Abstract
The therapeutic benefits of deep brain stimulation (DBS), a neurosurgical treatment for certain movement disorders and other neurologic conditions, are well documented, but DBS mechanisms remain largely unexplained. DBS is thought to modulate pathological neural activity. However, although astrocytes, the most numerous cell type in the brain, play a significant role in neurotransmission, chemical homeostasis and synaptic plasticity, their role in DBS has not been fully examined. To investigate astrocytic function in DBS, we applied DBS-like high frequency electrical stimulation for 24 hours to human astrocytes in vitro and analyzed single cell transcriptome mRNA profile. We found that DBS-like high frequency stimulation negatively impacts astrocyte metabolism and promotes the release of matricellular proteins, including IGFBP3, GREM1, IGFBP5, THBS1, and PAPPA. Our results suggest that astrocytes are involved in the long-term modulation of perineuronal environments and that they may influence persistent cell-to-cell interaction and help maintain neuromodulation over time.
Introduction
The therapeutic success of deep brain stimulation (DBS) for movement disorders has led to its consideration for a rapidly expanding set of neurologic and psychiatric conditions. This increased application makes it all the more critical that the molecular mechanisms underlying its therapeutic action be identified and characterized. Most such research has understandably focused on its effects on neurons because DBS was initially thought to silence pathologically hyperactive neurons at the site of stimulation (Benabid et al., 1987). Recent studies solidify the theory that DBS directly inhibits neuronal elements close to the stimulation site and elicits axonal activation and neurotransmitter release (Uc and Follett, 2007; Johnson et al., 2008; Shon et al., 2010). Moreover, mathematical models suggest that due to the disparate excitability of neural elements, both soma inhibition and axonal activation can be expected at the DBS electrode site (McIntyre and Grill, 1998). Together these studies suggest that DBS modulates a dynamic nervous system in which synaptic transmission and plasticity occur, although the exact mechanisms are not yet fully understood.
Neuronal theories fail to take into account the potential influence of astrocytes, which outnumber neurons and are actively involved in neuronal signaling through neuro-glia communication (Perea and Araque, 2005; Halassa and Haydon, 2010). Astrocytes make important contributions to neurotransmission, chemical homeostasis, synaptic plasticity, and cerebral circulation (Takano et al., 2006; Bekar et al., 2008; Tawfik et al., 2010). Interestingly, astrocytes also respond to DBS-like high-frequency stimulation (HFS) by altering important regulators of neuronal network activity and inducing release of glutamate, ATP, and adenosine (Pascual et al., 2005; Bekar et al., 2008; Chang et al., 2009; Tawfik et al., 2010). While little attention has been paid to their neuromodulatory impact, these results suggest novel roles for astrocytes in mediating therapeutic effect of DBS.
Here, using single cell RNA-sequencing (scRNA-seq) technology, we analyzed molecular mechanisms of HFS on human astrocytes. Similar to clinical DBS, long-term HFS changes astrocytic gene expression profiles extensively, and those changes are particularly prominent in extracellular components, which can modulate network activity and refine neuronal circuits. These findings have a significant impact on understanding the mechanisms of action underlying the therapeutic effects of DBS.
Experimental procedures
Cell culture and Electrical stimulation
Primary human astrocytes were purchased from Life Technologies (Ref. # K1884, Lot # 180839), and maintained on Geltrex-coated T-25 flask in DMEM supplemented with 10% FBS and N2 supplement (Thermo Fisher Scientific Inc). For electrical stimulation, cells were plated on poly-L-lysine (Sigma-Aldrich, P4707, 0.01% solution) coated 6-well plate. At 70–80% cell confluence, high frequency stimulation (HFS) (100 Hz, 100 μsec pulse width, 2.0–2.5V) was applied for 24 hours through a pair of platinum wires (Fig. 1A and B). The impedance between the wires was 171.4 +/− 38.1 Ω (90.4–216 Ω) in growth media.
