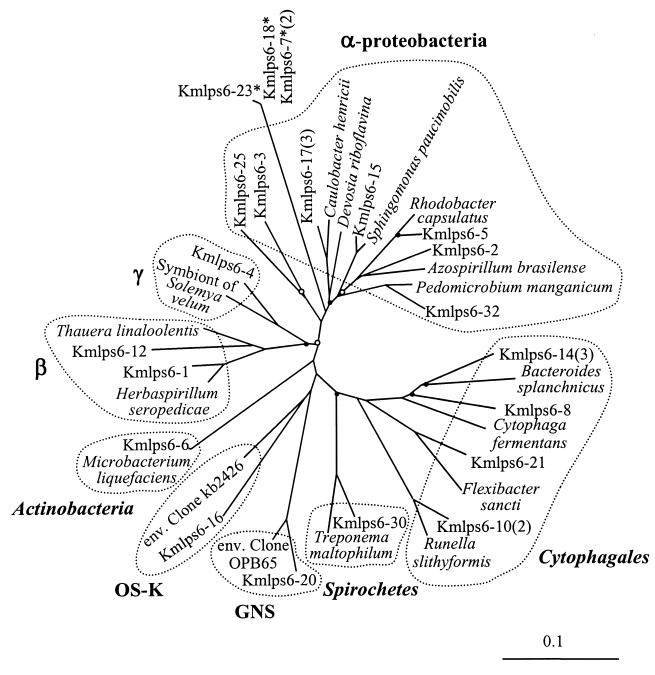
Phylogenetic tree (unrooted) showing the affiliations of the partial 16S rDNA sequences determined for the SEP-6 sample. The reference strains are C. henricii ATCC 29530, D. riboflavina IFO 13584, R. capsulatus C5, A. brasilense NCIMB 11860, P. manganicum ACM 3038, C. fermentans ATCC 19072, F. sancti ATCC 23092, R. slithyformis ATCC 29530, T. maltophilum BR, M. liquefaciens DSM 20638, H. seropedicae DSM 6445, and others indicated in the legend of Fig. 5. See the legend for Fig. 5 for further explanation.
TABLE 2.
Clusters of phylogenetically related RIS-RFLP phylotypes from the samples collected in September 1998
| RIS-RFLP phylotype | Occurrencea
|
Representative Kmlps clone(s)b | Affiliationc (reference strain) | Identity to reference strain (%) | Identity within cluster (%) | ||
|---|---|---|---|---|---|---|---|
| SEP-0 | SEP-3 | SEP-6 | |||||
| 1 | 9 | 2 | 0-1/3-15 | M. flagellatum KT1 | 94.4/95.1 | 98.4–99.5 | |
| 2 | 7 | 1 | 0-4/3-24 | M. flagellatum KT1 | 94.6/95.1 | ||
| 3 | 2 | 1 | 0-23/3-8 | M. flagellatum KT1 | 94.2/94.6 | ||
| 4 | 2 | 3-20 | M. flagellatum KT1 | 94.2 | |||
| 5 | 1 | 0-28 | A. lipoferum NCIMB 11861 | 96.4 | 99.1–99.5 | ||
| 6 | 1 | 1 | 0-16/3-13 | A. lipoferum NCIMB 11861 | 95.5/95.5 | ||
| 7 | 1 | 1 | 0-27/3-19 | A. lipoferum NCIMB 11861 | 96.2/96.2 | ||
| 8 | 1 | 2 | 0-12/3-2 | B. putredinis ATCC 29800 | 83.9/84.0 | 99.3–99.7 | |
| 9 | 1 | 0-2 | B. putredinis ATCC 29800 | 83.9 | |||
| 10 | 2 | 3-12 | B. putredinis ATCC 29800 | 84.0 | |||
| 11 | 1 | 3-31 | B. putredinis ATCC 29800 | 83.7 | |||
| 12 | 1 | 4 | 1 | 0-17/3-1/6-18 | Mitochondrion of R. americana ATCC 50394 | 91.1/90.7/91.3 | 98.7–99.7 |
| 13 | 2 | 3-7 | Mitochondrion of R. americana ATCC 50394 | 90.9 | |||
| 14 | 1 | 3-29 | Mitochondrion of R. americana ATCC 50394 | 90.9 | |||
| 15 | 2 | 6-7 | Mitochondrion of R. americana ATCC 50394 | 91.1 | |||
| 16 | 1 | 6-23 | Mitochondrion of R. americana ATCC 50394 | 90.7 | |||
Number of clones from each sample belonging to each phylotype.
