Keywords: zinc, pathogen, Candida, macrophage
Abstract
The zinc restriction and zinc toxicity are part of host defence, called nutritional immunity. The crucial role of zinc homeostasis in microbial survival within a host is established, but little is known about these processes in the opportunistic human fungal pathogen Candida parapsilosis. Our in silico predictions suggested the presence of at least six potential zinc transporters (ZnTs) in C. parapsilosis—orthologues of ZRC1, ZRT3 and ZRT101—but an orthologue of PRA1 zincophore was not found. In addition, we detected a species-specific gene expansion of the novel ZnT ZRT2, as we identified three orthologue genes in the genome of C. parapsilosis. Based on predictions, we created homozygous mutant strains of the potential ZnTs and characterized them. Despite the apparent gene expansion of ZRT2 in C. parapsilosis, only CpZRT21 was essential for growth in a zinc-depleted acidic environment, in addition we found that CpZrc1 is essential for zinc detoxification and also protects the fungi against the elimination of murine macrophages. Significantly, we demonstrated that C. parapsilosis forms zincosomes in a Zrc1-independent manner and zinc detoxification is mediated by the vacuolar importer CpZrc1. Our study defines the functions of C. parapsilosis ZnTs, including a species-specific survival and zinc detoxification system.
1. Introduction
Among trace elements, metal ions such as iron and zinc are vital for living organisms. These micronutrients are crucial for maintaining cellular homeostasis and approximately one-third of all proteins require metal cofactors to properly function. Metal elements are also essential for several metabolic processes such as DNA polymerase activity and transcription modifier mechanisms [1,2]. Zinc is the second most abundant trace metal after iron in humans. Zinc plays an important role in the function of both the innate and adaptive immune system, and it is required for the function of a wide variety of enzymes and other proteins including superoxide dismutases, alcohol dehydrogenases and transcription factors [3,4]. To survive within a mammalian host, human fungal pathogens use iron and zinc from the host's environment. This has been previously demonstrated by functional studies, where the absence of iron or zinc-acquisition-related factors (e.g. transporters and binding proteins) resulted in decreased virulence and growth defects of pathogenic fungi [5]. Limiting the accessibility of certain microelements is also an effective solution to control the growth of pathogens. For instance, the host is able to keep trace elements in a bound form using secreted metal-binding proteins, such as calprotectin, thereby limiting their uptake by pathogens, and thus inhibiting the extent of an infection [5]. On the other hand, zinc can also be toxic to pathogens. For example, murine macrophages are able to raise zinc concentration to a toxic level in the phagolysosome and effectively eliminate Mycobacterium tuberculosis in this compartment [6]. Furthermore, in the case of murine BMDMs, an IFN-I-dependent regulatory pathway can result in zinc toxification in the phagolysosome following Candida glabrata infection [7]. Taken together, pathogens face a ‘double-barrelled shotgun’ when encountering a potential host, as they need to overcome not only the limited accessibility of essential nutrients but, upon an effective host cellular response, potentially toxic levels of metal ions. These protective responses by the host are known as nutritional immunity [5].
In order to survive in the host, fungal pathogens require transport systems to access zinc ions from their environment as well as zinc detoxification machinery to protect against a deadly amount of zinc within phagolysosomes. Eukaryotic organisms encode multiple zinc transporters (ZnTs) to maintain cellular zinc homeostasis. Zrt-, Irt-like proteins (Zip) transporters transport zinc ions from the extracellular space to the cytoplasm [8], whereas ZnTs are responsible for the transport of zinc ions from the cytoplasm into intracellular organelles in the presence of high zinc. To date, the majority of information about fungal zinc homeostasis comes from Saccharomyces cerevisiae. In S. cerevisiae, several ZnT proteins (ScZrt1, ScZrt2, ScZrt3, ScZrc1 and ScFet4) have been described that, along with certain stress proteins, are regulated by the transcription factor Zap1 [9]. The ScZRT1, ScZRT2 and ScFET4 genes encode plasma membrane-localized transporters that are responsible for the transport of zinc ions from the environment to the cytoplasm. ScZrt1 acts as a high-affinity ZnT, while ScZrt2 and ScFet4 act as low-affinity transporters [9]. To prevent cellular toxicity, S. cerevisiae intracellular ZnTs actively transport accumulated zinc into a vacuole via ScZrc1 and ScCot1 [10]. Additionally, under zinc depletion, the vacuolar ScZrt3 transports zinc from the vacuole stores to the fungal cytoplasm [11].
Zinc uptake systems have also been studied in human fungal pathogens including Aspergillus fumigatus, Blastomyces dermatitidis, Histoplasma capsulatum, Candida dubliniensis and C. albicans. In C. albicans, the primary mode of uptake that has been described in this species takes place through the zincophore system, where the limited amount of zinc ions are bound by a zinc-binding protein or ‘zincophore’ [12]. The process is mediated by the secreted Pra1 (pH-regulated antigen 1) protein, which is able to bind zinc ions with high affinity in the host's environment and transport them to the fungal Zrt101 plasma membrane transporter. Citiulo et al. demonstrated that C. albicans can use zinc from the cytoplasm of endothelial cells when it is in the penetrating hyphae form [12]. In addition to the Pra1-Zrt101 synergic zinc uptake system, which functions at neutral–alkaline pH, C. albicans uses Zrt2 plasma membrane-located ZnTs to obtain zinc from acidic environments [13]. Crawford et al. showed that CaZRT1 expression is pH-dependent and supports the growth of C. albicans cells in the absence of CaZrt2 at neutral–alkaline pH. The deletion of CaZRT2 reduces fungal colonization of kidneys in mice [13]. C. albicans is able to store the accumulated zinc ions through a rapid compartmentalizing process into zincosomes, which is mediated by CaZrc1.
Over the past two decades, non-albicans Candida species have gained significant importance in medicine due to their increasing prevalence in life-threatening and frequently lethal infections. Of the non-albicans Candida, C. parapsilosis is often identified as the second or third most common cause of candidemia (5%–46%) and its prevalence is dependent on geographical regions and the examined patient groups [14]. This species particularly threatens low birth weight infants, the immunosuppressed and patients receiving long-term antibiotic treatment or chemotherapy [14]. While the main transmission route of C. albicans is thought to be vertical, with humans colonized at an early age (first months of life), the most common source of C. parapsilosis transmission is through the hands of healthcare workers or non-sterile clinical devices [15]. Several studies have reported that C. parapsilosis is less susceptible to echinocandins and resistance to azoles has also been reported [16,17]. C. parapsilosis also effectively adheres to and forms biofilms on catheters and other medical equipment [18]. Besides secreted hydrolytic enzyme and prostaglandin production, morphological switching with biofilm formation, metal ion acquisition has been suggested as an important prerequisite of invasion by this species [19–22].
Notably, perturbed iron homeostasis affects the virulence of C. parapsilosis [23,24]. Furthermore, the multi-copper oxidase encoded by FET3 also influences the species' morphology, biofilm formation and prostaglandin production, all of which are confirmed C. parapsilosis virulence factors. Similar to other pathogens, other metals and their transporters may also aid the virulence of C. parapsilosis. Here, we examine the ZnTs of C. parapsilosis in silico and have generated homozygous knock-out mutants of each. We further aimed to expose the generated mutant strains to various types of stressors and zinc-limiting conditions and to define their function in zinc transport of C. parapsilosis. In this study, we also aimed to uncover their role in host–pathogen interactions using in vitro macrophage and in vivo infection models.
2. Material and methods
2.1. Strain maintenance and cultivation
All of the strains used in this study were stored frozen at −80°C. Homozygous mutants were maintained on selective drop out (0.19% YNB-2% D-glucose) agar media supplemented with 100 unit ml−1 penicillin-streptomycin. Heterozygous mutant strains were maintained similarly, with L-Leucine (2.5 mg ml−1) added to the media as a supplement. The C. parapsilosis CLIB 214 wild-type strain was maintained on YPD (1% D-glucose, 1% peptone, 0.5% yeast extract) media containing 100 unit ml−1 penicillin-streptomycin. The gene reintegrated and green fluorescence protein (GFP)-labelled strains were maintained on YPD supplemented with 200 µg ml−1 nourseothricin.
Prior to the experiments, strains were cultured in YPD, then 100 µl of the cultivation was re-inoculated in 2 ml of YPD liquid medium containing 100 unit ml−1 penicillin-streptomycin at 30°C with overnight shaking (200 r.p.m.). Cells were then washed with 1 × PBS three times before adjusting to the working concentrations.
2.2. Generation of Candida parapsilosis mutant strains
All strains used for this study are summarized in the electronic supplementary material, table S1. Primers used in this study are summarized in the electronic supplementary material, table S2, plasmids in the electronic supplementary material, table S3. All mutant strains were verified with colony PCR and Southern hybridization.
Hetero- and homozygous mutants were generated according to the protocol described by Holland et al., using the C. parapsilosis CLIB 214 histidine and leucine auxotrophic strain, with modifications in the transformation method [25]. Two individual transformants were selected for each gene.
For reintegration, CpZRT21 and CpZRC1 were introduced into the CpNEUT5 L neutral locus using the Gateway cloning method as adapted to C. parapsilosis by Németh et al. [26]. For functional restoration, CpZRC1 with a CaTDH3 promoter was introduced into the mutant strain Δ/Δ CpZRC1 by transformation as described by Németh et al. [27].
For phagocytosis experiments, GFP-labelled CLIB 214 RI, Δ/Δ CpZRT21 and Δ/Δ CpZRC1 cells were used, where the CaTDH3-GFP fusion construction was integrated into the CpNEUT5 L neutral locus as described by Németh et al. [27].
To determine the intracellular localization of CpZrc1, we introduced Gly5-GFP on the C-terminus of the target gene according to the CRISPR-Cas9-based gene modification and transformation method described by Lombardi et al. [28].
2.3. Phenotypic characterization
Viability of each homozygous mutant was tested under the 30 different stress conditions listed in electronic supplementary material, table S3. Strains were prepared and washed as mentioned above. Cell concentration was adjusted to 104/10³/10²/10 cells/ in 5 µl. The growth of each strain was examined after 3 days of incubation at 30 C. 4 × 105 cells were plated into 96-well plates and growth kinetics was determined by spectrophotometry. Optical density was measured at 600 nm.