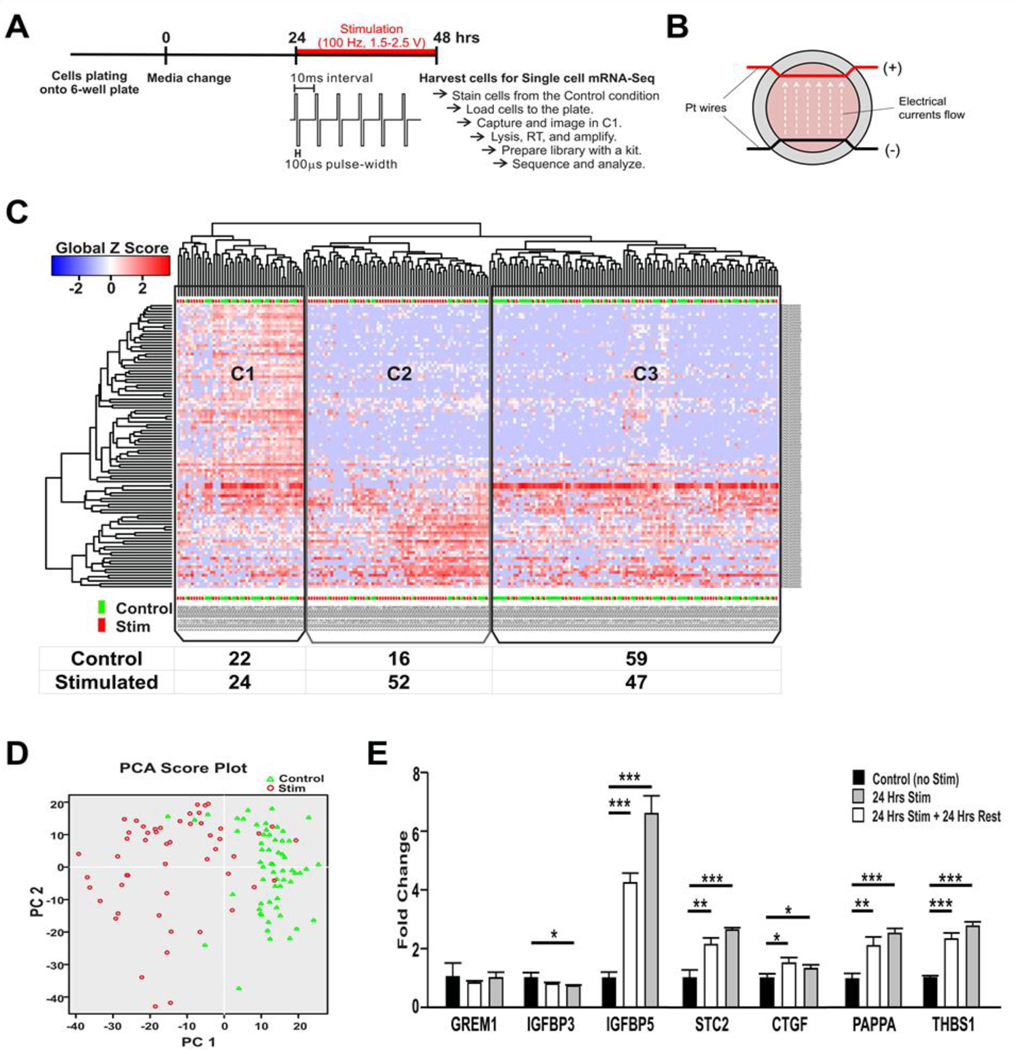
Figure 1.