Rows with multiple clones indicate representative phylotypes from each sample.
Organism in RDP with most similar rDNA sequence.
rDNA sequence analysis.
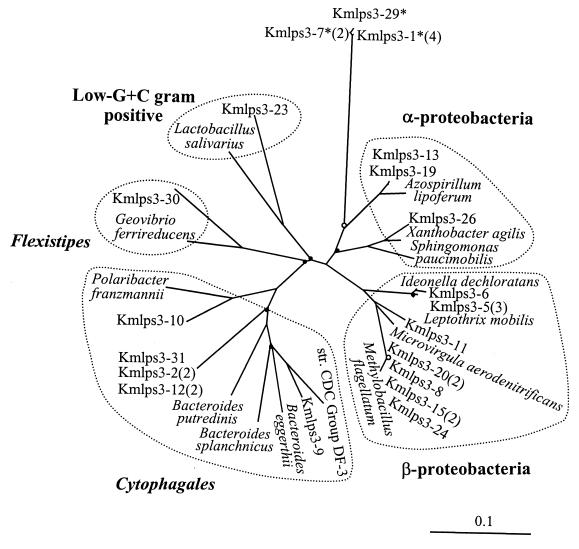
The rDNA-RIS fragments have about 600 bp of 16S rDNA (ca. 450 bp were sequenced in this study), which is sufficient for phylogenetic identification, at approximately the genus level, of organisms belonging to groups well represented in sequence databases. Overall, the greatest numbers of cloned partial rDNA sequences were associated with the Proteobacteria (28 sequences) and the Cytophagales (Flexibacter-Cytophaga-Bacteroides group) (12 sequences), which were found in all three samples. The sequences from samples SEP-0, SEP-3, and SEP-6, respectively, evidenced a clear trend toward increasing richness of higher taxa (Fig. 5 to 7), such as divisions, as defined by Hugenholtz et al. (21). Sequences from SEP-0, SEP-3, and SEP-6 were associated with two, four, and six bacterial divisions, respectively.
Several clusters of very similar sequences (98.4 to 99.7% sequence identity) were found in the samples (Table 2). Sequences closely affiliated with Methylobacillus flagellatum dominated sample SEP-0 and were abundant in SEP-3. Sequences closely affiliated with Azospirillum lipoferum and ones less closely affiliated with Bacteroides putredinis were found in both SEP-0 and SEP-3. Sample SEP-6 did not have sequences from the above three clusters. Finally, sequences affiliated with the mitochondrion of Reclinomonas americana were detected in all three samples, suggesting the occurrence of protozoa, particularly in the middle to the end of the system.
One sequence from sample SEP-3 (Kmlps3-11) is most similar to the 16S rDNA of DhA-73 (GenBank accession no. AF125876), a thermophilic, resin-acid-degrading β-proteobacterium isolated from a bioreactor treating pulp mill effluent (49). Three sequences from sample SEP-6 (Kmlps6-13, -6-26, and -6-29) are most similar to the fungal 18S rDNA (positions 1131 to 1653, Saccharomyces cerevisiae numbering) of Spiromyces aspiralis (94% identical), Monoblepharella elongata (93% identical), and Monoblepharella elongata (98% identical), respectively. Several of the determined sequences in each sample were not closely associated with any recognized genus, but all of the bacterial sequences appear to be associated with recognized divisions. In one case, a sequence was affiliated with a candidate division, OS-K (Fig. 7). Each sequence had greater than 83% identity to a reference strain sequence, while strains of different divisions typically have less than 80% identity (21).