2.4. Low zinc medium
To characterize ZnT mutant strains growth properties and tolerance to zinc, we used low zinc medium (LZM) supplemented with different concentrations (0.05, 1, 10, 15, 17.5, 20, 22.5, 25, 50 and 100 mM) of zinc from 0.1 or 2 M ZnSO4 stock solutions. To prepare the base LZM, we created the medium by adding all essential amino acids, nutrients and ions except zinc. We followed a modified protocol provided by Crawford et al. [13]. The unbuffered pH of our LZM was 7.5. To adjust to acidic/alkalic pH, citric acid (1 M) or sodium phosphate (0.2 M) was used, respectively.
2.5. Gene expression analysis
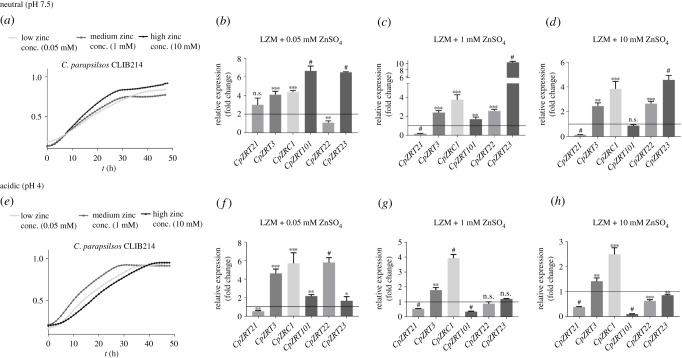
Strains were cultured in YPD+ 100 unit ml−1 penicillin-streptomycin at 30°C with shaking (200 rpm) overnight. Then, 4 × 105 cells were inoculated in the indicated media and cells were grown for 24 h. Cell harvesting and extraction was performed according to the manufacturer's protocols with the RiboPure RNA Purification Kit. Five hundred nanograms of RNA was transcribed by reverse transcription using the RevertAid first-strand cDNA synthesis kit protocol. Real-time PCR was performed by primers listed in electronic supplementary material, table S2 and the conditions were as follows: one cycle of denaturation for 3 min at 95°C; denaturation at 95°C for 10 s; 49 cycles of annealing at 60°C for 30 s, and elongation at 65°C for 30 s; with a final extension step at 72°C for 30 s. In the case of the determination of the predicted ZnT gene expressions in CLIB 214 wild-type strain Δ/Δ Ct threshold cycle (CT) analysis used, fold changes calculated from the normalized Cq values to the expression of CpTUB4 housekeeping gene in YPD first in case of each condition and then to each gene (figure 2). The fold changes of ZRT2 orthologues were calculated by Δ/Ct analysis, the ZnT gene Cq values were normalized to the values of CpTUB4 in acidic LZM medium in the case of each strain (figure 4).
Figure 2.
Gene expression changes in the expression of ZnTs under neutral (pH 7.5) and acidic (pH 4) conditions. C. parapsilosis CLIB 214 cells were grown in YPD, and then inoculated into liquid LZM with neutral or acidic pH supplemented with low (0.05 mM), medium (1 mM) and high (10 mM) concentrations of ZnSO4. (a,d) Growth kinetics of C. parapsilosis CLIB 214 cells in different zinc-supplemented LZM media. (b–d, e–g) Gene expression changes in ZnT genes. Data were obtained from triplicate experiments, and the expression of CpTUB4 in YPD was used as the internal control for Δ/Δ Ct threshold cycle (CT) analysis. For statistical analysis, the Mann–Whitney test was used, *p < 0.05; **p < 0.01; ***p < 0.001; #p < 0.0001.
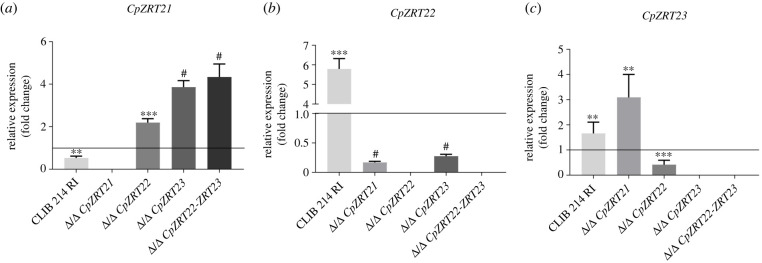
Figure 4.
Acidic pH and low zinc concentration-dependent expression changes of CpZRT2. (a–c) Gene expressions represent the relative value to the expression of CpTUB4 in acidic low zinc environment by Δ/Ct. Data show the means of triplicate experiments. For statistical analysis, Mann–Whitney test was used, *p < 0.05; **p < 0.01; ***p < 0.001; #p < 0.0001.
2.6. Fluorescence staining
For intracellular zinc visualization, the Zinquin fluorescent probe was used. Overnight cultured yeast cells were inoculated in 1 x PBS, stained with 25 µM Zinquin and incubated for 40 min protected from light. The cells were then washed two times in 1 x PBS and prepared for flow cytometry or microscopy. Zinquin+ population was determined by subtracting the autofluorescence signal of the unstained cells using the gating strategy shown in electronic supplementary material, figure S5A.
To visualize the location of the GFP-tagged CpZrc1, the fungal cell wall, nucleus and vacuole were stained. Concanavalin-TRITC (final cc: 100 µg ml−1) or CalcoFluor White (final cc: 5 µg ml−1) were used for cell wall staining, the nucleus was labelled with DAPI (final cc: 50 µg ml−1), and the fungal vacuoles were stained with FM4-64 (final cc: 10 µg ml−1) in 200 µl final staining volume for 40 min at 30°C with shaking (200 r.p.m.) and protection from light. Afterwards, cells were washed in 1 x PBS two times and prepared for microscopic analysis.
2.7. Maintenance of J774.2 murine macrophages
J774.2 murine macrophages were cultured in Dulbecco's Modified Eagle Medium (DMEM, Lonza) supplemented with 100 unit ml−1 penicillin-streptomycin and 10 V/V % fetal bovine serum and incubated at 37°C, with 5% CO2. The medium was changed on confluent cultures every 3 days and immediately prior to the experiments.
2.8. Phagocytosis assay
J774.2 macrophages (1 × 106) in 24-well plates were infected with the GFP-tagged strain or Alexa Fluor 647 (Invitrogen) stained (according to the protocol provided by Invitrogen) mutant strains in a 5 : 1 ratio (yeast cell: macrophage). After 0.5, 1, 2 and 3 h of coincubation, wells were washed with 1 x PBS, and then macrophages were collected. Macrophage singlet cells (104) were examined with flow cytometry analysis (Amnis) per strain. Phagocytosis + macrophage population was determined by subtracting the autofluorescence signal of the uninfected J774.2 control (electronic supplementary material, figure S5B).
2.9. Fungal elimination assays
The killing efficiency of C. parapsilosis strains by J774.2 macrophages after 3 h of coincubation was determined by two approaches using an MOI of 5 : 1.
For the flow cytometry-based killing assay, 106 J774.2 were challenged with 5 × 106 yeasts/well in 24-well plates. Control wells contained yeast cells only. After 3 h of incubation, cells were collected and washed two times with FUN1 buffer (10 mM NaHEPES, 2 m/V % glucose), and then macrophage disruption was achieved by passage of the collected cells through insulin syringes. In order to separate yeast cells and macrophages debris, the fungal cell wall specific Calcofluor White was used together with the FUN1 dye to detect live fungal cells. The combined staining was set as follows: 195 µl of cell suspension (approx. 5 × 106 cells) in FUN1 buffer + 1 µl FUN1 (100 µM) + 4 µl CalcoFluor White (2.5 mg ml−1). After 40 min of staining at room temperature (in the dark), cells were washed two times with FUN1 buffer, and live cells were enumerated by flow cytometry. Control wells contained only fungal cells in DMEM to determine 100% of live yeast cells. CalcoFluor White+ and FUN1 + populations were determined using the gating strategy added in electronic supplementary material, figure S5C.
A CFU-based fungal elimination assay was performed according to a standardized protocol with minor changes [29]. J774.2 cells (2 × 105) were infected with 106 yeast cells in 96-wells plates. Control wells contained only Candida cells in a similar amount. Three hours of post-infection, cells were collected and forced through an insulin needle. Suspensions was plated on YPD+1% penicillin-streptomycin plates and incubated at 30°C for 3 days. Killing efficiency by macrophages was determined as previously described by Németh et al. [29].
2.10. Galleria mellonella infection
Galleria mellonella survival experiments were performed as described previously [29]. The larvae were infected with 10 µl of Candida suspension containing 6 × 106 yeast cells in 1 x PBS via their pro-legs using a 26 G needle. A group of larvae was inoculated with PBS as control, another group remained untreated. Larvae were incubated at 25°C, survival was monitored daily for 14 days.
2.11. Statistical analysis
Statistical significance was determined by unpaired t-tests or Mann–Whitney tests using the GraphPad Prism v. 7.0 software. The non-parametric Mann–Whitney tests routinely used to improve the analysis of the gene expression data to detect the differences between the median value of the groups with more power and strict approach than the other tests [30]. Differences between groups were considered statistically significant at p < 0.05.