Identification of the effect of HFS on cultured human astrocytes using single cell RNA-Seq and verification of the single cell analysis using qRT-PCR. (A) Experimental design. Culture media was changed one day before the stimulation. HFS (100 Hz, 100 μsec pulse width, 1.5–2.5 V) was applied for 24 hrs. (B) Schematic of the stimulation device. A pair of platinum wires were placed in parallel through the entire chamber of a 6-well plate. The wire was coated with silicon, and only the part across the wall was exposed. The bare wire nearly touched the bottom of the culture plate and the media fully covered the wire. (C) Heatmap of expression (Global Z-score) obtained by unsupervised two dimensional hierarchical clustering analysis using the most significant 100 genes. According to the gene expression profile, three clusters were identified. In C1, cells from both stimulated and control groups were evenly distributed. Cells in C2 and C3 were mainly from the stimulated and the control groups, respectively. (D) Principal component analysis (PCA) plot using all expressed genes of 111 cells (52 stimulated cells identified in C2 and 59 control cells identified in C3). PCA confirmed that gene expression profiling modulated by stimulation is strong enough to divide cells into two groups (stimulated vs. control). (E) qPCR assay to verify the data obtained by scRNA-Seq. Nine candidate genes were identified and confirmed by qPCR assay independently (***, p<0.001. **, p<0.01, *, p<0.05). Abbreviaions: GREM1, gremlin1; IGFBP, insulin like growth factor binding protein; CYR61, cysteine rich angiogenic inducer 61 (has been identified as CTGF-2 and also IGFBP10); STC2, stanniocalcin2; CTGF, connective tissue growth factor (has been identified as IGFBP8); TGFB2, Transforming growth factor beta 2; PAPPA, pregnancy-associated plasma protein A; THBS1, thrombospondin1.
Single cell whole mRNA sequencing and Data analysis
After stimulation, cells were loaded onto the Fluidigm C1 HT chip (Fluidigm Corporation, CA) at a concentration of 500 cells/μl, stained with Live/Dead cell viability/cytotoxicity kit (Thermo Fisher Scientific, MA), and imaged by phase contrast and fluorescence microscopy to assess the number and the viability of cells per capture site. After captured in the Fluidigm C1 system, a full length of double stranded cDNA was generated using SMARTer Ultra Low RNA kit for Illumina (Takara Bio, CA), and cDNA was diluted to 250 pg and constructed libraries using the Illumina Nextera XT DNA Sample Preparation kit (Illumina, CA). Libraries were quantified by a High Sensitivity DNA analysis kit (Agilent, CA) and Qubit dsDNA BR Assay kits (Thermo Fisher Scientific, MA). All 251 libraries were pooled and sequenced 100 bp paired-end on Illumina HiSeq 2500 Rapid Run. Additional analysis recommendations from Illumina include using only read 2 (R2) reads from paired-end FASTQ files for downstream alignment and expression analysis and trimming the polyA stretch from the 3’ end of the R2 reads. After preprocessing the R2 reads based on these recommendations, analysis was performed using the Mayo MAPRSeq RNA-Seq analysis workflow (v2.0.0). As a part of this workflow, FASTQC (v0.10.1) was used to perform quality control of the R2 reads. Post quality control, the Tophat (v2.0.12) aligner was used to perform alignment of reads to the hg19 build of the human reference genome. FASTQ formatted raw files for each sample were mapped and aligned in reference to hg19. After alignment, the BAM files were sorted and reformatted using Samtools (v0.1.18). The final BAM files were used as input for Subread’s featureCounts (v1.4.4) for counting reads mapping to genes and exons, as defined by Ensembl annotations. The BAM files were also used as input for RSeQC (v2.3.2) for generating various plots and files for further quantification and visualization. In total, 137 cells in stimulated group and 114 cells in control group were captured.