Within the above sequence clusters, the distinct RIS-RFLP phylotypes originated from RISs of the same length (i.e., the same clone insert size). There were only four exceptions to this generalization (Kmlps3-20, -0-28, -0-27, and -3-12), within their respective clusters (Table 2). Three pairs of clones with distinct RIS-RFLP phylotypes had identical partial rDNA sequences (Kmlps3-15 and -3-24, -3-2 and -3-12, and -0-17 and -6-7). These pairs presumably represent distinct rrn operons occuring in one organism or distinct strains of very closely related organisms. Seven RIS-RFLP phylotypes were found in more than one sample (Table 2). In one of these seven cases, the representative clones had identical rDNA sequences (Kmlps0-27 and -3-19). In the other six cases, the sequence similarity is very high. Some of the sequence differences could be due to sequencing errors. In all cases, identical RIS-RFLP patterns corresponded to identical or nearly identical rDNA sequences. Thus, the sequence analysis confirmed interpretation of RIS-RFLP analyses by indicating the phylogenetic relevance of the RIS-RFLP phylotypes.
DISCUSSION
RIS-LP allowed determination of percent similarity in pairwise comparisons of bacterial communities based on banding profiles from electrophoresis of rDNA-RIS amplicons. This comparison improved upon previous methods in which size-separated RIS amplicons were qualitatively compared (1, 8, 14, 37). By incorporating the relative abundance of amplicons, the percent similarity determination also improved upon previously used similarity coefficients based only upon the absence or presence of DNA bands, such as those resulting from DGGE separation of 16S rDNA amplicons (28, 31, 32). The progressive community transition observed on both sampling dates (Fig. 3) is consistent with the physicochemical data (discussed below). This pattern suggests that the community percent similarities accurately reflect community structure and are related to community function.
The progressive community transition as the wastewater passed through the lagoon (Fig. 3, Table 1) can be viewed as a community succession. This “spatial succession” does have a temporal component if considered as a function of the hydraulic retention time of wastewater in the lagoon (Fig. 1). This succession is likely related to the changing physicochemical conditions in the system (Fig. 2). The succession was partly allogenic, a consequence of external factors. For example, the decreasing temperature in the lagoon would have a selective effect on the community. The succession was also partly autogenic, a consequence of the community's activities. For example, the reduction of degradable organic carbon depleted certain substrates, including methanol and resin acids. Degradation of organic carbon depleted DO to very low levels at the beginning of the system. Microbial activity may have affected the pH. Also, grazing by protozoan members of the community likely affected community structure. Thus, the changing physicochemical conditions in the lagoon were both a cause and a result of the community succession.
Certain physicochemical factors appeared to be particularly import in effecting community changes. On both sampling dates, the most abrupt change in the community occurred where the temperature dropped from 39 to 35°C (Fig. 2 and 3). This community change is consistent with the fact that most mesophilic bacteria have temperature maxima near 35°C. It appears that this threshold may have been a major factor in determining community structure. In February 1999, a second major change in the community also occurred where the DO first began to substantially increase (to rise above 0.5 mg/liter) in the lagoon. This change suggests that DO may have been another important determinant of community structure.
RIS-LP analysis had less resolution than RIS-RFLP analysis, since some single RIS-LP bands yielded multiple RIS-RFLP phylotypes. The resolution of RIS-LP analysis could be increased by improved separation techniques, such as the two-dimensional gel electrophoresis that was previously used for mutational analysis (25). We conclude that RIS-LP analysis, by the current method, permits meaningful comparisons of community similarity, but it does not permit determination of community structure with the accuracy of the more laborious analysis of RIS-RFLP or rDNA sequences. RIS-LP analysis is particularly useful to determine which samples of a large set merit further comparison by more laborious methods, as was done in this study.
The available evidence supports the conclusion that the community's spatial succession in the lagoon involved increasing phylogenetic diversity from the beginning to the end of the lagoon. RIS-RFLP analysis indicated a clear trend of increasing diversity in the clone libraries, from the September 1998 samples, from the beginning to the end of the lagoon (Table 1, Fig. 4). The parallel analysis of these samples makes it likely that any potential biases in the method would affect the samples similarly. While the quantitative data (e.g., diversity indices) may not apply directly to the community structure, they do permit meaningful comparisons between the samples. The relative differences between the clone libraries probably do reflect a real trend in the diversity of the community.