3. Results
3.1. Identification of potential zinc transporters in Candida parapsilosis
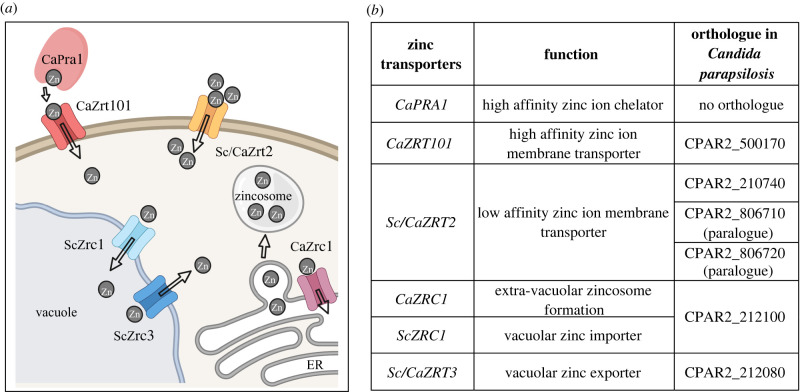
We performed in silico analyses to identify ZnTs in C. parapsilosis (figure 1). BLASTp analysis using C. albicans Zrt101, Zrt2, Zrt3 and Zrc1 as query identified six orthologues of known C. albicans and S. cerevisiae-ZnTs by nucleotide similarity (figure 1b). Notably, the orthologue of the CaPra1 zincophore zinc-binding protein was not identified in the genome of C. parapsilosis [12]. On the other hand, several putative zinc ion membrane transporters were detected. CPAR2_500170 was identified as the orthologous gene of the CaZRT101, which, in C. albicans, acts in conjunction with Pra1. We identified three transporters with high sequence similarity to Sc/CaZRT2; these were CPAR2_210740, CPAR2_806710 and CPAR2_806720, which we named ZRT21, ZRT22 and ZRT23, respectively. Of these, CPAR2_210740 exhibited the highest sequence similarity to C. albicans zinc importer Zrt2. CPAR2_806710 and CPAR2_806720 appeared to be the result of an ancestral ZRT2 gene duplication followed by recent tandem duplication in C. parapsilosis. CPAR2_212100 was identified as the orthologue of CaZRC1 and ScZRC1, which function in zincosome and vacuolar zinc import, respectively. Both previously investigated Zrc1 transporters play important roles in zinc detoxification in these fungi. We also identified the orthologue of the vacuolar zinc exporter, Sc/CaZRT3, as CPAR2_212080 in the C. parapsilosis genome.
Figure 1.
In silico ZnT analysis. (a) ZnTs in S. cerevisiae and C. albicans. (b) The target genes selected for KO generation in C. parapsilosis. Image (a) created with BioRender.com.
3.2. Zinc concentration-dependent expression of zinc transporters
Next, we aimed to investigate the expression patterns of the identified ZnT genes. We first determined low (0.05 mM), medium (1 mM) and high (10 mM) levels of zinc that could support the growth of C. parapsilosis CLIB 214 strain in LZM, pH 7.5 basal medium (figure 2a) and pH 4 (figure 2e). In order to compare the expression of genes under different zinc regimes, the expression of each transporter gene was analysed by Δ/Δ Ct where the internal control was the expression of CpTUB4 in the wild-type strain in YPD medium. CpZRT101 was expressed at its highest levels in low zinc media at neutral pH and progressively downregulated by the addition of zinc (figure 2b–d). At acidic pH, ZRT101 was expressed at much lower levels, but again downregulated by the addition of exogenous zinc. Surprisingly, CpZRT21 was transcribed at far lower levels, and with a higher expression at neutral pH than acidic. CpZRT22 showed the highest transcription at low zinc and low pH while its paralogue, CpZRT23, exhibited the highest expression at medium zinc levels at neutral pH and was downregulated at acidic pH. We also measured ZRC1 and ZRT3, which encode intracellular organellar zinc importers and exporters, respectively. At neutral pH ZRC1 was modestly upregulated in response to increasing zinc, as might be expected for a gene involved in zinc detoxification. Finally, ZRT3, which encodes a vacuolar zinc exporter in other species, did not exhibit considerable differential regulation, except at low zinc and acidic pH, under which it was induced.
3.3. Generation of knock-out mutants and their phenotypical characterizations
To study the potential role of the identified ZnTs in C. parapsilosis zinc homeostasis, we generated homozygous deletion mutants of each of the six genes identified in figure 1. Two independent null mutants were created per gene, using the auxotrophy complementation method developed by Holland et al. [25]. The two independent mutants of each homozygous KO strains showed the same phenotype in all cases. To generate reintegrant strains, CpZRT21 and CpZRC1 were introduced into the CpNEUT5 L neutral locus of each null mutant strain. The reintegration of a single copy of CpZRC1 did not complement the phenotype of Δ/Δ CpZRC1, so CpZRC1 was placed under control of the CaTDH3 promoter in the CpNEUT5 L region.
3.4. Characterization of the zinc uptake system
3.4.1. Zinc concentration and pH-dependent growth of Δ/Δ CpZRT21
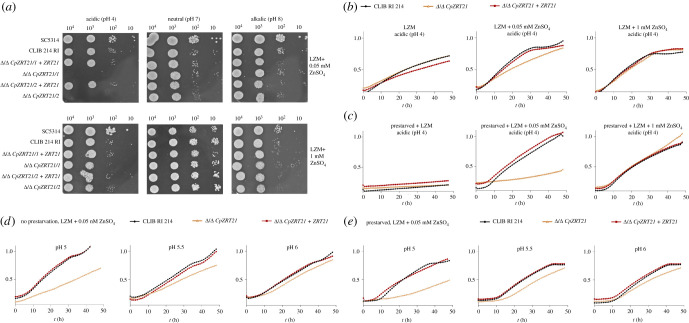
Based on our phylogenetic analysis of zinc importers in C. parapsilosis (figure 1), the most striking feature of this species was the gene expansion of ZRT2. This transporter is critical for zinc uptake and growth of C. albicans in acidic environments. We therefore sought to functionally characterize the orthologue of Zrt2 in C. parapsilosis (which we have termed Zrt21), as well as its paralogues—Zrt22 and Zrt23. Independent Δ/Δ CpZRT21, Δ/Δ CpZRT22 and Δ/Δ CpZRT23 strains, as well as a double mutant lacking both ZRT22 and ZRT23, were generated and assayed for growth at acidic, neutral and alkaline environments. Only deletion of ZRT21 rendered C. parapsilosis unable to grow on low zinc agar plates at acidic pH (figure 3a). In liquid media, wild-type, Δ/Δ CpZRT21 and Δ/Δ CpZRT21+ CpZRT21 grew relatively poorly in unsupplemented LZM and growth was enhanced by the addition of zinc, with the Δ/Δ CpZRT21 mutant exhibiting a moderate growth defect in comparison to wild-type and revertant in LZM + 0.05 mM zinc. We therefore extended this analysis by first preculturing cells under zinc limitation to deplete intracellular zinc reserves before the inoculation of growth assays. Figure 3c shows that when pre-starved of zinc, all strains failed to grow upon subsequent inoculation of LZM without zinc. Comparing the growth of normal- versus zinc pre-starved cells in unsupplemented LZM suggests that C. parapsilosis may be using intracellular zinc stores. When zinc pre-starved cells were used to inoculate LZM + 0.05 mM zinc, the Δ/Δ CpZRT21 mutant exhibited a severe growth defect in comparison to wild-type and revertant. This suggests that, like its C. albicans orthologue Zrt2, Zrt21 plays a vital role in zinc uptake in acidic media (figure 3a,c), while the transporter is dispensable for growth at neutral–alkaline pH (figure 3a). We therefore performed growth analysis at discrete acidic pH values supplemented with 0.5 mM zinc either or without zinc pre-starvation to probe the granularity of Zrt21-dependent growth. Figure 3d,e shows that Δ/Δ CpZRT21 growth was still strongly reduced at pH 5.0, had a more moderate growth defect at pH 5.5, and grew at close to wild-type rates at pH 6.0. These data show that, as pH increases towards neutrality, Zrt21 becomes redundant.
Figure 3.
The enhanced growth of the C. parapsilosis Δ/Δ CpZRT21 mutant upon alkalinization. (a) Spot assays were performed with the CpZRT21 null mutants on acidic (pH = 4), neutral (pH = 7) and alkalic (pH = 8) solid media with different zinc concentrations. (b) Figures represent the growth kinetics of the indicated strains under acidic (pH4) environmental conditions in different zinc-supplemented media as indicated. (c) For zinc pre-starvation, strains were pre-cultured in liquid YPD medium, and then inoculated into a liquid LZM medium for 24 h. Cells were washed with 1 mM EDTA before the growth test. (d) Growth assays in discrete pH valued (pH 5–pH 6) LZM medium supplemented with 0.05 mM zinc without zinc pre-starvation and (e) with LZM pre-starvation. The spot assays were performed three times. Growth curves represent mean data of three independent experiments performed in triplicates.
3.4.2. Expression compensation of CpZRT2 genes
Because of the complexity of the zinc uptake repertoire of C. parapsilosis, and the essentiality of ZRT21 in acidic environments, we next assessed the effect of ZRT21, ZRT22 and ZRT23 deletion on the expression of these genes. For this analysis, we selected liquid LZM pH 4, supplemented with 0.05 mM zinc, inoculated with YPD pre-grown cells as this condition permitted the growth of all strains (figure 3) and in which we inferred the existence of Zrt21-dependent zinc uptake from the surrounding media (compare Δ/Δ CpZRT21 figure 3b versus figure 3c, central panels). The deletion of the CpZRT22 and CpZRT23 (and also the double disruption of the genes) resulted in an increase in the expression of CpZRT21 (figure 4a). In addition, the absence of CpZRT21 repressed the expression of CpZRT22 but increased the transcript level of CpZRT23. Finally, the expression of CpZRT22 and CpZRT23 decreased in the Δ/Δ CpZRT23 and Δ/Δ CpZRT22 mutants, respectively (figure 4b,c). These data point towards a complex regulatory interplay among these zinc import genes in C. parapsilosis.
3.5. Functional characterization of CpZRC1
3.5.1. CpZRC1 is essential for zinc detoxification
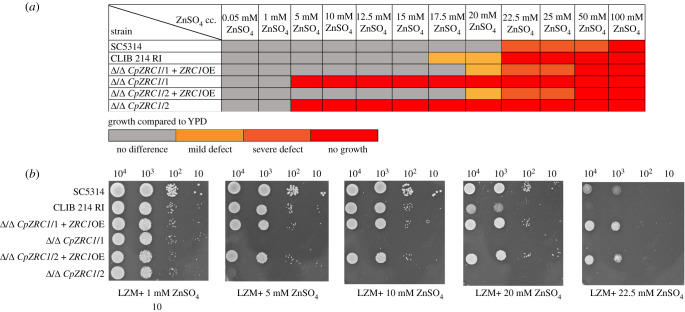
We first characterized zinc tolerance in C. parapsilosis in comparison to C. albicans. Figure 5 shows that when cultured on LZM agar plates, C. parapsilosis wild-type exhibited robust growth with up to 10 mM zinc supplementation. At 20 mM zinc, growth of C. parapsilosis but not C. albicans was inhibited, and at greater than or equal to 22.5 mM zinc, colony formation of C. parapsilosis was blocked completely. The deletion of ZRC1 had no impact on C. parapsilosis on LZM media containing 1 mM zinc; however, at 5 mM and above, no growth was observed for the Δ/Δ CpZRC1 mutant. Similar results were observed in liquid media: C. parapsilosis wild-type tolerated up to 17.5 mM zinc in liquid LZM media, with growth substantially reduced at 20 mM and above whereas the growth of the Δ/Δ CpZRC1 mutant was inhibited at 5 mM and above (figure 6). Again, TDH3 promoter-driven overexpression of ZRC1 permitted robust fungal growth at zinc levels well beyond that tolerated by wild-type C. parapsilosis (figures 5 and 6).