After converting to RPKM values, tertiary analysis was performed using Singlular package (Fluidigm, CA). Fourteen cells in the stimulated group and seventeen cells in the control group were excluded in the outlier analysis step. On average, approximately 500,000 reads were generated for each cell with an average of 80% reads mapped to the genome. Gene detection rates averaged 4,800 genes (count >2) and total detected genes (count >2) were 9439. To show the separation of cell population by HFS, unsupervised hierarchical clustering was performed using the 100 most variant genes by ANOVA analysis. Differentially expressed genes were identified using the log2 transformed RPKM values between two groups. We considered genes to have significant expression changes if the fold change was ≥ ±2 and P < 0.05 as determined by ANOVA (Supplement data 1). To further understand the biological meaning of the data set, Gene Set Enrichment Analysis (GSEA, Broad Institute) was carried out using 6483 genes (log2 mean value > 1) by permutation test 1,000 times with gene sets. Catalog C5 Gene Ontology, Hallmarks, Naba, and Reactome gene sets in Molecular Signatures Database v5.2 (MsigDB) were used to find the most significant pathway (false discovery rate (FDR) q-val <0.05) within data set (Supplement data 2).
Quantitative RT-PCR (qRT-PCR)
To validate single cell RNA-Seq Data, qRT-PCR was performed. Cells were prepared with the same protocol used for single cell RNA-Seq experiment (Fig. 1A) or after additional 24 hours incubation. Total RNA was isolated using the RNeasy Mini Kit (QIAGEN, Germany) and cDNA was generated using SuperScript III Reverse Transcriptase (Thermo Fisher Scientific, MA) according to the manufacturer’s protocol. PCR reaction was performed using a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, CA) with SYBR Green Master Mix (Bio-Rad, CA). RNA was initially denatured for 5 min followed by 40 cycles of denaturing at 95°C for 15 sec, and annealing/elongation at 60°C for 1 min. The primer sequences for validation of putative target genes are listed in the Supplement data 3.
Results
To characterize the role of astrocytes in DBS in the context of molecular effects, we applied HFS for 24 hours onto cultured human astrocytes and performed scRNA-Seq, which allows one to characterize and study sub-populations of cells and dissect the differences among the heterogeneity of cell populations. To identify characteristics of cells based on transcriptome profiling, we performed an unsupervised hierarchical clustering approach using the 100 most variable genes between stimulated and unstimulated conditions. We found that the cells separated out into three clusters, which included: C1: Noise cluster (cells with high expression of cell cycle genes); C2: Stimulated state cluster (stimulated cells); and C3: Normal state cluster (control (unstimulated) cells) (Fig.1.C). About 20% of cells in both the control and stimulated conditions (total of 46 cells: 22 out of 97 cells in the control condition and 24 out of 123 cells in the stimulated condition) showed high expression of genes involved in cell cycles reflecting the in vitro culture condition (Supplement data 4).
One of the major sources of biological noise is the cell cycle, during which a cell increases in size, replicates its DNA, and splits into daughter cells (Barron and Li, 2016). For this reason, all cells in the C1 Noise cluster were excluded from analysis. We found that HFS was able to shift the cell population from C3 (Normal state) to C2 (Stimulated state). As a result, 52 cells in C2 and 59 cells in C3 were selected as a stimulated cell subset and a non-stimulated control cell subset, respectively, for further analysis. That is, only 42% of the stimulated cells were found in the C2 cluster, which highlights the benefit of single cell analysis. Had we used the conventional bulk-tissue based RNA-Seq approach, their expression changes could have been diluted or quelled.
Additionally, we applied principal component analysis (PCA) to confirm the accuracy of our selection criteria by including all expressed genes (> 2 reads). The PCA plot showed a clear separation between subsets of stimulated and control cells (Fig1.D). We next identified 190 genes that were the most differentially expressed between two subsets (fold change > ±2, Supplement data 1). Among them, 97 genes, including many extracelluar matrix-associated protein genes, such as IGFBP3, GREM1, CTGF, STC2, SERPINE1, B4GALT1, TGM2, IGFBP5, THBS, PAPPA were up-regulated, while 93 genes were downregulated; most of the downregulated genes were related to energy metabolism and included COX5B, ATP5C1, ETFB, and TIMM8B.