This trend of increasing diversity through the course of the lagoon is also consistent with what one would expect on the basis of the physicochemical data. Relatively low phylogenetic diversity at the beginning of the lagoon (Table 1; Fig. 5) is consistent with the relatively high temperature, low DO level, and high concentrations of organic compounds, some of which may be toxic (Fig. 2). This situation has the characteristics of a physicochemically controlled community, which is predicted to have relatively low diversity (3, 4). Such communities are considered typical of disturbed environments, including polluted environments. Succession leading to increasing diversity in the middle and end of the lagoon (Table 1; Fig. 6 and 7) is consistent with the moderating conditions (Fig. 2). This situation has the characteristics of a biologically controlled community, which is predicted to have relatively high diversity and increasing population interactions.
FIG. 6.
Phylogenetic tree (unrooted) showing the affiliations of the partial 16S rDNA sequences determined from the SEP-3 sample. The reference strains are X. agilis SA35, I. dechloratans CCUG 30898, L. mobilis DSM 10617, M. aerodenitrificans SGL Y2, P. franzmannii 301, G. ferrireducens. PAL-1, L. salivarius subsp. salivarilus ATCC 11741, and others indicated in the legend of Fig. 5. See the legend for Fig. 5 for further explanation.
The inverse relationship between temperature and microbial diversity in the lagoon is consistent with a number of other reports. A trend very similar to that in the lagoon was reported for a marine hydrothermal vent; as water flowed away from the vent and cooled, the microbial diversity in the water increased (41). Similarly, microbial succession in compost systems involved increasing diversity as temperature decreased during the latter part of the process (7, 36). A comparison of communities in treatment systems for pharmaceutical wastewater revealed less diversity in those at high temperature (50 to 58°C) than in those at moderate temperature (28 to 32°C) (26). A comparison of communities in anaerobic systems treating a synthetic wastewater suggested that one at 55°C was less diverse than one at 35°C (40).
In some cases, phylogenetic association permitted plausible inferences about the functions of community members in the aerated lagoon. The 16S rDNA analysis indicates that organisms in two phylogenetic clusters were among the pioneer populations in the lagoon (Table 2; Fig. 5 and 6). The niches of these two groups can be surmised on the basis of the physicochemical conditions of the lagoon and the physiological characteristics of their closest known relatives. The first cluster appeared to be dominant at the beginning and relatively abundant in the middle of the lagoon. This cluster is closely related to Methylobacillus flagellatum, which is an obligate methylotroph (9). Pulp mill effluents have a high methanol content, comprising up to 15% of biological oxygen demand (6). Thus, the first cluster likely represents a population of thermophilic methylotrophs tolerant of low levels of DO. The second cluster appeared to be relatively abundant at the beginning and middle of the lagoon. This cluster is closely related to Azospirillum lipoferum, which is a nitrogen fixer (12). Pulp mill effluents have low nitrogen content, because of the low C:N ratio of wood. Nitrogen fixation has been reported in aerated lagoons treating pulp mill effluents (10, 11) and in stabilization basins of activated sludge systems treating such effluents (16). Many free-living nitrogen-fixing bacteria are microaerophilic. Several microaerophilic free-living nitrogen-fixing bacteria have been enriched from this lagoon (unpublished data). Thus, the second cluster likely represents a population of thermophilic, microaerophilic nitrogen fixers.
The 16S rDNA sequence analysis confirmed that RIS-RFLP phylotype diversity observed in the lagoon was related to phylogenetic diversity at all taxonomic levels (Fig. 5 to 7). It has been suggested that consideration of higher taxa is an important aspect of evaluating diversity (44). The increasing bacterial phylogenetic diversity over the course of the lagoon in September 1998 suggests a concomitant increase in functional diversity. Accordingly, the presence of phototrophic bacteria at the end of the lagoon is suggested by an rDNA sequence affiliated with the green nonsulfur bacterium (GNS) division (Fig. 7). The rDNA sequences affiliated with mitochondrial rDNA of Reclinomomas americana suggest the presence of predatory protozoa (Table 2). The presence of fungi is suggested by rDNA sequences affiliated with the 18S rDNA of aquatic fungi. Such fungi may use relatively recalcitrant components of pulp mill effluent, such as cellulose and lignin.