Figure 5.
Viability of the generated mutant strains in the presence of different zinc concentrations. The homozygous deletion mutants were applied for spot assays and their growth was examined in the presence of 31 different stress conditions. The viability of CpZRC1 deletion mutants altered significantly in high zinc concentrations. (a) Summary of the results of all performed experiments. (b) Representative images of spot assays were performed with the ZRC1 strains.
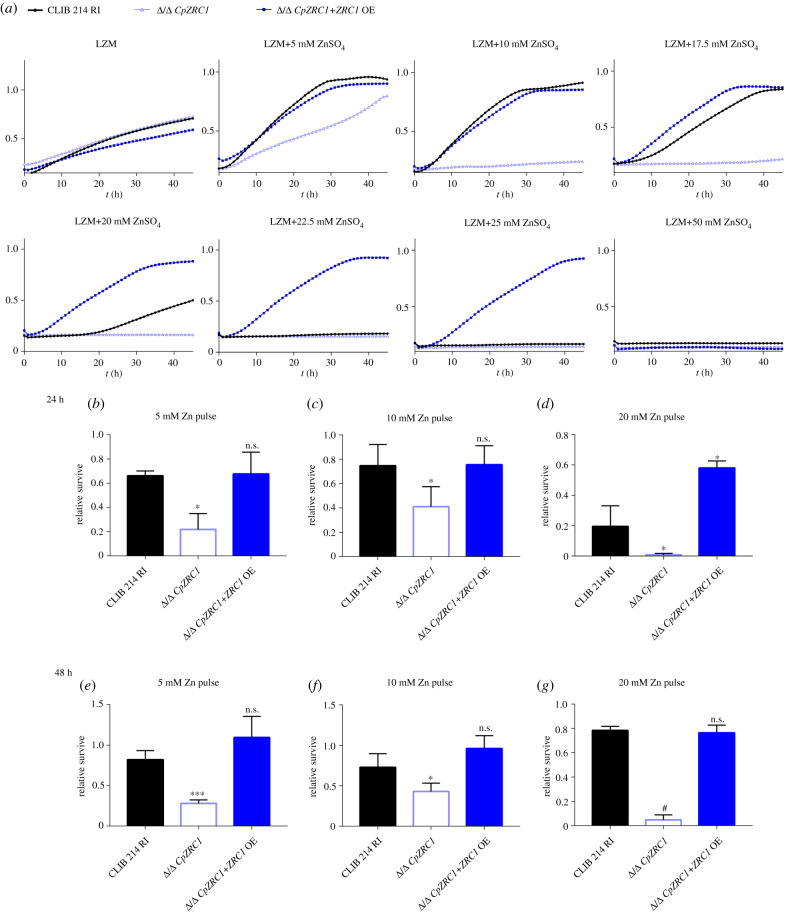
Figure 6.
CpZRC1 is essential for zinc detoxification. (a) Growth kinetics of Δ/Δ CpZRC1 strains in different zinc-containing liquid media. (b–g) Results of zinc survival assays. After 5, 10 and 20 mM zinc pulses for 24 and 48 h, 500 cells were plated on YPD then viable cells were counted. The growth curves represent the results of three independent experiments performed in triplicates. Relative survival was determined from three independent experiments performed in triplicates. For statistical analysis, unpaired t-tests were used, *p < 0.05; **p < 0.01; ***p < 0.001; #p < 0.0001.
Besides reduced growth (figures 5 and 6), the viability of Δ/Δ CpZRC1 cells also decreased at greater than or equal to 5 mM ZnSO4 concentrations, as reflected by the significantly lower number of colonies on YPD plates recovered following 5, 10 and 20 mM zinc pulses for both 24 and 48 h (figure 6b–g). Notably, the overexpression of CpZRC1 overcame this affect both on solid plates and in liquid medium. Interestingly, both the growth and survival of the Δ/Δ CpZRC1 +ZRC1OE strain exceeded that of the wild-type strain's, both on solid plates and in liquid medium supplemented with higher amounts of zinc (20, 22,5, 25 mM ZnSO4) (figures 5 and 6).
3.5.2. Zincosome formation in Candida parapsilosis
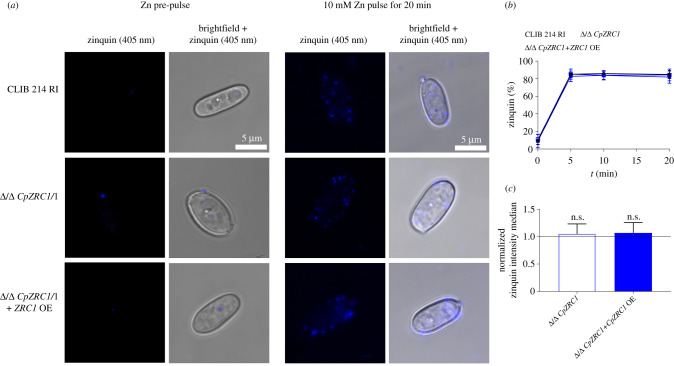
Results of the phenotypic analyses suggested that CpZRC1 is essential for zinc detoxification in C. parapsilosis. Saccharomyces cerevisiae Zrc1 is a vacuolar zinc importer, and C. albicans Zrc1 is responsible for zincosome (intracellular zinc store) formation [11,13]. Thus, we aimed to examine whether Zrc1 has a similar function in C. parapsilosis. For this purpose, we used Zinquin, a dye that is used to stain zincosome zinc. Candida parapsilosis strains were challenged with a 10 mM zinc for 5, 10 and 20 min. After zinc treatment, the intracellular localization of Zinquin was determined via confocal microscopy (figure 7a). In addition, the percentage of Zinquin+ population by flow cytometry (figure 7b) and the median of Zinquin signal intensity (figure 7c) by flow cytometry. The wild-type C. parapsilosis, Δ/Δ CpZRC1, Δ/Δ CpZRC1 + ZRC1OE strains were able to form zincosomes equally in a rapid way following stimulation with 10 mM of zinc treatment after 5–20 min.
Figure 7.
Zincosome formation in C. parapsilosis is CpZrc1 independent. Strains were cultured in liquid YPD medium, then inoculated into LZM liquid medium supplemented with high level (10 mM) of zinc. Zinquin fluorescent dye was used to visualize intracellular zincosomes. (a) Images represent the localization of zincosomes after 20 min of 10 mM zinc pulse. (b) Percentage of Zinquin+ cells at indicated timepoints. (c) Figure represents the intensity median of Zinquin in cells after 20 min by flow cytometer. Experiments were performed three times. For statistical analysis, unpaired t-tests were used, p < 0.05.
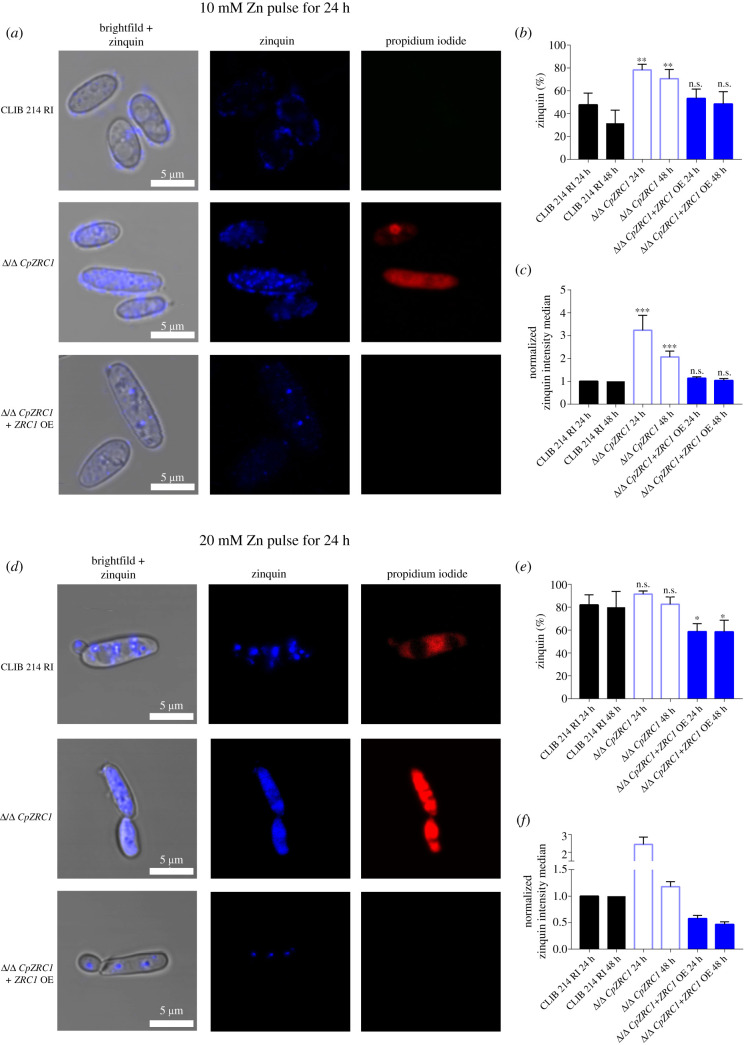
Next, the effect of prolonged zinc treatment was tested. Cells were treated with 10 or 20 mM zinc for 24 h followed by Zinquin staining and propidium iodide to assess cell viability (figure 8a,d). Interestingly, the Δ/Δ CpZRC1 strain exhibited more intense Zinquin staining than wild-type and revertant cells. This contrasts with C. albicans Δ/Δ CpZRC1 cells, which exhibit reduced zincosomes and Zinquin staining [13]. In the absence of CpZRC1, the 10 mM concentration of zinc pulse for 24 and 48 h increased the Zinquin+ population and Zinquin signal intensity but also reduced viability (figure 8a–c,e,f). In line with our previous observations, the 20 mM zinc pulse proved to be toxic for wild-type cells, while the Δ/Δ CpZRC1 + ZRC1OE strain was able to survive (figure 8d). Furthermore, in the wild-type and CpZRC1 mutant strain, the Zinquin+ population increased and the zincosomes appeared to leak resulting in homogeneous Zinquin signals (figure 8d,e). The reintegrant Δ/Δ CpZRC1 + ZRC1OE strain showed a phenotype similar to that of the wild-type only in the case of 10 mM zinc treatment.