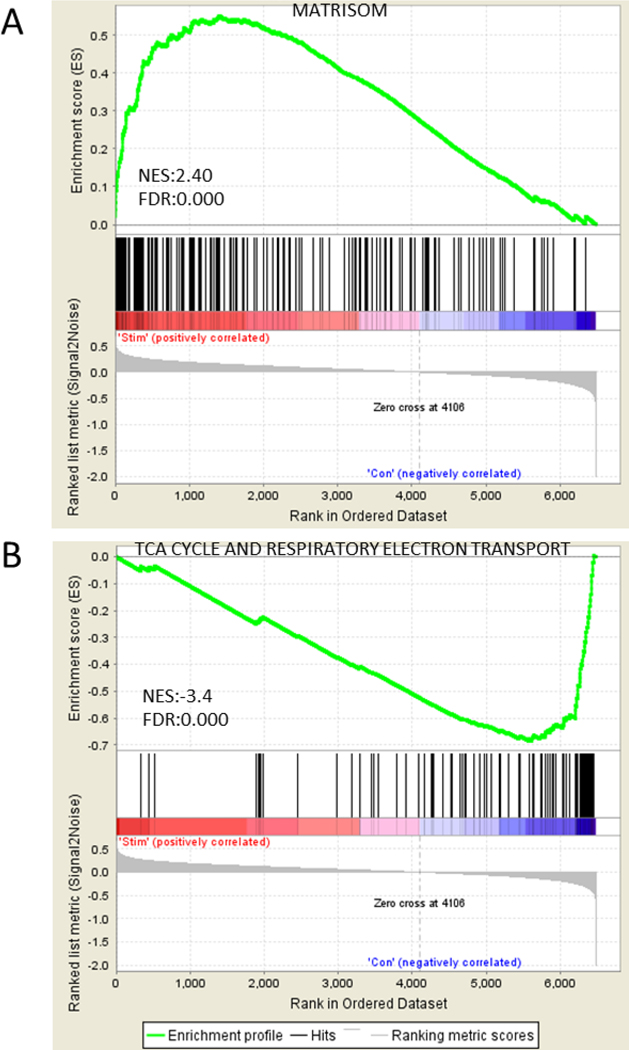
We next applied gene set enrichment analysis (GSEA) to explore HFS-induced physiological changes. According to the GSEA, the extracellular matrix protein gene set (matrisome) was significantly enriched and positively correlated with the stimulated subset (FDR < 0.000, NES, 2.4), while citric acid (TCA) cycle and respiratory electron transport gene sets were significantly enriched and negatively correlated with the control subset (FDR < 0.000, NES, −3.4) (Fig. 2). In addition, GO terms for molecular functions were significantly enriched in cell adhesion molecule binding. For biological processes, the enriched GO terms were extracellular matrix structure organization, and for the cellular component, the enriched GO terms were cell surface and endoplasmic reticulum rumen. Importantly, most enriched gene sets in down-regulated genes were associated with cellular metabolic functions of endoplasmic reticulum (ER), mitochondria, and ribosome biogenesis (Table 1 & Supplement data 2). However, the reactive astrocyte associated gene sets, including reactive oxygen species (ROS) generation and pro-inflammatory cytokines, were not activated by HFS.
Figure 2.
The representative enrichment plots for the gene sets of both stimulated and control groups by GSEA. (A) Matrisom gene set was significantly enriched positively in the stimulated group. (B) TCA cycle and respiratory electron transport gene set was significantly enriched negatively in the control group. Top of enrichment plot, the running enrichment score for the gene set as the analysis walks down the ranked list. Middle of enrichment plot, the location of genes from the each gene set within the ranked list. Bottom of enrichment plot, plot of the ranked list of 6483 genes. Y axis, value of the ranking metric; X axis, the rank for 6483 genes. NES, Normalized enrichment score. FDR, false discovery rate.