The bacterial community in the lagoon changed over a period of 5 months. During this time, the change in the bacterial community was large relative to the spatial differences in the community on either sampling date (Fig. 3). Substantial allogenic factors may have contributed to this temporal change. These factors include the ambient temperature, which had a substantial effect on the temperature gradient in the lagoon and the temperature at the end of the lagoon (Fig. 2). Temperature would have had both a direct selective effect on the bacterial community as well as indirect effects, including altering the hydrodynamics (such as convection), heat transfer, mass transfer, and oxygen solubility. Another potentially important factor was the chemical composition of the effluent entering the lagoon. This composition would have changed whenever the pulp and paper mill changed the tree species that were pulped or changed the pulping process in order to make various products.
ACKNOWLEDGMENTS
We thank Maari Hirvi of Weyerhaeuser Canada for collecting the samples and measuring some of the physicochemical properties of those samples.
This work was supported by the Sustainable Forest Management Network.
REFERENCES
- 1.Acinas S G, Antón J, Rodríguez-Valera F. Diversity of free-living and attached bacteria in offshore western Mediterranean waters as depicted by analysis of genes encoding 16S rRNA. Appl Environ Microbiol. 1999;65:514–522. doi: 10.1128/aem.65.2.514-522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.American Water Works Association, Water Pollution Control Federation, and American Public Health Association. Standard methods for the examination of water and wastewater. 19th ed. Washington, D.C.: American Public Health Association; 1995. [Google Scholar]
- 3.Atlas R M. Use of microbial diversity measurement to assess environmental stress. In: Klug M J, Reddy C A, editors. Current perspectives in microbial ecology. Washington, D.C.: American Society for Microbiology; 1984. pp. 540–545. [Google Scholar]
- 4.Atlas R M, Horowitz A, Krichevsky M, Bej A K. Response of microbial populations to environmental disturbance. Microb Ecol. 1991;22:249–256. doi: 10.1007/BF02540227. [DOI] [PubMed] [Google Scholar]
- 5.Atlas R M, Bartha R. Microbial communities and ecosystems. In: Atlas R M, Bartha R, editors. Microbial ecology: fundamentals and applications. Redwood, Calif: The Benjamin/Cummings Publishing Company; 1993. p. 142. [Google Scholar]
- 6.Blackwell B R, MacKay W B, Murray F E, Oldham W K. Review of kraft foul condensates. TAPPI J. 1979;62:33–37. [Google Scholar]
- 7.Blanc M, Marilley L, Beffa T, Aragno M. Thermophilic bacterial communities in hot composts as revealed by most probable number counts and molecular (16S rDNA) methods. FEMS Microbiol Ecol. 1999;28:141–149. [Google Scholar]
- 8.Borneman J, Triplett E W. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol. 1997;63:2647–2653. doi: 10.1128/aem.63.7.2647-2653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bratina B J, Brusseau G A, Hanson R S. Use of 16S rRNA analysis to investigate phylogeny of methylotrophic bacteria. Int J Syst Bacteriol. 1992;42:645–648. doi: 10.1099/00207713-42-4-645. [DOI] [PubMed] [Google Scholar]
- 10.Bruce M E, Clark T A. Klebsiella and nitrogen fixation in pulp and paper mill effluents and treatment systems. Appita J. 1994;47:231–237. [Google Scholar]
- 11.Clark T A, Dare P H, Bruce M E. Nitrogen fixation in an aerated stabilization basin treating bleached kraft mill wastewater. Water Environ Res. 1997;69:1039–1046. [Google Scholar]
- 12.Fani R, Bandi C, Bazzicalupo M, Ceccherini M T, Fancelli S, Gallori E, Gerace L, Grifoni A, Miclaus N, Damiani G. Phylogeny of the genus Azospirillum based on 16S rDNA sequence. FEMS Microbiol Lett. 1995;129:195–200. doi: 10.1111/j.1574-6968.1995.tb07579.x. [DOI] [PubMed] [Google Scholar]