Figure 8.
Zincosome formation under toxic zinc levels. Strains were grown in liquid YPD medium and then inoculated into LZM liquid medium supplemented with 10 mM or 20 mM zinc. Cells were then stained with Zinquin (zincosomes) and Propidium Iodide (viability). (a,d) Images represent CpZRC1 strains challenged with 10 mM and 20 mM zinc for 24 h. (b,e) The Zinquin+ population was determined after zinc treatment. (c,f) Zinquin intensity median presented after the zinc pulse. Experiments were performed three times using flow cytometer. For statistical analysis, unpaired t-tests were used, *p < 0.05; **p < 0.01; ***p < 0.001.
3.5.3. Intracellular localization of CpZrc1
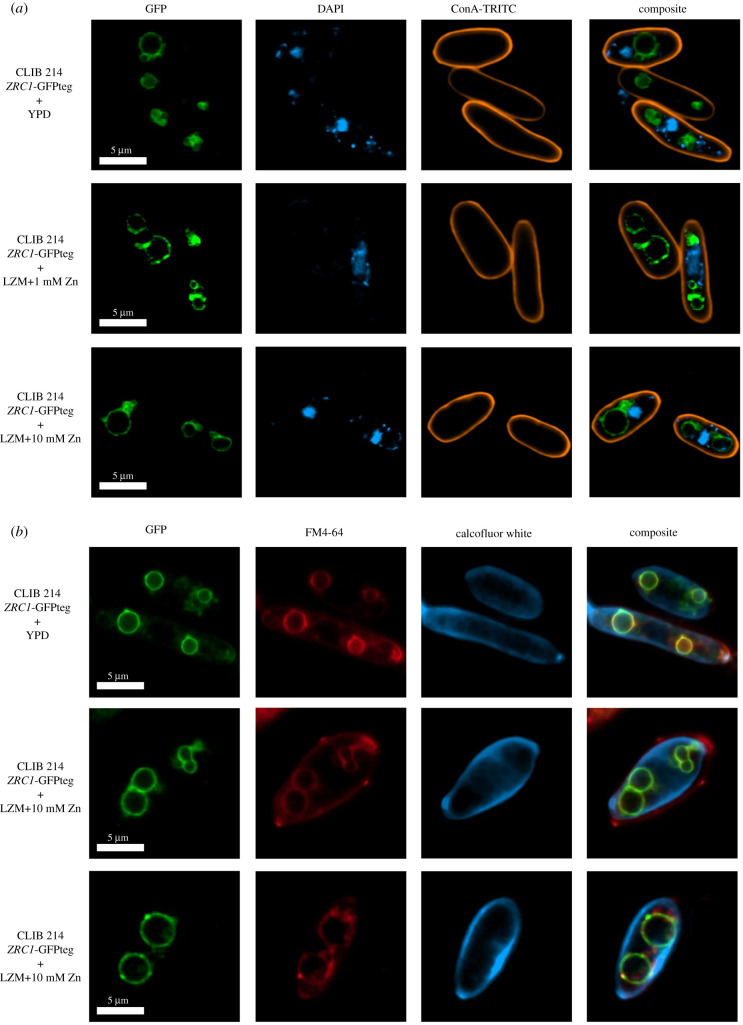
To visualize the localization of CpZrc1, we tagged the C-terminus of CpZRC1 with GFP. The generated strain was inoculated in YPD or LZM liquid medium supplemented with 1 mM (medium level) or 10 mM (high level) of zinc and incubated for 24 h. After incubation, ConcanavalinA-TRITC (orange) and DAPI (blue) were applied as cell wall and nucleus staining (figure 9a). In addition, cell wall dye CalcoFluor White (blue) and vacuole stain FM4-64 (red) were also used to characterize CpZrc1 localization (figure 9b). Confocal microscopy revealed that CpZrc1 localizes outside of the nucleus, was absent from the cell wall, and instead co-localizes with the FM4-64 signal in the vacuolar membrane. The concentration of zinc supplementation did not affect the signal of GFP-tagged CpZrc1.
Figure 9.
CpZrc1 co-localizes to vacuolar membrane. The CLIB 214 ZRC1-GFP-tagged strains were grown in liquid YPD medium and then inoculated in liquid YPD or LZM medium containing 1 mM/ 10 mM zinc. Cells were incubated for 24 h. (a) Then, cells were co-stained with ConcanavalinA-TRITC (cell wall) and DAPI (nucleus). (b) Cells also stained with FM4-64 (vacuole) and CalcoFluor White (cell wall) dye. Representative pictures of the intracellular CpZrc1 localization were taken by a confocal microscope. The experiment was performed twice.
3.6. Role of CpZRT21 and CpZRC1 in virulence
3.6.1. Phagocytosis is not affected by CpZRT21 and CpZRC1
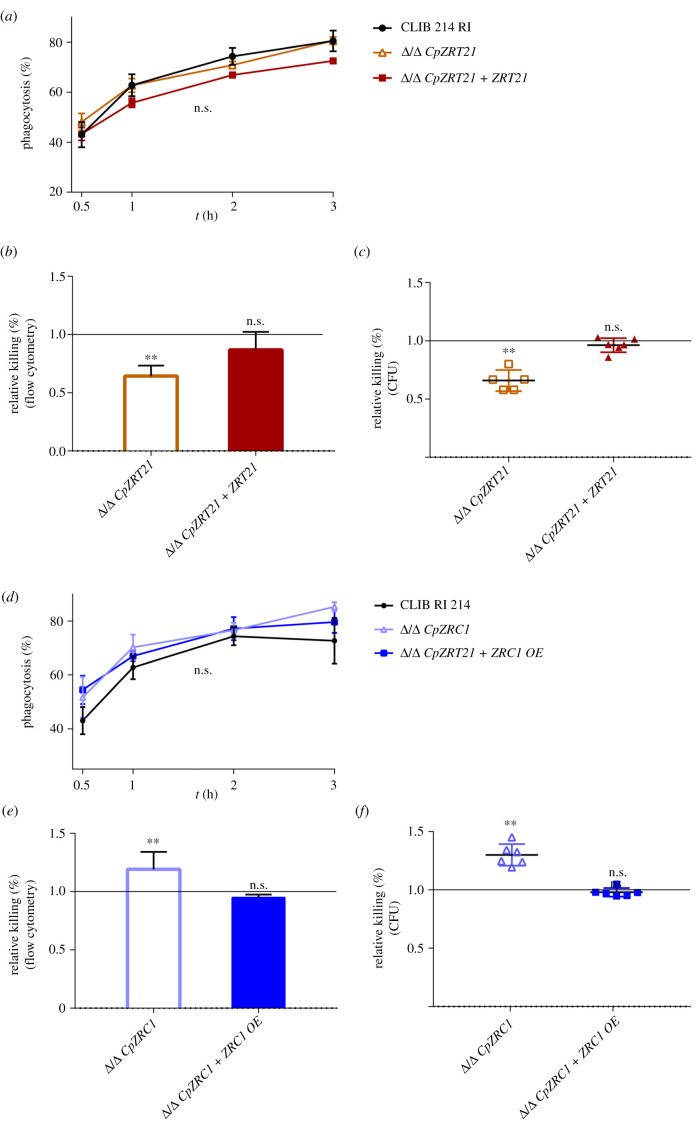
Macrophages play key roles in controlling pathogenic fungi. Interestingly, these phagocytes can mount either zinc-starvation or intoxification responses against phagocytosed fungi [7,31]. We therefore assessed the interaction between C. parapsilosis ZnT mutant strains and J774.2 murine macrophages. We first examined phagocytosis efficiency 0.5, 1-, 2- and 3-hours of post-infection using flow cytometry analysis. Each strain was labelled with Alexafluor 647 succinimidyl ester in order to determine the phagocytosis+ macrophage population after the indicated time points. We found no significant difference between the uptake of ZRT21, ZRC1 mutants and the wild-type strain regardless of the selected time points (figure 10a,d).
Figure 10.
The absence of CpZRT21 increases C. parapsilosis susceptibility to killing whereas CpZrc1 protects against elimination by J774.2 macrophages. (a,d) J774.2 murine macrophages and the GFP-tagged or AlexaFluor647-stained C. parapsilosis strains were co-incubated for the indicated time points, and then phagocytosis kinetics was determined by flow cytometry. (b,e) The killing efficiency of J774.2 macrophages was determined after 3 h. Calcofluor White and the two-colour fluorescent FUN1 were applied as a co-staining method for determining yeast cell viability. (c,f) shows the results of killing assay performed by CFU counting method from YPD plates. The MOI = 1 : 5 was used for the experiments. Data were normalized to the elimination rate of the wild-type. Experiments were performed at least three times. For statistical analysis, unpaired t-tests were used, *p < 0.05; **p < 0.01.
3.6.2. CpZRT21 and CpZRC1 influence fungal killing by murine macrophages
Fungal cell elimination efficiency by J774.2 macrophages was determined 3 h post-infection by flow cytometry analysis, using FUN1 and Calcofluor White staining. Killing efficiency was also measured by CFU counting of recovered yeast cells 3 h after yeast-J774.2 interactions. The results of both methods revealed that deletion of CpZRT21 led to less efficient fungi elimination (figure 10b,c). By contrast, the lack of CpZRC1 led to more efficient killing by macrophages (figure 10e,f). The reintegrant strains showed a phenotype similar to that of the wild-type. Because Zrc1 is essential for zinc detoxification, these data suggest that C. parapsilosis may experience zinc toxicity within J774.2 macrophages.