Table. 1.
Most significantly enriched gene sets using GO gene set In the human astrocytes by HFS.
| Enriched gene sets | Response in the stimulated cells | NES | NOM p-value | FDR q-value |
|---|---|---|---|---|
|
| ||||
| GO_CELL_SURFACE | Up | 2.27 | 0 | 0.002 |
| GO_ENDOPLASMIC_RETICULUM_LUMEN | Up | 2.2 | 0 | 0 |
| GO_EXTRACELLULAR_STRUCTURE_ORGANIZATION | Up | 2.7 | 0 | 0.001 |
| GO_CELL_ADHESION_MOLECULE_BINDING | Up | 2.4 | 0 | 0 |
| GO_MULTI_ORGANISM_METABOLIC_PROCESS | Down | −3.75 | 0 | 0 |
| GO_INNER_MITOCHONDRIAL_MEMBRANE_PROTEIN_COMPLEX | Down | −3.19 | 0 | 0 |
| GO_RIBOSOME_BIOGENESIS | Down | −3.22 | 0 | 0 |
| GO_TRANSLATIONAL_INITIATION | Down | −3.68 | 0 | 0 |
NES: normalized enrichment score; NOM p-val: nominal p value; FDR: false discovery rate
We further verified the single cell RNA-Seq data using qRT-PCR independently. Seven up-regulated genes were selected based on their known involvement in neuro-glia interactions and the altered expression of these genes was confirmed. The single cell RNA-Seq results were consistently reproduced in the bulk cell population using qRT-PCR (Fig1.E).
Discussion
In this study, we investigated the effects of DBS-like HFS on cultured human astrocytes using single-cell mRNA-Seq analysis. Our results suggest that DBS-like HFS downregulates astrocytic metabolism while promoting the secretion of matricellular proteins at the individual level of cells. Our approach overcame the inherent heterogeneity challenges which can arise due to differences in the response kinetics of individual cells to the same stimulation. Single cell RNA-Seq allowed us to isolate the subpopulation of interest and to investigate mechanisms explaining differences between subpopulations. We identified and validated that the most substantial changes among significantly upregulated genes were for proteins found in extracellular components, especially matricellular proteins (e.g.,IGFBP3, GREM1, CTGF, STC2, SERPINE1, B4GALT1, TGM2, IGFBP5, THBS1, PAPPA ).
It is known that matricellular proteins can be secreted into the perineuronal net and modulate neural networks by interacting with cell-surface receptors, proteases, hormones, and other bioeffector molecules (Bornstein, 2009). For example, IGFBP3, IGFBP5, THBS1, and PAPPA are related to the IGF1 pathway (Nam et al., 2000; Laursen et al., 2001), GREM1 is involved in the BMP pathway (Brazil et al., 2015), and CTGF is related to Wnt, BMP and TGF-β pathways (Abreu et al., 2002; Luo et al., 2004). In addition, STC2 (Zhang et al., 2000; Ito et al., 2004) and TGM2 (Cho et al., 2010; Basso et al., 2012) have been found to attenuate neuronal cell death, and a study on skeletal muscle showed that electrical stimulation can have an immediate and acute negative impact on cellular metabolism, impairing cell viability and even inducing cell death (Thelen et al., 1997). This result suggests a novel astrocyte-mediated mechanism in the therapeutic effects of DBS. In contrast, reactive astrocyte associated gene sets, including reactive oxygen species (ROS) generation and pro-inflammatory cytokines were not activated by HFS.
Previous investigations of DBS mechanisms have focused either on local neuronal activity changes around the DBS electrode or on its global effects on neuronal networks through synaptic transmission. These studies aimed to characterize the acute effect of DBS on neurons because electrical stimulation of a neuron with a uniformly excitable membrane can have direct effects on neuronal activity. There is little doubt that DBS acts on neurons to attenuate clinical symptoms. For example, DBS targeting neurons of the subthalamic nucleus can have an immediate effect on tremor and certain motor symptoms of Parkinson’s disease. However, for disorders such as dystonia, drug-resistant depression, or obsessive compulsive disorder, DBS can take weeks to months to effect symptom relief. These long-term DBS effects are equally important and call for investigation of the role of other cell types, such as astrocytes, that have non-excitable membranes.