- 13.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 14.Fisher M M, Triplett E W. Automated approaches for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl Environ Microbiol. 1999;65:4630–4636. doi: 10.1128/aem.65.10.4630-4636.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García-Martínez J, Acinas S G, Antón A I, Rodríguez-Valera F. Use of the 16S–23S ribosomal genes spacer region in studies of prokaryotic diversity. J Microbiol Methods. 1999;36:55–64. doi: 10.1016/s0167-7012(99)00011-1. [DOI] [PubMed] [Google Scholar]
- 16.Gauthier F, Neufeld J D, Driscoll B T, Archibald F S. Coliform bacteria and nitrogen fixation in pulp and paper mill effluent treatment systems. Appl Environ Microbiol. 2000;66:5155–5160. doi: 10.1128/aem.66.12.5155-5160.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gürtler V. Typing of Clostridium difficile strains by PCR-amplification of variable length 16S–23S rDNA spacer regions. J Gen Microbiol. 1993;139:3089–3097. doi: 10.1099/00221287-139-12-3089. [DOI] [PubMed] [Google Scholar]
- 18.Gürtler V, Barrie H D. Typing of Staphylococcus aureus strains by PCR-amplification of variable-length 16S–23S rDNA spacer regions: characterization of spacer sequences. Microbiology. 1995;141:1255–1265. doi: 10.1099/13500872-141-5-1255. [DOI] [PubMed] [Google Scholar]
- 19.Gürtler V, Stanisich V A. New approaches to typing and identification of bacteria using the 16S–23S rDNA spacer region. Microbiology. 1996;142:3–16. doi: 10.1099/13500872-142-1-3. [DOI] [PubMed] [Google Scholar]
- 20.Hartmann R, Ulbrich K N, Erdmann V A. An unusual rRNA operon constellation: in Thermus thermophilus HB8 the 23S/5S rRNA operon is a separate entity from the 16S rRNA operon. Biochimie. 1987;69:1097–1104. doi: 10.1016/0300-9084(87)90009-5. [DOI] [PubMed] [Google Scholar]
- 21.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hurlbert S H. The nonconcept of species diversity: a critique and alternative parameters. Ecology. 1971;52:577–586. doi: 10.2307/1934145. [DOI] [PubMed] [Google Scholar]
- 23.Jensen M A, Webster J A, Straus N. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl Environ Microbiol. 1993;59:945–952. doi: 10.1128/aem.59.4.945-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolisms. New York, N.Y: Academic Press, Inc.; 1969. p. 21. [Google Scholar]
- 25.Lam W L, Lee T-S, Gilbert W. Active transposition in zebrafish. Proc Natl Acad Sci USA. 1996;93:10870–10875. doi: 10.1073/pnas.93.20.10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LaPara T M, Nakutsu C H, Pantea L, Alleman J E. Phylogenetic analysis of bacterial communities in mesophilic and thermophilic bioreactors treating pharmaceutical wastewater. Appl Environ Microbiol. 2000;66:3951–3959. doi: 10.1128/aem.66.9.3951-3959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liesack W, Stackebrandt E. Evidence for unlinked rrn operons in the planctomycete Pirellula marina. J Bacteriol. 1989;171:5025–5030. doi: 10.1128/jb.171.9.5025-5030.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindström E S. Bacterioplankton community composition in a boreal forest lake. FEMS Microb Ecol. 1998;27:163–174. [Google Scholar]
- 29.Maidak B L, Cole J R, Parker C T, Jr, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mueller J C, Leach J M, Walden C C. Detoxification of bleached kraft mill effluents—a manageable problem. TAPPI J. 1977;60:135–137. [Google Scholar]
- 31.Murray A E, Hollibaugh J T, Orrego C. Phylogenetic composition of bacterioplankton from two California estuaries compared by denaturing gradient gel electrophoresis of 16S rDNA fragments. Appl Environ Microbiol. 1996;62:2676–2680. doi: 10.1128/aem.62.7.2676-2680.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murray A E, Preston C M, Massana R, Taylor L T, Blakis A, Wu K, DeLong E F. Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters near Anvers Island, Antarctica. Appl Environ Microbiol. 1998;64:2585–2595. doi: 10.1128/aem.64.7.2585-2595.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicholas K B, Nicholas H B, Jr, Deerfield D W., II GeneDoc: analysis and visualization of genetic variation. EMBNEW News. 1997;4:14. [Google Scholar]