3.6.3. CpZRT21 and CpZRC1 is not essential for in vivo fungal colonization of Galleria mellonella
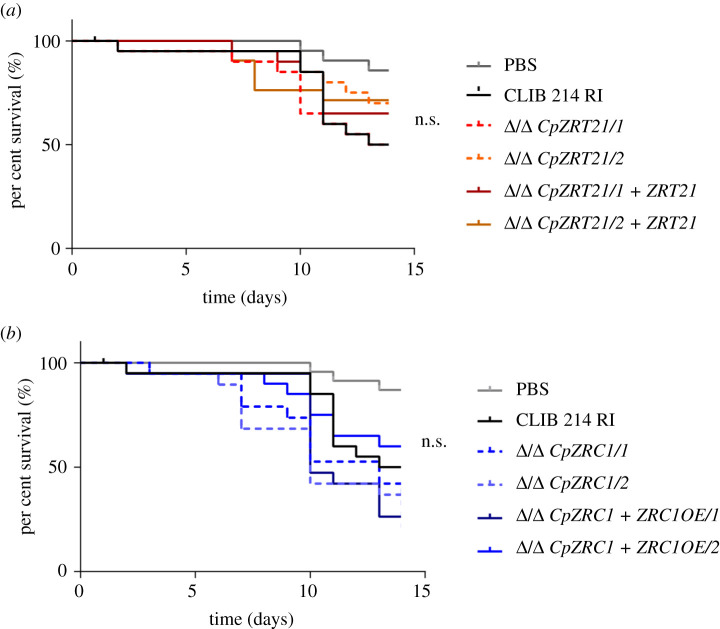
Next, we investigate the role of the described ZnTs of C. parapsilosis in the Galleria mellonella insect infection model. Larvae were infected with the wild-type, Δ/Δ CpZRT21 and Δ/Δ CpZRC1 strains then the survival rate was monitored for 14 days. We found no significant difference in survival of the larvae between the mutant strains and wild-type in this model (figure 11).
Figure 11.
In vivo Galleria mellonella infection with wild-type, CpZRT21 and CpZRC1 mutant strains. After 7 days of acclimatization, Galleria mellonella larvae were infected with the indicated fungal strains. The surviving capability of the larvae was determined in the next 14 days. Twenty larvae were used per strain. For statistical analysis, the Mantel–Cox test was used, *p < 0.05; **p < 0.01; ***p < 0.001.
4. Discussion
Zinc is essential for cellular life and its uptake by fungal pathogens is crucial for maintaining both viability and pathogenicity [3]. In this study, we aimed to assess the role of zinc uptake and homeostasis in the human fungal pathogen C. parapsilosis and to examine their role in virulence. Based on in silico predictions, transporters for zinc uptake (Sc/CaZrt2, CaZrt101), detoxification (Sc/CaZrc1) and vacuolar export (ScZrt3) were identified in C. parapsilosis, similarly to C. albicans and S. cerevisiae. However, we did not find a potentially functional CaPra1 orthologue in the C. parapsilosis genome.
The availability of zinc is low within the host due to nutritional immunity. In fungal pathogens, zinc uptake required for survival and also host colonization is mediated by Zip transporter families [4]. Our in silico analyses predicted four such cellular zinc importer related to C. albicans Zrt101 and Zrt2. CpZrt101 showed similarity with the CaZrt101 membrane-localized ZnT and its transcriptional regulation was similar to that of CaZRT101, exhibiting increased expression at neutral than acidic pH. This is interesting because the orthologue of Candida ZRT101 in the distantly related mould Aspergillus fumigatus zrfC is also positively regulated by neutral pH. This indicates that pH-dependent regulation of this ZnT may be conserved across the ascomycota phylum of fungi. In this context, it is worthwhile noting that although Zrt101 is missing from the model yeast S. cerevisiae, orthologues are present in most ascomycete fungi [12].
The orthologue of the Zrt101-associated zincophore, the CaPra1 zincophore, was not identified in the genome of C. parapsilosis as previously reported [12]. By contrast, three genes (CpZRT21, CpZRT22 and CpZRT23) showed high similarity with the acid-adapted transporter Sc/CaZRT2. CpZRT22 and CpZRT23 appear to be the result of an ancestral duplication of the conserved CaZRT2/CpZRT21, followed by a subsequent tandem duplication [32]. PRA1 loss and ZRT2 expansion in C. parapsilosis is noteworthy from ecological perspective. This yeast is a common colonizer of human skin which is mildly acidic. Because Pra1 is adapted for zinc uptake at neutral–alkaline pH and Zrt2 at acidic, the genetic arrangements observed in C. parapsilosis may be associated with such pH adaptation.
Our gene expression analysis revealed that CpZRT21 was transcribed at lower levels at acidic versus neutral pH. This is in contrast with the ZRT21 orthologue in C. albicans (ZRT2) that is more highly expressed at acidic than neutral–alkaline pH [14,33]. Nevertheless, CpZRT21 did exhibit downregulation upon zinc supplementation at both pH values, as expected for a gene involved in zinc acquisition, and as previously observed for CaZRT2.
Notably, all four of these previously described yeast species encode only two Zip-type plasma membrane zinc importers each. CpZRT22 exhibited the expression profile of a canonical acid-adapted zinc uptake with increased transcripts level at depleted zinc and low pH values. Its paralogue gene CpZRT23 showed the highest expression at neutral pH, and it was downregulated significantly by lowering the pH in the medium. Interestingly, in acidic media, CpZRT23 was more highly expressed at medium (1 mM) zinc than at either low (0.05 mM) or high (10 mM) zinc. This bimodal pattern in response to zinc is reminiscent of the low-affinity zinc importer ZRT2 from S. cerevisiae [34]. All of the Zrt2-like genes' transcript levels altered in response to zinc.
Together, these phylogenetic and gene expression observations indicate that the zinc import machinery of C. parapsilosis is more complex than that of the other studied yeast species.
The previously investigated yeast species S. cerevisiae, C. gattii C. albicans and C. dubliniensis each encode only two Zip-type cellular zinc importers. In S. cerevisiae, C. gattii and C. dubliniensis, the Zap1 transcription factor is responsible for the regulation of Zip importers in response to environmental zinc levels [25–27]. Candida albicans Zap1 also regulates the zinc regulon in this species [35]. Thus, a similar regulator to Zap1 might also be present in C. parapsilosis. Indeed, a BLASTp search using C. albicans Zap1 as query identified a predicted transcription factor with high-sequence similarity (1 × 10−176) in C. parapsilosis called CPAR2_403080.
In liquid media, C. parapsilosis grew poorly under zinc limitation and growth was restored upon zinc supplementation. Δ/Δ CpZRT21 exhibited only a minor growth defect compared to wild-type and revertant in LZM + 0.05 mM zinc. The pre-culture for these growth assays was YPD, which contains approximately 29 µM zinc [36]. We reasoned that the fungus may have stored some of the zinc from this pre-culture as has been previously shown for S. cerevisiae [11]. To test this hypothesis, cells were first pre-cultured under zinc-limiting conditions before inoculation. We found that cells from a zinc-depleted pre-culture were unable to grow when inoculated into LZM (figure 3). This indicates that C. parapsilosis, like S. cerevisiae, may have the capacity to generate and use intracellular stores (left-hand panel of figure 3b,c). When cells from a zinc-depleted pre-culture were used to inoculate LZM + 0.05 mM zinc, wild-type and Δ/Δ CpZRT21 +ZRT21 strains grew well, but Δ/Δ CpZRT21 growth was severely inhibited. This suggests that Zrt21 is essential for zinc uptake and growth when the cells must secure low levels (0.05 mM) of zinc from the environment. Finally, supplementation with excess zinc (1 mM) bypassed the requirement for Zrt21.
Interestingly, of the four identified ZnTs, only CpZRT21 showed a growth defect under zinc-depleted acidic environmental conditions. However, the transcription of the CpZRT21, CpZRT22 and CpZRT23 in this media revealed interesting patterns of expression. Compared to wild-type cells, the deletion of ZRT22 and ZRT23 either alone or in combination resulted in the upregulation of ZRT21. Such compensatory upregulation is common among genes in the same micronutrient uptake pathway. The deletion of ZRT22 downregulated ZRT23 while the absence of ZRT23 downregulated ZRT22. These two genes are, however, located at the same genetic locus, and we cannot rule out some perturbation of regulation due to gene deletion itself. The most interesting observation was that the deletion of ZRT21 (which is located on a different chromosome than CpZRT22/23) resulted in the simultaneous downregulation of CpZRT22 and upregulation of CpZRT23. While it is possible that the zinc restriction experienced by Δ/Δ CpZRT21 cells may contribute towards the observed differential regulation of CpZRT22 and CpZRT23, we think this is unlikely as CpZRT22 was progressively upregulated under low zinc conditions in acidic media whereas CpZRT23 expression was only modestly higher. These results suggest that the Zrt2-based zinc uptake in C. parapsilosis regulated by CpZRT21, CpZRT22 and CpZRT23 might be under the same co-regulation process. It could explain why neither the single deletion nor the double deletion of CpZRT22 and CpZRT23 resulted in a loss of viability. Hence, CpZrt21 may compensate for the lack of ZRT22 and ZRT23. In the future, this hypothesis could be confirmed with the generation and thorough characterization of a Δ/Δ CpZRT21/ZRT22/ZRT23 triple mutant strain.