Our findings identified important astrocytic involvement in DBS mechanisms and established single cell RNA-Seq as a valid approach for studying DBS-induced astrocytic changes. One limitation of our study was that our use of human cells necessitated the use of an in vitro culture system. The next immediate step would be to perform these same tests in an in vivo animal model of HFS.
Overall, our results suggest that DBS-like HFS induces minimal negative changes in cellular metabolism and that this conditional change promotes astrocytes to secret multiple types of matricellular proteins into the perineuronal net. The astrocyte-secreted matricellular proteins may influence environmental and cellular signaling efficiency through extracellular matrix formation by modulating synapse development, changing the binding affinity of neuronal receptors, and inducing neuronal plasticity. This observation warrants further investigation into the role of astrocytes as a powerful contributor in the sustained therapeutic effect brought about by DBS.
Supplementary Material
Acknowledgements
We thank Dr. Penelope Duffy for her help in preparing this manuscript. This study was funded in part by the National Institutes of Health, National Institute of Neurological Disorders and Strokes (NS 88260). John M. Nasseff, Sr. Career Development Award was granted to SYC.
References
- Abreu JG, Ketpura NI, Reversade B, De Robertis EM (2002) Connective-tissue growth factor (CTGF) modulates cell signalling by BMP and TGF-beta. Nat Cell Biol 4:599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron M, Li J (2016) Identifying and removing the cell-cycle effect from single-cell RNA-Sequencing data. Sci Rep 6:33892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso M, Berlin J, Xia L, Sleiman SF, Ko B, Haskew-Layton R, Kim E, Antonyak MA, Cerione RA, Iismaa SE, Willis D, Cho S, Ratan RR (2012) Transglutaminase inhibition protects against oxidative stress-induced neuronal death downstream of pathological ERK activation. J Neurosci 32:6561–6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekar L, Libionka W, Tian GF, Xu Q, Torres A, Wang X, Lovatt D, Williams E, Takano T, Schnermann J, Bakos R, Nedergaard M (2008) Adenosine is crucial for deep brain stimulation-mediated attenuation of tremor. Nat Med 14:75–80. [DOI] [PubMed] [Google Scholar]
- Benabid AL, Pollak P, Louveau A, Henry S, de Rougemont J (1987) Combined (thalamotomy and stimulation) stereotactic surgery of the VIM thalamic nucleus for bilateral Parkinson disease. Appl Neurophysiol 50:344–346. [DOI] [PubMed] [Google Scholar]
- Bornstein P (2009) Matricellular proteins: an overview. J Cell Commun Signal 3:163–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazil DP, Church RH, Surae S, Godson C, Martin F (2015) BMP signalling: agony and antagony in the family. Trends Cell Biol 25:249–264. [DOI] [PubMed] [Google Scholar]
- Chang SY, Shon YM, Agnesi F, Lee KH (2009) Microthalamotomy effect during deep brain stimulation: potential involvement of adenosine and glutamate efflux. Conf Proc IEEE Eng Med Biol Soc 2009:3294–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SY, Lee JH, Bae HD, Jeong EM, Jang GY, Kim CW, Shin DM, Jeon JH, Kim IG (2010) Transglutaminase 2 inhibits apoptosis induced by calcium- overload through down-regulation of Bax. Exp Mol Med 42:639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halassa MM, Haydon PG (2010) Integrated brain circuits: astrocytic networks modulate neuronal activity and