- 34.Normand P, Ponsonnet C, Nesme X, Neyra M, Simonet P. ITS analysis of prokaryotes. In: Akkermans A D L, Van Elsas J D, De Bruijn F J, editors. Molecular microbial ecology manual. Boston, Mass: Kluwer Academic Publishers; 1996. pp. 1–12. [Google Scholar]
- 35.Palys T, Nakamura L K, Cohan F M. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int J Syst Bacteriol. 1997;47:1145–1156. doi: 10.1099/00207713-47-4-1145. [DOI] [PubMed] [Google Scholar]
- 36.Peters S, Koschinsky S, Schwieger F, Tebbe C. Succession of microbial communities during hot composting as detected by PCR-single-strand-conformation polymorphism-based genetic profiles of small-submunit rRNA genes. Appl Environ Microbiol. 2000;66:930–936. doi: 10.1128/aem.66.3.930-936.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robleto E A, Borneman J, Triplett E W. Effects of bacterial antibiotic production on rhizosphere microbial communities from a culture-independent perspective. Appl Environ Microbiol. 1998;64:5020–5022. doi: 10.1128/aem.64.12.5020-5022.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rouhbakhsh D, Baumann P. Characterization of a putative 23S–5S rRNA operon of Buchnera aphidicola (endosymbiont of aphids) unlinked to the 16S rRNA-encoding gene. Gene. 1995;155:107–112. doi: 10.1016/0378-1119(94)00910-k. [DOI] [PubMed] [Google Scholar]
- 39.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 40.Sekiguchi Y, Kamagata Y, Syutsubo K, Ohashi A, Harada H, Nakamura K. Phylogenetic diversity of mesophilic and thermophilic granular sludges determined by 16S rDNA gene analysis. Microbiology. 1998;44:2655–2665. doi: 10.1099/00221287-144-9-2655. [DOI] [PubMed] [Google Scholar]
- 41.Sievert S M, Brinkhoff T, Muyzer G, Ziebis W, Kuever J. Spatial heterogeneity of bacterial populations along an environmental gradient at a shallow submarine hydrothermal vent near Milos Island (Greece) Appl Environ Microbiol. 1999;65:3834–3842. doi: 10.1128/aem.65.9.3834-3842.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simberloff D. Use of rarefaction and related methods in ecology. In: Dickson K L, Cairns J J, Livingston R J, editors. Biological data in water pollution assessment: quantitative and statistical analyses. Philadelphia, Pa: American Society for Testing and Materials; 1978. pp. 150–165. [Google Scholar]
- 43.Stubbs S L J, Brazier J S, O'Neill G L, Duerden B I. PCR targeted to the 16S–23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J Clin Microbiol. 1999;37:461–463. doi: 10.1128/jcm.37.2.461-463.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vane-Wright R I, Humphries C J, Williams P H. What to protect systematics and the agony of choice. Biol Conserv. 1991;55:235–254. [Google Scholar]
- 45.Werker A G, Hall E R. Limitations for biological removal of resin acids from pulp mill effluent. Water Sci Technol. 1999;40(11–12):281–288. [Google Scholar]
- 46.Yang C-H, Crowley D E. Rhizosphere microbial community structure in relation to root location and plant iron nutritional status. Appl Environ Microbiol. 2000;66:345–351. doi: 10.1128/aem.66.1.345-351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu Z, Martin V J, Mohn W W. Occurrence of two resin acid-degrading bacteria and a gene encoding resin acid biodegradation in pulp and paper mill effluent biotreatment systems assayed by PCR. Microb Ecol. 1999;38:114–125. doi: 10.1007/s002489900163. [DOI] [PubMed] [Google Scholar]
- 48.Yu Z, Mohn W W. Killing two birds with one stone: simultaneous DNA and RNA extraction from activated sludge biomass. Can J Microbiol. 1999;45:269–272. [Google Scholar]
- 49.Yu Z, Mohn W W. Isolation and characterization of thermophilic bacteria capable of degrading dehydroabietic acid. Can J Microbiol. 1999;45:513–520. doi: 10.1139/w99-028. [DOI] [PubMed] [Google Scholar]