In a host niche, microorganisms can encounter potentially toxic levels of zinc following phagocytosis by immune cells. Recent studies established that macrophages are able to harm pathogens via sequestration of a toxic amount of zinc ions in the phagolysosome to kill or limit their growth [6,7]. Notably, several pathogenic species possess strategies to detoxify zinc to survive such an antimicrobial activity, and this ability is considered to be an important virulence factor [6,13]. Based on the orthology relations with S. cerevisiae and C. albicans, we predicted the zinc detoxification transporter Zrc1 to be CPAR2_212100 in C. parapsilosis. It should be noted that all ZRC1 orthologues studied to date [13,37–39] play important roles in zinc detoxification. At acidic pH, CpZRC1 expression increased with decreasing zinc levels. While this may appear counterintuitive, it has been described for other fungi and may represent an adaptive mechanism to protect cells against future zinc shock. The fact that we observed this phenomenon at acidic, but not neutral/alkaline pH, may be due to the much greater solubility, and thus bioavailability, of zinc ions at lower pH [40]. We also found that deletion of CpZRC1 resulted in a viability defect at 5 mM zinc-containing conditions, which also supports the zinc detoxification role of CpZrc1. Notably, the reintegration of one copy of CpZRC1 with its native promoter did not complement the wild-type phenotype, which led us to overexpress CpZRC1 using CaTDH3 promoter in the Δ/Δ CpZRC1 mutant strain. The overexpression of CpZRC1 restored the wild-type phenotype at 5–10 mM zinc-containing environment. At greater than or equal to 17.5 mM zinc concentrations, CpZRC1 overexpression even aided fungal survival. The zinc survival assay also supported the zinc detoxification role of CpZrc1: the deletion of CpZRC1 resulted in growth deficiency after 24 and 48 h of 5 to 20 mM of zinc treatment. Zrc1 operates two different zinc detoxification processes in S. cerevisiae and C. albicans. In S. cerevisiae, ScZrc1 is responsible for vacuolar zinc import, while it defends against high amounts of intracellular zinc via CaZrc1-dependent zincosome formation in C. albicans [13,41]. To determine the function of Zrc1 in C. parapsilosis, a zincosome-specific fluorescent stain was used. The toxic (10 mM) amount of zinc treatment resulted in rapid zincosome formation in C. parapsilosis after 5–20 min of incubation, and the Δ/Δ CpZRC1 mutant was also able to form the same amount of zincosomes as the wild-type. The 10 mM zinc treatment for 24 and 48 h was toxic for the Δ/Δ CpZRC1 strain, which correlated with the high amount of zincosomes and dead cells detected under the same circumstances by confocal microscopy and flow cytometer. Interestingly, while the 20 mM zinc treatment (which is already toxic for the wild-type) resulted in high amount of zincosome formation in the wild-type and Δ/Δ CpZRC1 strains, the Zinquin level in the Δ/Δ CpZRC1 + ZRC1OE strain was reduced. In C. parapsilosis, zincosome formation appeared overwhelmed in the absence of CpZrc1, so zincosomes were overloaded with zinc ions in the presence of high levels of zinc. We hypothesized that CpZrc1 either has another zinc detoxification target organelle or (in contrast with C. albicans) it is localized on the membrane of vacuoles. Indeed, C-terminal tagging of Zrc1 with GFP and confocal microscopy with cell wall, nuclear and vacuolar co-staining revealed predominantly the vacuolar localization of CpZrc1. Therefore, despite being phylogenetically more closely related to C. albicans, C. parapsilosis Zrc1 subcellular localization more closely resembles that of S. cerevisiae and C. neoformans [38,41].
Acquisition of zinc in environments where the micronutrient is limited conditions and defence against toxic zinc concentrations are both important for pathogenic fungi to survive and successfully colonize a host. In order to investigate the interaction between the host and the ZnT mutants, J774.2 murine macrophages, G. mellonella larvae were used to assess the mutant strain's virulence properties both in vitro and in vivo. Phagocytosis of the Δ/Δ CpZRT21 and Δ/Δ CpZRC1 strains by the J774.2 cells was similar to that of the wild-type in all time points. However, Δ/Δ CpZRC1 cells were cleared more effectively by the macrophages, while Δ/Δ CpZRT21 cells were less effectively killed. Macrophages are able to increase phagolysosomal zinc to aid the clearance of pathogens. In the case of M. tuberculosis, zinc rapidly accumulates in the phagolysosomes [6]. A recent study also revealed a zinc-based killing process by murine bone marrow-derived macrophages during C. glabrata infection [7]. The behaviour of our zinc uptake and zinc detoxification defective mutants suggests that C. parapsilosis cells might trigger a similar effect once taken up by J774.2 macrophages. The Δ/Δ CpZRT21 mutant may be partially defended by the excess zinc as it is lacking the major plasma membrane zinc importer. Simultaneously, increased killing of the Δ/Δ CpZRC1 cells, which lack the vacuolar membrane-localized zinc detoxification system, indicate that C. parapsilosis may be experiencing zinc poisoning within these immune cells.
Galleria mellonella larvae can use as a model of humoral and cellular innate immune responses. According to Trevijano-Contador & Zaragoza, Galleria larvae are suitable for the investigation of innate immunity and its response to fungal infections [42]. Galleria mellonella were used to uncover the virulence properties of many mutant strains and also to detect the effect of nutrient deprivation on the sensitivity of the larvae for C. albicans infection [43,44] These above-mentioned results showed a strong correlation with mammalian in vivo model approaches, so we used this insect organism as a substitute for vertebrate in vivo experiments.
To uncover the effects of CpZRT21 and CpZRC1 deletion in vivo, we infected larvae with the indicated strains and monitored their survival for 14 days. During the in vivo experiments, we detected no virulence alterations among the generated mutant strains in a 14-day-long survival experiment using G. mellonella larvae. Overall, we can assume that the lack of CpZrt21 membrane ZnT and CpZrc1 vacuolar zinc importer do not affect the immune response and the survival rate of the healthy nutrient-supplied Galleria mellonella larvae to C. parapsilosis infection. Additionally, a more in-depth analysis is considered to uncover the response and the survival rate of the nutrient deprivation in Galleria mellonella during Candida parapsilosis infection.
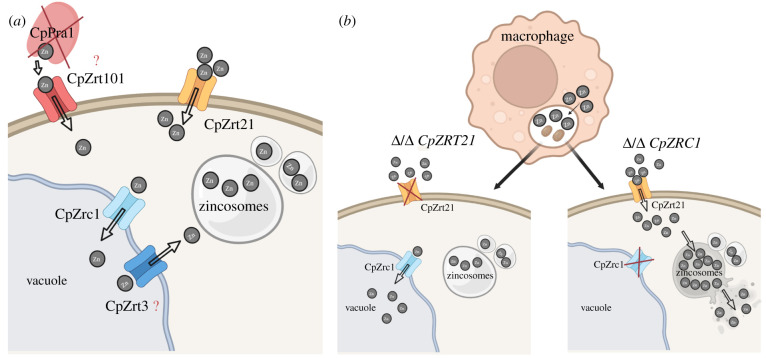
In summary, we have identified and characterized for the first time five ZnTs in C. parapsilosis (figure 12a). Similar to C. albicans, growth of C. parapsilosis in an acidic environment is mediated exclusively by the membrane ZnT CpZrt21. However, in this species, two additional Zrt2 orthologues are also present that may be under the same regulation as CpZRT21 in the setting of zinc-limited conditions. We revealed that C. parapsilosis forms zincosomes under high zinc-supplemented environmental conditions, although, in contrast with C. albicans, in a Zrc1-independent way. Instead identified a C. parapsilosis vacuolar-localized ZnT, CpZrc1, which is essential for zinc detoxification and also protects C. parapsilosis cells against killing by murine macrophages. Based on our findings, we can hypothesize that, once C. parapsilosis cells are phagocytosed by macrophages, zinc ions accumulate in the phagolysosome which may contribute towards fungal killing (figure 12b).
Figure 12.
Summary of the zinc homeostasis in C. parapsilosis. (a) shows the described zinc uptake and zinc storing mechanisms of C. parapsilosis. (b) Effects of the hypothetical zinc poisoning-based C. parapsilosis elimination by macrophages in the absence of CpZRT21 or CpZRC1. Images created with BioRender.com.
Data accessibility
All data are available in the manuscript or as electronic supplementary material [45].
Authors' contributions
T.T.: conceptualization, data curation, investigation, methodology, writing—original draft and writing—review and editing; M.T.N.: conceptualization, data curation and methodology; F.B.: conceptualization, investigation, methodology and writing—review and editing; C.V.: conceptualization, methodology and writing—review and editing; F.J.: data curation, investigation and methodology; D.W.: conceptualization, data curation, investigation, writing—original draft and writing—review and editing; A.G.: conceptualization, data curation, investigation, methodology, supervision, validation, writing—original draft and writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
D.W. is supported by a Wellcome Trust Senior Research Fellowship (grant no. 214317/Z/18/Z) and the MRC and University of Exeter (grant no. MR/N006364/2).