behavior. Annu Rev Physiol 72:335–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito D, Walker JR, Thompson CS, Moroz I, Lin W, Veselits ML, Hakim AM, Fienberg AA, Thinakaran G (2004) Characterization of stanniocalcin 2, a novel target of the mammalian unfolded protein response with cytoprotective properties. Mol Cell Biol 24:9456–9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MD, Miocinovic S, McIntyre CC, Vitek JL (2008) Mechanisms and targets of deep brain stimulation in movement disorders. Neurotherapeutics : the journal of the American Society for Experimental NeuroTherapeutics 5:294–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen LS, Overgaard MT, Soe R, Boldt HB, Sottrup-Jensen L, Giudice LC, Conover CA, Oxvig C (2001) Pregnancy-associated plasma protein-A (PAPP-A) cleaves insulin-like growth factor binding protein (IGFBP)-5 independent of IGF: implications for the mechanism of IGFBP-4 proteolysis by PAPP-A. FEBS Lett 504:36–40. [DOI] [PubMed] [Google Scholar]
- Luo Q, Kang Q, Si WK, Jiang W, Park JK, Peng Y, Li XM, Luu HH, Luo J, Montag AG, Haydon RC, He TC (2004) Connective tissue growth factor (CTGF) is regulated by Wnt and bone morphogenetic proteins signaling in osteoblast differentiation of mesenchymal stem cells. J Biol Chem 279:55958–55968. [DOI] [PubMed] [Google Scholar]
- McIntyre CC, Grill WM (1998) Sensitivity analysis of a model of mammalian neural membrane. Biol Cybern 79:29–37. [DOI] [PubMed] [Google Scholar]
- Nam TJ, Busby WH, Rees C, Clemmons DR (2000) Thrombospondin and osteopontin bind to insulin-like growth factor (IGF)-binding protein-5 leading to an alteration in IGF-I-stimulated cell growth. Endocrinology 141:1100–1106. [DOI] [PubMed] [Google Scholar]
- Pascual O, Casper KB, Kubera C, Zhang J, Revilla-Sanchez R, Sul JY, Takano H, Moss SJ, McCarthy K, Haydon PG (2005) Astrocytic purinergic signaling coordinates synaptic networks. Science 310:113–116. [DOI] [PubMed] [Google Scholar]
- Perea G, Araque A (2005) Glial calcium signaling and neuron-glia communication. Cell Calcium 38:375–382. [DOI] [PubMed] [Google Scholar]
- Shon YM, Lee KH, Goerss SJ, Kim IY, Kimble C, Van Gompel JJ, Bennet K, Blaha CD, Chang SY (2010) High frequency stimulation of the subthalamic nucleus evokes striatal dopamine release in a large animal model of human DBS neurosurgery. Neurosci Lett 475:136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T, Tian GF, Peng W, Lou N, Libionka W, Han X, Nedergaard M (2006) Astrocyte-mediated control of cerebral blood flow. Nat Neurosci 9:260–267. [DOI] [PubMed] [Google Scholar]
- Tawfik VL, Chang SY, Hitti FL, Roberts DW, Leiter JC, Jovanovic S, Lee KH (2010) Deep brain stimulation results in local glutamate and adenosine release: investigation into the role of astrocytes. Neurosurgery 67:367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen MH, Simonides WS, van Hardeveld C (1997) Electrical stimulation of C2C12 myotubes induces contractions and represses thyroid-hormone-dependent transcription of the fast-type sarcoplasmic-reticulum Ca2+-ATPase gene. Biochem J 321 ( Pt 3):845–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uc EY, Follett KA (2007) Deep brain stimulation in movement disorders. Semin Neurol 27:170–182. [DOI] [PubMed] [Google Scholar]
- Zhang KZ, Lindsberg PJ, Tatlisumak T, Kaste M, Olsen HS, Andersson LC (2000) Stanniocalcin: A molecular guard of neurons during cerebral ischemia. P Natl Acad Sci USA 97:3637–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.