References
- 1.Waldron KJ, Rutherford JC, Ford D, Robinson NJ. 2009. Metalloproteins and metal sensing. Nature 460, 823-830. ( 10.1038/nature08300) [DOI] [PubMed] [Google Scholar]
- 2.Colvin RA, Holmes WR, Fontaine CP, Maret W. 2010. Cytosolic zinc buffering and muffling: their role in intracellular zinc homeostasis. Metallomics 2, 306. ( 10.1039/b926662c) [DOI] [PubMed] [Google Scholar]
- 3.Andreini C, Bertini I, Rosato A. 2009. Metalloproteomes: a bioinformatic approach. Accounts Chem. Res. 42, 1471-1479. ( 10.1021/ar900015x) [DOI] [PubMed] [Google Scholar]
- 4.Eide DJ. 2006. Zinc transporters and the cellular trafficking of zinc. Biochim. et Biophys. Acta Mol. Cell Res. 1763, 711-722. ( 10.1016/j.bbamcr.2006.03.005) [DOI] [PubMed] [Google Scholar]
- 5.Hood MI, Skaar EP. 2012. Nutritional immunity: transition metals at the pathogen-host interface. Nat. Rev. Microbiol. 10, 525-537. ( 10.1038/nrmicro2836) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Botella H, et al. 2011. Mycobacterial P1-Type ATPases mediate resistance to zinc poisoning in human macrophages. Cell Host Microbe 10, 248-259. ( 10.1016/j.chom.2011.08.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riedelberger M, et al. 2020. Type I interferons ameliorate zinc intoxication of Candida glabrata by macrophages and promote fungal immune evasion. iScience 23, 101121. ( 10.1016/j.isci.2020.101121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liuzzi JP, Lichten LA, Rivera S, Blanchard RK, Aydemir TB, Knutson MD, Ganz T, Cousins RJ. 2005. Interleukin-6 regulates the zinc transporter Zip14 in.liver and contributes to the hypozincemia of the acute-phase response. Proc. Natl Acad. Sci. USA 102, 6843-6848. ( 10.1073/pnas.0502257102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao H, Eide D. 1996. The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc. Natl Acad. Sci. USA 93, 2454-2458. ( 10.1073/pnas.93.6.2454) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyabe S, Izawa S, Inoue Y. 2001. The Zrc1 is involved in zinc transport system between vacuole and cytosol in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 282, 79-83. ( 10.1006/bbrc.2001.4522) [DOI] [PubMed] [Google Scholar]
- 11.Simm C, Lahner B, Salt D, LeFurgey A, Ingram P, Yandell B, Eide DJ. 2007. Saccharomyces cerevisiae vacuole in zinc storage and intracellular zinc distribution. Eukaryotic Cell 6, 1166-1177. ( 10.1128/EC.00077-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Citiulo F, Jacobsen ID, Miramón P, Schild L, Brunke S, Zipfel P, Brock M, Hube B, Wilson D. 2012. Candida albicans scavenges host zinc via Pra1 during endothelial invasion. PLoS Pathogens 8, e1002777. ( 10.1371/journal.ppat.1002777) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crawford AC, et al. 2018. Biphasic zinc compartmentalisation in a human fungal pathogen. PLoS Pathogens 14, 1-28. ( 10.1371/journal.ppat.1007013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tóth R, et al. 2019. Candida parapsilosis: from genes to the bedside. Clin. Microbiol. Rev. 32, e00111-18. ( 10.1128/CMR.00111-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lupetti A, Tavanti A, Davini P, Ghelardi E, Corsini V, Merusi I, Boldrini A, Campa M, Senesi S. 2002. Horizontal transmission of Candida parapsilosis candidemia in a neonatal intensive care unit. J. Clin. Microbiol. 40, 2363-2369. ( 10.1128/JCM.40.7.2363-2369.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Govender NP, et al. 2016. Emergence of azole-resistant Candida parapsilosis causing bloodstream infection: results from laboratory-based sentinel surveillance in South Africa. J. Antimicrobial Chemotherapy 71, 1994-2004. ( 10.1093/jac/dkw091) [DOI] [PubMed] [Google Scholar]
- 17.Meletiadis J, Curfs-Breuker I, Meis JF, Mouton JW. 2017. In vitro antifungal susceptibility testing of Candida isolates with the EUCAST methodology, a new method for ECOFF determination. Antimicrobial Agents Chemotherapy 61, e02372–16. ( 10.1128/AAC.02372-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shu Y, Yu S, Zha L, Fu P, Cui T. 2017. Catheter-related fungal endocarditis caused by Candida parapsilosis in a hemodialysis patient. Hemodialysis Int. 21, E66-E68. ( 10.1111/hdi.12566) [DOI] [PubMed] [Google Scholar]
- 19.Horváth P, Nosanchuk JD, Hamari Z, Vágvölgyi C, Gácser A. 2012. The identification of gene duplication and the role of secreted aspartyl proteinase 1 in Candida parapsilosis virulence. J. Infectious Dis. 205, 923-933. ( 10.1093/infdis/jir873) [DOI] [PubMed] [Google Scholar]
- 20.Chakraborty T, Thuer E, Heijink M, Tóth R, Bodai L, Vágvölgyi C. 2018. Eicosanoid biosynthesis influences the virulence of Candida parapsilosis. Virulence 9, 1019-1035. ( 10.1080/21505594.2018.1475797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papp C, et al. 2018. Echinocandin-induced microevolution of Candida parapsilosis influences virulence and abiotic stress tolerance. mSphere 3, e00547-18. ( 10.1128/msphere.00547-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papp C, et al. 2020. Triazole evolution of Candida parapsilosis results in cross-resistance to other antifungal drugs, influences stress responses, and alters virulence in an antifungal drug-dependent manner. mSphere 5, e00821-20. ( 10.1128/msphere.00821-20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chakraborty T, Tóth Z, Tóth R, Vágvölgyi C, Gácser A. 2020. Iron metabolism, pseudohypha production, and biofilm formation through a multicopper oxidase in the human-pathogenic fungus Candida parapsilosis. mSphere 5, e00227-20. ( 10.1128/msphere.00227-20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chakraborty T, Tóth R, Gácser A. 2019. Eicosanoid production by Candida parapsilosis and other pathogenic yeasts. Virulence 10, 970-975. ( 10.1080/21505594.2018.1559674) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holland LM, et al. 2014. Comparative phenotypic analysis of the major fungal pathogens Candida parapsilosis and Candida albicans. PLoS Pathogens 10, e1004365. ( 10.1371/journal.ppat.1004365) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nemeth T, Papp C, Vagvolgyi C, Chakraborty T, Gacser A. 2020. Identification and characterization of a neutral locus for knock-in purposes in C. parapsilosis. Front. Microbiol. 11, 1194. ( 10.3389/fmicb.2020.01194) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Németh T, Nosanchuk JD, Vagvolgyi C, Gacser A. 2021. Enhancing the chemical transformation of Candida parapsilosis. Virulence 12, 937-950. ( 10.1080/21505594.2021.1893008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lombardi L, Salzberg LI, Cinnéide EÓ, O'Brien C, Butler G. 2019. Large-scale generation of mutant strains in Candida parapsilosis using CRISPR-Cas9. Access Microbiol. 1, 695. ( 10.1099/acmi.ac2019.po0436) [DOI] [Google Scholar]
- 29.Németh T, et al. 2013. Characterization of virulence properties in the C. parapsilosis sensu lato species. PLoS ONE 8, e68704. ( 10.1371/journal.pone.0068704) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vengatesan K, Mahajan SB, Sanjeevikumar P, Mangrule R, Kala V, Pragadeeswaran. 2018. Performance analysis of gene expression data using Mann–Whitney U test. In pp. 701-709. ( 10.1007/978-981-10-4762-6_67) [DOI] [Google Scholar]
- 31.Rossi DCP, et al. 2021. A metabolic inhibitor arms macrophages to kill intracellular fungal pathogens by manipulating zinc homeostasis. J. Clin. Investigation 131, e147268. ( 10.1172/JCI147268) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson D. 2015. An evolutionary perspective on zinc uptake by human fungal pathogens. Metallomics 7, 979-985. ( 10.1039/c4mt00331d) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bensen ES, Martin SJ, Li M, Berman J, Davis DA. 2004. Transcriptional profiling in Candida albicans reveals new adaptive responses to extracellular pH and functions for Rim101p. Mol. Microbiol. 54, 1335-1354. ( 10.1111/j.1365-2958.2004.04350.x) [DOI] [PubMed] [Google Scholar]
- 34.Bird AJ, Blankman E, Stillman DJ, Eide DJ, Winge DR. 2004. The Zap1 transcriptional activator also acts as a repressor by binding downstream of the TATA box in ZRT2. EMBO J. 23, 1123-1132. ( 10.1038/sj.emboj.7600122) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nobile CJ, Nett JE, Hernday AD, Homann OR, Deneault JS, Nantel A, Andes DR, Johnson AD, Mitchell AP. 2009. Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol. 7, e1000133. ( 10.1371/journal.pbio.1000133) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wehmeier S, Morrison E, Plato A, Raab A, Feldmann J, Bedekovic T, Wilson D, Brand AC. 2020. Multi trace element profiling in pathogenic and non-pathogenic fungi. Fungal Biol. 124, 516-524. ( 10.1016/j.funbio.2020.03.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacDiarmid CW, Milanick MA, Eide DJ. 2003. Induction of the ZRC1 metal tolerance gene in zinc-limited yeast confers resistance to zinc shock. J. Biol. Chem. 278, 15 065-15 072. ( 10.1074/jbc.M300568200) [DOI] [PubMed] [Google Scholar]
- 38.Cho M, Hu G, Caza M, Horianopoulos LC, Kronstad JW, Jung WH. 2018. Vacuolar zinc transporter Zrc1 is required for detoxification of excess intracellular zinc in the human fungal pathogen Cryptococcus neoformans. J. Microbiol. 56, 65-71. ( 10.1007/s12275-018-7475-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clemens S, Bloss T, Vess C, Neumann D, Nies DH, Zur Nieden U. 2002. A transporter in the endoplasmic reticulum of Schizosaccharomyces pombe cells mediates zinc storage and differentially affects transition metal tolerance. J. Biol. Chem. 277, 18 215-18 221. ( 10.1074/jbc.M201031200) [DOI] [PubMed] [Google Scholar]
- 40.Łoboda D, Rowińska-Zyrek M. 2017. Zinc binding sites in Pra1, a zincophore from: Candida albicans. Dalton Trans. 46, 13 695-13 703. ( 10.1039/c7dt01675a) [DOI] [PubMed] [Google Scholar]
- 41.MacDiarmid CW. 2000. Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J. 19, 2845-2855. ( 10.1093/emboj/19.12.2845) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trevijano-Contador N, Zaragoza O. 2018. Immune response of Galleria mellonella against human fungal pathogens. J. Fungi 5, 3. ( 10.3390/jof5010003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Desalermos A, Tan X, Rajamuthiah R, Arvanitis M, Wang Y, Li D, Kourkoumpetis TK, Fuchs BB, Mylonakis E. 2015. A multi-host approach for the systematic analysis of virulence factors in Cryptococcus neoformans. J. Infect. Dis. 211, 298-305. ( 10.1093/infdis/jiu441) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Banville N, Browne N, Kavanagh K. 2012. Effect of nutrient deprivation on the susceptibility of Galleria mellonella larvae to infection. Virulence 3, 497-503. ( 10.4161/viru.21972) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takács T, Németh MT, Bohner F, Vágvölgyi C, Jankovics F, Wilson D, Gácser A. 2022. Characterization and functional analysis of zinc trafficking in the human fungal pathogen Candida parapsilosis. Figshare. ( 10.6084/m9.figshare.c.6075355) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Takács T, Németh MT, Bohner F, Vágvölgyi C, Jankovics F, Wilson D, Gácser A. 2022. Characterization and functional analysis of zinc trafficking in the human fungal pathogen Candida parapsilosis. Figshare. ( 10.6084/m9.figshare.c.6075355) [DOI] [PMC free article] [PubMed]
Data Availability Statement
All data are available in the manuscript or as electronic supplementary material [45].